Abstract
The wide uptake of next-generation sequencing and other ultra-high throughput technologies by life scientists with a diverse range of interests, spanning fundamental biological research, medicine, agriculture and environmental science, has led to unprecedented growth in the amount of data generated. It has also put the need for unrestricted access to biological data at the centre of biology. The European Bioinformatics Institute (EMBL-EBI) is unique in Europe and is one of only two organisations worldwide providing access to a comprehensive, integrated set of these collections. Here, we describe how the EMBL-EBI’s biomolecular databases are evolving to cope with increasing levels of submission, a growing and diversifying user base, and the demand for new types of data. All of the resources described here can be accessed from the EMBL-EBI website: http://www.ebi.ac.uk
INTRODUCTION
New DNA sequencing methods are revolutionising biology, with impacts throughout the pure and applied life-sciences. In the last five years there have been spectacular improvements in the speed, capacity and affordability of genome sequencing (1). This has made it feasible to perform large-scale studies of broad application to medicine, agriculture and environmental science. These will enable humankind to gain a deeper understanding of human variability (www.1000genomes.org), unravel the links between genetic variation and disease (www.wtccc.org.uk), identify and select for high yield and disease resistance in agricultural crops (2), and catalogue biodiversity (http://www.barcodinglife.org) with the aim of improving species conservation.
The European Bioinformatics Institute, part of the European Molecular Biology Laboratory (EMBL-EBI), has a mandate to provide biomolecular data resources of universal relevance to biological and medical research. Although its focus is European, its impact is global and it is the European node in several worldwide collaborative initiatives to collect, organise and disseminate data for the life-sciences. Demand for access to these data is high and continues to grow, averaging 3.5 million web requests on the EMBL-EBI website each day. Approximately 300 000 unique users visit the EMBL-EBI website every month, and close to a million jobs per month are performed using the EMBL-EBI’s web-services.
The roots of the EMBL-EBI’s data collection lie in the world’s first public database of DNA sequences, developed at the European Molecular Biology Laboratory in Heidelberg in the early 1980s. To this day, the European Nucleotide Archive (ENA) (3) is a central pillar of the EMBL-EBI, not only because DNA is the code of life and a reference point for objects in other databases, but also because it set precedents that have become established principles for the management of biological research data. These include: providing open and unrestricted access to the data; setting up data sharing agreements among globally important data collections; and working with journal publishers to ensure that data forming the basis of scientific publications are submitted to appropriate databases and become part of the public record of science (Box 1).
Box 1.The EMBL-EBI’s principles of service provision.
Accessibility—We are the custodians (not the owners) of biological data provided by the community, and progress in biological research depends on completely open access to these data. All our data and tools are therefore freely available to the research community, without restriction.
Compatibility—The EMBL-EBI has possibly done more than any other organisation in the world to promote the adoption of standards in bioinformatics; the development of these standards promotes data sharing.
Comprehensive data sets—Where several publicly available repositories exist, we have negotiated data-sharing agreements to ensure that our resources are comprehensive and up-to-date. We also negotiate with publishers to ensure that, wherever practicable, biological data are placed in a public repository as part of the publication process and cross-referenced in the relevant publication.
Portability—If practical our datasets are available for download. In many cases the entire software system can be downloaded and installed locally.
Quality—Our databases are enhanced through annotation: features of the objects stored in them are extracted from other sources, defined and interpreted. Much of our annotation is performed by highly qualified biologists, and automated annotation is subjected to rigorous quality control. In many cases we also exploit expertise outside of the EBI for specialist annotation.
