Abstract
Histone modifications play important roles in chromatin remodeling, gene transcriptional regulation, stem cell maintenance and differentiation. Alterations in histone modifications may be linked to human diseases especially cancer. Histone modifications including methylation, acetylation and ubiquitylation probed by ChIP-seq, ChIP-chip and qChIP have become widely available. Mining and integration of histone modification data can be beneficial to novel biological discoveries. There has been no comprehensive data repository that is exclusive for human histone modifications. Therefore, we developed a relatively comprehensive database for human histone modifications. Human Histone Modification Database (HHMD, http://bioinfo.hrbmu.edu.cn/hhmd) focuses on the storage and integration of histone modification datasets that were obtained from laboratory experiments. The latest release of HHMD incorporates 43 location-specific histone modifications in human. To facilitate data extraction, flexible search options are built in HHMD. It can be searched by histone modification, gene ID, functional categories, chromosome location and cancer name. HHMD also includes a user-friendly visualization tool named HisModView, by which genome-wide histone modification map can be shown. HisModView facilitates the acquisition and visualization of histone modifications. The database also has manually curated information of histone modification dysregulation in nine human cancers.
INTRODUCTION
Eukaryotic DNA is packaged into chromatin incorporating repeating nucleosomes by wrapping DNA around core histones (H2A, H2B, H3 and H4). In mammalian cells, the N-terminal tail of histone is subject to many chemical modifications such as methylation, acetylation, ubiquitylation, phosphorylation and ADP-ribosylation. Histone modifications provide accessible targets for effectors such as histone methyltransferase and acetyltransferase (1) and have different impact on chromatin structure and gene transcription, depending on the types and locations of the modifications (2,3). Currently, methyl-, acetyl- and ubiquityl-groups among various modification types have been studied mainly by ChIP-based technologies. For specific loci at the N-terminal tails of arginines and lysines of histone, up to three methyl-groups can be added. Histone methylation types at different locations have been ascribed to either activating or repressive functions (4). For example, H3K4me3 is positively correlated with gene expression, while H3K9me3 is implicated in heterochromatin formation and gene silencing (5). The normal pattern of histone modifications is vital for chromatin stability and transcriptional regulation (5). Disturbed changes of histone modifications may be correlated with cancer (6). RARβ2 promoter is silenced by H3K27me3 enrichment specifically in prostate cancer without DNA hypermethylation dependence (7). ChIP-based experiments including ChIP-seq, ChIP-chip and qChIP are efficient at probing histone modifications, and have produced a large amount of histone modification data (8). It is useful to have a repository of such data so that in-depth data mining can be performed.
There have been a few resources for histone or histone modifications, such as the ChromatinDB (9), Histone Database (10–12), SysPTM (13) and HistoneHits (14). ChromatinDB is genome-wide resource of histone modifications in Saccharomyces cerevisiae. The Histone Database focuses on histone sequences and structures in many species. Although providing post-translational modification (PTM) information and modified residues information of histone sequences and structures, the Histone Database lacks large-scale profiles of histone modifications for functional evaluation. The major difference between SysPTM and HHMD is that SysPTM is a curated PTM platform for online query and analysis while HHMD is a repository of large-scale histone modification data and also has built-in functions for analysis and visualization. HistoneHits is a database for systematic collections of histone mutants in yeast (14). To the best of our knowledge, there has been no specialized database that focuses on histone modifications in mammals, which hinders further systematic and in-depth data mining. Therefore, there is need to build such a database that dedicates to the storage and analysis of experimental histone modification data. A database of this kind would be beneficial to histone modification studies such as identification of differential histone modification regions (D-HMRs) for a given set of histone modifications.
Cancer has been considered as a complex disease, which involves both genetic and epigenetic alteration. Until now there have been several comprehensive projects dedicated to cancer studies, including The Cancer Genome Atlas (TCGA) (15) and a large amount of researches studying cancer from epigenomic perspective (6,16,17). Although DNA methylation undergoes significant changes, other epigenetic changes such as histone modifications also reflect the tumorigenesis process (6,18). Global aberrant histone modification patterns in tumorigenesis provide novel potential of molecular screening for cancer prevention, diagnosis and treatment (19,20). Tools for histone modification data integration and analysis are still needed for cancer biomarker identification.
We developed the human histone modification database (HHMD), which is available at http://bioinfo.hrbmu.edu.cn/hhmd or http://www.hhmd.org. HHMD focuses on the storage and integration of histone modification information from experimental data. The latest release of HHMD provides genomic context (hg18) for histone modification alignment, which can be used to make comparisons between genomic and epigenomic data.
HHMD incorporates a set of tools for querying histone modifications. Five search options were provided for advanced searches, namely histone modification, gene ID, functional categories, chromosome location and cancer name. Furthermore, HHMD provides a visualization tool—HisModView. It has the capability of investigating histone modifications in a genomic context by superimposing histone modification data on DNA methylation, GC contents and gene information. HHMD may be a useful resource for researchers who are interested in epigenetic regulation and computational epigenetics in human and other species. With the in-house data and built-in tools in HHMD, it makes D-HMRs identification between cancer and control possible, and therefore benefits cancer biomarker identifications.
DATA COLLECTION
HHMD contains four types of data: (i) high-throughput histone modifications, (ii) MeDIP methylation, (iii) curated information of aberrant histone modifications, genes and cancers and (iv) GC contents, RefSeq gene (21) and other genomic annotations.
A total of 43 histone modification types (Table 1) classified by histone types have been included in the current release of HHMD. Specifically, there are 228 high-throughput histone modification datasets by manual confirmation. Each dataset is assigned a unique HM ID (e.g. HM-26). All histone modifications were collected from biological experiments, including high-throughput datasets and curated information from literature. All the high-throughput datasets were probed from ChIP-based technology. Among these datasets, 81 were from ChIP-chip, 87 from ChIP-seq and 55 from qChIP. Summarized ChIP-seq data files derived from tag-based bed files with window size of 200 bp were used to speed up HHMD. Histone H3, the most sequenced type, associates with 155 datasets. There are 50 datasets for H4, 11 for H2A and six for histone H2B. Regarding modification types, 122 datasets are methylation-related, 87 of acetylation-related and 1 of ubiquitylation-related. High-throughput datasets were collected from various institutes and websites (Table 2). An interface form for submission of new histone modification information was also provided in HHMD.
Table 1.
List of histone modifications for 228 high-throughput datasets
| Histone type | Histone modifications |
|---|---|
| H2Ab | H2AK5ac, H2AK9ac, H2AZ |
| H2Bb | H2BK120ac, H2BK12ac, H2BK20ac, H2BK5ac, H2BK5me1, UbH2Ba |
| H3b | H3K14ac, H3K18ac, H3K23ac, H3K27ac, H3K27me1, H3K27me2, H3K27me3, H3K36ac, H3K36me1, H3K36me3, H3K4ac, H3K4me1, H3K4me2, H3K4me3, H3K79me1, H3K79me2, H3K79me3, H3K9ac, H3K9me1, H3K9me2, H3K9me3, H3R2me1, H3R2me2, H3aca |
| H4b | H4K12ac, H4K16ac, H4K20me1, H4K20me3, H4K5ac, H4K8ac, H4K91ac, H4Kaca,H4R3me2, H4aca |
aNon-specific antibodies are used.
bThe list is classified according to histone types.
Table 2.
List of institutes and websites of high-throughput datasets used in this database
| Institute/project | Website |
|---|---|
| UCSC | http://genome.ucsc.edu/cgi-bin/hgTables |
| ENCODE | http://hgdownload.cse.ucsc.edu/goldenPath/hg18/wgEncode BroadChipSeq/ |
| Canada’s Michael Smith Genome Sciences Centre | http://www.bcgsc.ca/data/histone-modification |
| Laboratory of Molecular Immunology, National Heart, Lung and Blood Institute, NIH | http://dir.nhlbi.nih.gov/Papers/lmi/epigenomes/hgtcell.aspx |
To investigate the relationships between histone modifications and DNA methylation, HHMD integrated a panel of large-scale methylation data from MeDIP (22). The methylation data comprises information from 16 tissues including GM06990 cell line data (23).
The functional and genomic annotations of the genes in HHMD were obtained from various databases [i.e. NCBI (24), UCSC (25), GO (Gene Ontology) (26), UniProt (14), Affymetrix probe ID, KEGG (27), RefSeq Protein (28), OMIM (29), GI, UniGene (30), PIR (31) and Ensembl (32)].