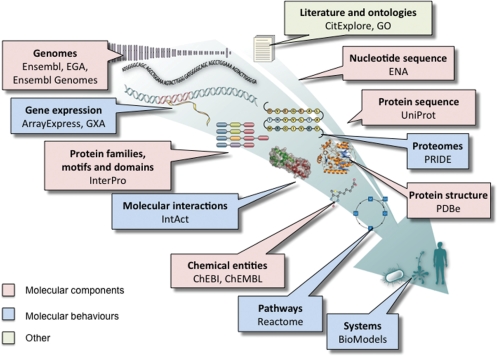
The EMBL-EBI was established as an Outstation of EMBL in during the 1990s. It was already collaborating with the Swiss Institute of Bioinformatics to develop the Swiss-Prot and TrEMBL Protein sequence databases, and soon began collaborating with the Protein Data Bank on protein structure data. Up until this point, life-science research still focused on studying one gene or one protein at a time, but the completion of the first full genome sequences marked an important turning point: whole genome analysis became possible; high-throughput technologies for studying transcripts, proteins, small molecules and structures on a genome-wide scale began to be widely adopted, and the era of reductionism began to make way for systems biology. Our desire to keep pace with researchers’ demand for open access to new data types, coupled with the need to standardise data in different databases, has driven a significant expansion of the EMBL-EBI’s core set of databases (Figure 1). These ‘core’ databases are those that aim to provide complete collections of generic value to life-science. They fall roughly into two categories: those describing the molecular components of biological systems (nucleotide sequences, protein sequences, macromolecular structures and small molecules, for example) and those describing their ‘behaviours’ or the outcomes of those behaviours (transcription, translation and interaction, for example). In addition to the core databases, the EMBL-EBI also hosts a large number of specialist data resources. Two examples are reviewed in this database issue and illustrate the variety of applications and communities that the EMBL-EBI’s databases support: the European Mutant Mouse Archive (4), a partner in the International Mouse Phenotyping Consortium, provides information on mouse mutant strains, enabling life-science researchers to link phenotypic information to mutations, and to source mouse lines for their research. The non-redundant patent-sequence database collection (5) was created through a long-standing collaboration between the European Patent Office and the EMBL-EBI. It provides open access to sequences associated with patent applications—of broad utility not only to intellectual property specialists searching for prior art, but also to the research community as a whole, because sequences published in patent applications may not be published elsewhere, and the patent applications may contain unique scientific information.
Figure 1.
The EMBL-EBI’s core data resources, colour coded according to whether they focus on molecular entities or molecular behaviours.
Here we provide a bird’s eye view of the EMBL-EBI’s core biomolecular database collection, describing the rationale for recent launches and major developments, and providing a starting point for users. Several reviews of individual EMBL-EBI data resources are provided in this issue and are cross-referenced here.
GENOMES
A natural reference point for biology
The genome is a concept at the heart of biology. Since the first complete genome was sequenced in the mid 1990s, over 1000 more have been sequenced, annotated and submitted to the public databases; but these represent only a small proportion of the total number expected in the near term. Ultra-high throughput sequencing technologies are generating genome sequences at a rapidly accelerating rate, both to gap-fill portions of the taxonomy where no genome sequence has yet been deciphered and to generate data for variation in populations of species of particular interest. These technologies are also being used to generate gene regulation and expression data on a genome-wide scale.
The EMBL-EBI, in collaboration with the Wellcome Trust Sanger Institute, developed the Ensembl Genome Browser (6) in 2000. Ensembl’s original purpose was to facilitate navigation and analysis of the human genome, focusing on the annotation of known genes and predicting the location of previously uncharacterised ones. This methodology was extended to other important model organisms, including brewer’s yeast and fruit fly. As the list of sequenced organisms grew, a strategic decision was taken to focus on chordates owing to their ability to help us understand human biology and evolution. Over the past 10 years, as the genomes of more organisms have been sequenced, Ensembl’s coverage has grown to some 51 genomes. Year 2009 has witnessed addition of the first reptile genome—that of the anole lizard—filling an important evolutionary gap by adding the final vertebrate class to Ensembl’s collection. Inter-species comparative genomics experiments using the Ensembl system have provided some important insights into previously overlooked regions of the genome [for example, the discovery that the human and chicken genomes shared large stretches of conservation in non-coding regions (7)].
A unified view of the tree of life
The falling cost of genome sequencing makes it feasible that the genomes of all species of significant scientific interest will be sequenced in the near future, and that projects to sequence many individuals of the same species will follow. Such endeavours are already underway for humans (www.1000genomes.org), Arabidopsis (8), Plasmodium (http://www.genome.gov/26523588) and Drosophila (http://www.dpgp.org). One of the EMBL-EBI’s major achievements in 2009 has been the successful application of the Ensembl system to the rest of the taxonomic tree: Ensembl Genomes (9) provides a companion service to Ensembl in the form of five new sites: Ensembl Bacteria, Ensembl Protists, Ensembl Fungi, Ensembl Plants and Ensembl Metazoa. Ensembl Genomes replaces several pre-existing EMBL-EBI resources (Integr8, Genome Reviews and ASTD), thereby unifying services and simplifying data access for users. The launch of Ensembl Genomes provides a consistent framework for inter-species analyses across the whole of taxonomic space.
A common set of user interfaces, including a graphical genome browser, FTP, BLAST search, a query-optimised data warehouse, programmatic access and a Perl API, is provided for all domains, mirroring what was previously available for vertebrates through Ensembl. Data types incorporated include annotation of (protein and non-protein coding) genes, cross-references to external resources and high-throughput experimental data (for example, data from large-scale studies of gene expression and polymorphism visualised in their genomic context). Pre-computed comparative analyses, both within defined clades and across the wider taxonomy, and sequence alignments and gene trees resulting from these are also available.