To elucidate the interplay of histone modifications and cancer types, two strategies were introduced. First, we collected a panel of aberrant histone modifications in various human cancer types from the literature by manual curation. The current version compiled 833 curated relationships involving 17 human histone modifications and 555 genes that are related to nine human cancers. Among these relationships, 588 were from ChIP-chip and 237 were from ChIP-PCR. The nine cancers are gastric, ovarian, colon, leukemia, prostate, lymphoid, breast, lung and pancreatic cancer. HHMD has imbedded search functions that can be used to query human transcript IDs (21) and official gene symbols. Second, HHMD includes several high throughput aberrant histone modification data such as K562, GM06990 and HeLa cell lines. An interface was also built for submission of cancer-related histone modification information.
DATABASE USAGE
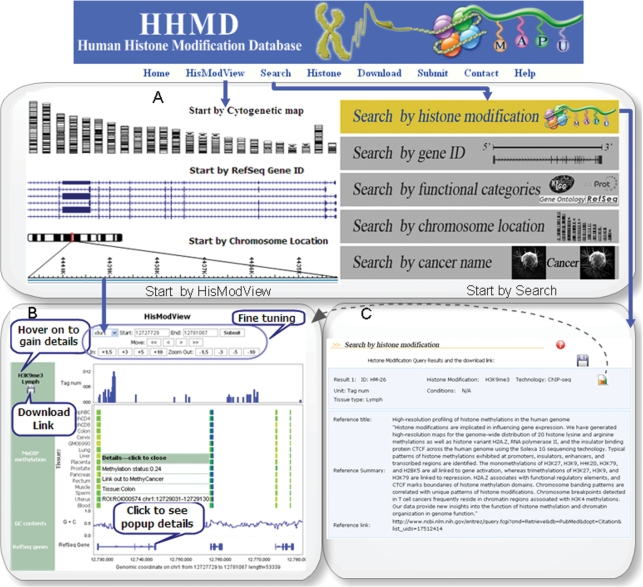
HHMD is a highly cross-linked database, which facilitates data acquisition and visualization. The overview of HHMD and two result pages are shown in Figure 1. Figure 1A presents three starting points of HisModView and five search options. To visually understand the data in HHMD, HisModView and searching tools are fully cross-linked. Search results can be downloaded from result pages of HisModView (Figure 1B), or reviewed offline (Figure 1C). In this way, users can analyze data more efficiently. For example, users can start by searching cancer name and visualize the genes in HisModView and then download the specific histone modification data in defined genomic range for further review.
Figure 1.
Screenshot showing the interrelation of tools in HHMD. Users can start search by ‘Search’ menu or start directly from ‘HisModView’ menu. (A) Screenshot showing the starting points of the searching and visualization tools. HHMD offers five advanced search options, namely search by histone modification, search by gene ID, search by functional categories, search by chromosome location and by cancer name. (B) Screenshot showing the HisModView tool. Users can view histone modifications aligned by human genomic coordinates (hg18). Meanwhile, genomic annotations such as GC contents and RefSeq genes and MeDIP methylation data can also be visualized. Users can fine-tune the genomic regions by left/right shifting or zooming in/out. Users can hover on the the texts of histone modification types to gain further details. Users are allowed to download the histone modifications data of the present range. Users who are interested in genomic annotations can get details from pop-up menus. (C) Screenshot showing the ‘Search by histone modification result’ when specifying HM ID: HM-26. Users can either download the histone modifications data or visualize the data in HisModView.
HHMD supports flexible query for various histone modifications and related genomic and functional annotations by providing five search tools. Taking the histone modification search as an example, users can specify the query options such as cytogenetic position, histone modification, technology, tissue, etc. For new users, histone modification search is the suggested option. If interested in specific datasets, they should query by HM IDs. For example, users can enter the page ‘search by histone modification’ and input ‘HM-26’ in the textbox labeled ‘Histone modification search by ID:’, and then proceed. The sample result page is shown in Figure 1C, where a download icon and a HisModView icon are available. Functional categories search is a specific module dedicated to study the relationships between histone modification patterns and functional classifications. Users who are interested in the histone modification distribution for genes of similar functions may find this module helpful. For example, users interested in KEGG pathway: hsa00030 can select ‘H3K4me3’ or other options from the pull-down menu ‘Histone modification:’, then type ‘hsa00030’ in the search field of KEGG ID and leave others as default. In this case, a report of six genes annotated with hsa00030 and 26 histone modification summaries will be returned.
As an efficient visualization tool, HisModView allows users to browse histone modifications and genomic annotations in the context of human genome (hg18). A snapshot of HisModView is shown in Figure 1B. Users can start HisModView from ‘Start by Cytogenetic map’, ‘Start by RefSeq Gene ID’ or ‘Start by Chromosome Location’. The HisModView result page has four sections, namely histone modification, MeDIP methylation, GC contents and RefSeq gene annotations. Labels and histograms of histone modifications are displayed in histone modification section for each genomic track. For each label, the description of the tissue and histone modification type is available and can be displayed by moving the mouse over the label. To study the relationships between histone modifications and DNA methylation, we have integrated a panel of methylation data from 16 tissues generated by MeDIP into the HisModView tool. In this section, methylation data is represented as matrices, each element within represents one ROI [region of interest, defined by Rakyan et al. (23)] and color coding represents different methylation level (from yellow to blue, represents from 0 to 100% methylation). Links to another epigenetic database for Homo Sapiens: MethyCancer (33) are available. The description of ROI, tissue type and the methylation level are also available in the popup menus. A detailed annotation report will be brought up by clicking any of the gene structures in RefSeq genes section. The gene structures such as introns and exons can be characterized for a specific gene and the range for viewing can be adjusted by centering on a specific gene.
DATABASE IMPLEMENTATION
HHMD was developed using J2EE. It was built using JSP, Struts and the Java connection pool Proxool. HHMD is running on an Apache Tomcat web server and a MySQL server. The scripts for data analysis were written in JAVA, which are available on HHMD website.
DISCUSSION AND FUTURE DEVELOPMENT
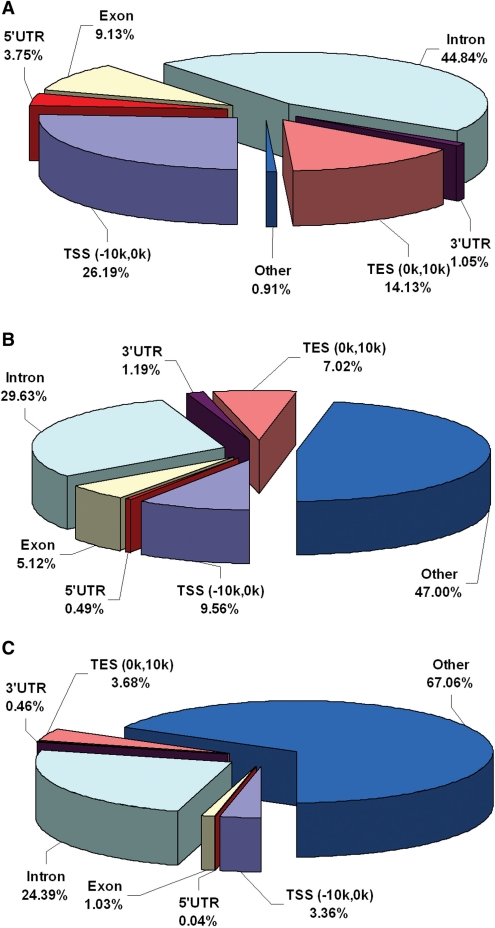
Recent studies of histone modification co-localization suggested that the co-localized histone modifications may mark functionally important regions. Co-localized histone modifications can provide specific cubic targets for biological effectors (34,35). To date, co-localization of H3K4me3 (activating) and H3K27me3 (repressive) is the most studied co-localized pair of markers, which has been ascribed to the developmental control of ES cells (36,37). Yet, identification of more co-localized histone modification pairs with functionalities is still of great interest. We used H3K4me3 and H3K9me3 profiles (38,39) in HHMD to further study the relationships between histone modification co-localization and gene function. H3K4me3 is a histone modification type associated with relaxed chromatin structure and active transcription, while H3K9me3 is a marker associated with heterochromatin formation, gene imprinting and repressive transcription (3,4). Significant imbalance of co-localized H3K4me3 and H3K9me3 is also suggested to have influences on developmental processes. We summarized the distribution of co-localization of H3K4me3 and H3K9me3 (repressive) from resting CD4+T cells to explore histone modification co-localization patterns in genomic context. As shown in Figure 2A–C, H3K4me3 and H3K9me3 co-localized regions (200 bp window size) reveal intermediate genomic pattern which seems to be contributed by both H3K4me3 and H3K9me3. The observation is in good agreement with the findings that methylation of H3 Lys4 and Lys9 play the contrary roles in chromatin regulation (35,40). The genes occupied by co-localized H3K4me3 and H3K9me3 are exemplified in Supplementary files, from which we find that protocadherin alpha gene cluster is marked by such co-localization.
Figure 2.
The pie charts show the genomic distributions of histone modifications in co-localization study of H3K4me3 and H3K9me3. (A) Genomic distributions of only H3K4me3 localization regions. (B) Genomic distributions of co-localized regions for H3K4me3 and H3K9me3. (C) Genomic distributions of H3K9me3 localization regions.
HHMD is predisposed to integrate genomic and epigenomic annotations from publicly available databases. Genomic annotations such as RefSeq genes, GC contents and various epigenomic annotations including histone modifications and DNA methylation data were compiled in HHMD. Users can access the data by searching and investigating histone modifications using HisModView and offline analysis tools.
HisModView is a visualization tool for data profiling and comparison, which makes the identification of variable histone modification regions in multiple tissues feasible. Virtual analysis tools such as D-HMRs identification and other analysis functions are to be integrated in a later release. A standalone version of HHMD that supports complex calculation will be released.
As a resource to study the potential function of histone modification markers, HHMD could be extended with utilities for identification of cancer-related histone modification markers for candidate genes. We will continue to investigate the relationships between histone modifications and other diseases in addition to cancer. Since histone modifications in other species are also accumulating, we will extend the research scope to build specific databases for histone modifications in other species as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Natural Science Foundation of China (Grant No. 30871394); the National High Tech Development Project of China, the 863 Program (Grant No. 2007AA02Z329); the National Basic Research Program of China, the 973 Program (Grant No. 2008CB517302); the Natural Science Foundation of Heilongjiang Province (Grant No. D2007-35); the Innovation and Technology special Fund for researchers of Harbin (Grant No. RC2007LX003004). Funding for open access charge: National Natural Science Foundation of China (Grant No. 30871394).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Yaoping Lei and Dr Diansong Zhou for revising the manuscript.
REFERENCES
- 1.Cheng X, Blumenthal RM. Mammalian DNA methyltransferases: a structural perspective. Structure. 2008;16:341–350. doi: 10.1016/j.str.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang Z, Schones DE, Zhao K. Characterization of human epigenomes. Curr. Opin. Genet. Dev. 2009;19:127–134. doi: 10.1016/j.gde.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Leeuwen F, van Steensel B. Histone modifications: from genome-wide maps to functional insights. Genome Biol. 2005;6:113. doi: 10.1186/gb-2005-6-6-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouzarides T. Histone methylation in transcriptional control. Curr. Opin. Genet. Dev. 2002;12:198–209. doi: 10.1016/s0959-437x(02)00287-3. [DOI] [PubMed] [Google Scholar]
- 5.Schones DE, Zhao K. Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 2008;9:179–191. doi: 10.1038/nrg2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 7.Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, Yamochi T, Urano T, Furukawa K, Kwabi-Addo B, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat. Genet. 2008;40:741–750. doi: 10.1038/ng.159. [DOI] [PubMed] [Google Scholar]
- 8.Bock C, Lengauer T. Computational epigenetics. Bioinformatics. 2008;24:1–10. doi: 10.1093/bioinformatics/btm546. [DOI] [PubMed] [Google Scholar]
- 9.O'C;onnor TR, Wyrick JJ. ChromatinDB: a database of genome-wide histone modification patterns for Saccharomyces cerevisiae. Bioinformatics. 2007;23:1828–1830. doi: 10.1093/bioinformatics/btm236. [DOI] [PubMed] [Google Scholar]
- 10.Baxevanis AD, Landsman D. Histone Sequence Database: new histone fold family members. Nucleic Acids Res. 1998;26:372–375. doi: 10.1093/nar/26.1.372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan SA, Aravind L, Makalowska I, Baxevanis AD, Landsman D. The histone database: a comprehensive WWW resource for histones and histone fold-containing proteins. Nucleic Acids Res. 2000;28:320–322. doi: 10.1093/nar/28.1.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marino-Ramirez L, Hsu B, Baxevanis AD, Landsman D. The Histone Database: a comprehensive resource for histones and histone fold-containing proteins. Proteins. 2006;62:838–842. doi: 10.1002/prot.20814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y. SysPTM – a systematic resource for proteomic research of post-translational modifications. Mol. Cell Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang H, Maertens AM, Hyland EM, Dai J, Norris A, Boeke JD, Bader JS. HistoneHits: a database for histone mutations and their phenotypes. Genome Res. 2009;19:674–681. doi: 10.1101/gr.083402.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang YC, Huang HC, Juan HF. MeInfoText: associated gene methylation and cancer information from text mining. BMC Bioinformatics. 2008;9:22. doi: 10.1186/1471-2105-9-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ongenaert M, Van Neste L, De Meyer T, Menschaert G, Bekaert S, Van Criekinge W. PubMeth: a cancer methylation database combining text-mining and expert annotation. Nucleic Acids Res. 2008;36:D842–D846. doi: 10.1093/nar/gkm788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Toyota M, Suzuki H, Yamashita T, Hirata K, Imai K, Tokino T, Shinomura Y. Cancer epigenomics: implications of DNA methylation in personalized cancer therapy. Cancer Sci. 2009;100:787–791. doi: 10.1111/j.1349-7006.2009.01095.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu. Rev. Med. 2008;59:267–280. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 21.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vucic EA, Wilson IM, Campbell JM, Lam WL. Methylation analysis by DNA immunoprecipitation (MeDIP) Methods Mol. Biol. 2009;556:141–153. doi: 10.1007/978-1-60327-192-9_10. [DOI] [PubMed] [Google Scholar]
- 23.Rakyan VK, Down TA, Thorne NP, Flicek P, Kulesha E, Graf S, Tomazou EM, Backdahl L, Johnson N, Herberth M, et al. An integrated resource for genome-wide identification and analysis of human tissue-specific differentially methylated regions (tDMRs) Genome Res. 2008;18:1518–1529. doi: 10.1101/gr.077479.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC enome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrell D, Dimmer E, Huntley RP, Binns D, O'D;onovan C, Apweiler R. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's; online Mendelian inheritance in man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuler GD, Boguski MS, Stewart EA, Stein LD, Gyapay G, Rice K, White RE, Rodriguez-Tome P, Aggarwal A, Bajorek E, et al. A gene map of the human genome. Science. 1996;274:540–546. [PubMed] [Google Scholar]
- 31.Srinivasarao GY, Yeh LS, Marzec CR, Orcutt BC, Barker WC, Pfeiffer F. Database of protein sequence alignments: PIR-ALN. Nucleic Acids Res. 1999;27:284–285. doi: 10.1093/nar/27.1.284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He X, Chang S, Zhang J, Zhao Q, Xiang H, Kusonmano K, Yang L, Sun ZS, Yang H, Wang J. MethyCancer: the database of human DNA methylation and cancer. Nucleic Acids Res. 2008;36:D836–D841. doi: 10.1093/nar/gkm730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischle W, Wang Y, Allis CD. Histone and chromatin cross-talk. Curr. Opin. Cell Biol. 2003;15:172–183. doi: 10.1016/s0955-0674(03)00013-9. [DOI] [PubMed] [Google Scholar]
- 35.Cosgrove MS, Wolberger C. How does the histone code work? Biochem. Cell Biol. 2005;83:468–476. doi: 10.1139/o05-137. [DOI] [PubMed] [Google Scholar]
- 36.Zhao XD, Han X, Chew JL, Liu J, Chiu KP, Choo A, Orlov YL, Sung WK, Shahab A, Kuznetsov VA, et al. Whole-genome mapping of histone H3 Lys4 and 27 trimethylations reveals distinct genomic compartments in human embryonic stem cells. Cell Stem Cell. 2007;1:286–298. doi: 10.1016/j.stem.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 37.Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 38.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 39.Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr. Opin. Cell Biol. 2002;14:286–298. doi: 10.1016/s0955-0674(02)00335-6. [DOI] [PubMed] [Google Scholar]