With such a broad scope, Ensembl Genomes is dependent on the contribution of the scientific community; the EBI can provide an infrastructure and a pan-taxonomic perspective, but the biological expertise is widely dispersed. Many of the databases within Ensembl Genomes are produced by, or in close collaboration with, specialist resources with domain-specific expertise (WormBase, VectorBase and Gramene, for example), and we are working actively to increase these relationships as new species are introduced into the site.
From genotype to phenotype
Distinguishing the genetic differences between individuals of the same species and linking these genotypic differences to phenotypic differences provide important leads for medical and agricultural research. The EMBL-EBI launched the European Genome-phenome Archive (EGA)—a repository for all types of potentially identifiable data types including the array-based genotype data from genome-wide association studies—in July 2008. The EGA stores the raw data from many types of experiments including case control studies, cancer sequencing and population studies. Available data types include single nucleotide polymorphism (SNP) and copy number variation (CNV) genotypes, whole genome sequence and phenotype data. Each data type is stored at the EGA using methods designed to ensure that the storage and distribution is done in accordance with the consent and confidentiality agreements that the research participants agreed to at the time of entry into the study.
Coping with the deluge of next-generation sequencing data
Next-generation sequencing has led to previously unimaginable amounts of data being deposited in the public nucleotide sequence databases. The ENA has been established (3) at the EBI to consolidate existing major sequence resources, namely, the European Trace Archive, previously maintained at the Wellcome Trust Sanger Institute, the EMBL Nucleotide Sequence Database (EMBL-Bank) and the Sequence Read Archive (SRA), the newly established repository for raw data from next generation sequencing platforms. Significant technology developments at ENA have led to improved submission and data access tools.
The ENA comprises three parts: ENA-Annotation, ENA-Assembly and ENA-Reads. ENA-Annotation contains detailed functional annotation, for example of individual, well characterised coding sequences. ENA-Assembly is designed for efficient storage of sequence assemblies. Finally, ENA-Reads is optimised for the efficient storage of sequence trace information. These include capillary trace sequences (the Trace Archive) and next-generation reads (SRA, which is the fastest growing part of the ENA). Entries from different data classes are connected through high-level sample and project information.
A new, automated sequence-read submission tool makes regular submission of next-generation sequencing data very straightforward. For manual submissions of annotated, assembled sequences, a template-based sequence submission system has been developed. Submitters can choose from a set of templates tailor-made for each major annotation scenario, can upload large-scale annotations prepared in third party annotation tools, and can also design their own templates.
The most recent innovation in ENA is a newly launched browser that provides integrated access to data from all parts of ENA, including top-level project records, taxonomic information, assembled sequence, functional annotation, assembly information and metadata for trace and next-generation reads. All accessions and many stored fields have been made available for search through the EB-eye search tool. Novel sequence similarity search tools optimised for short reads delivered by next-generation platforms are under development.
TRANSCRIPTOMES
Genome-wide gene expression assays, originally using microarrays and more recently high-throughput sequencing, can either answer specific questions (for example, which genes are differentially expressed in healthy versus diseased liver) or provide reference data sets (for example allowing gene expression patterns in different tissues, or at different developmental stages, to be compared). Large-scale expression datasets can be used to answer questions unrelated to the study for which the data were originally generated. For example, a gene expression study that reveals differentially expressed genes characteristic of a particular type of cancer may also reveal candidate genes for therapeutics development, or shed light on regulatory mechanisms perturbed in that form of cancer.
The ArrayExpress Archive was launched by the EMBL-EBI in 2002 as the world’s first open-access, standards-compliant repository for high-throughput transcriptomics assays. In compliance with the MIAME initiative (10), most scientific journals now require publication-related microarray gene expression data to be deposited in ArrayExpress (11) or the NCBI’s Gene Expression Omnibus (GEO) (12). Data from over 10 000 studies are available from these archives, but using these data to answer biological questions is not straightforward.
The EMBL-EBI launched the Gene Expression Atlas (GXA) (13) to simplify the analysis of gene expression data. Users can pose gene-centric queries, to find out under which conditions (or where in the organism) a gene of interest is differentially expressed. Alternatively, they can pose condition-centric queries, to find out which genes are differentially expressed in a particular condition or site. Both types of query can be combined to focus on particular genes and their role in a specific condition; for example, GXA makes it straightforward to search for members of the Wnt signalling pathway that are expressed in colorectal adenocarcinoma.
GXA takes a subset of the data from the ArrayExpress Archive, including data imported from GEO (12) and subjects it to rigorous curation. Mapping of genes to the latest genome-builds ensures that each gene in GXA has an unambiguous reference point. Mapping of conditions to a purpose-built ontology—the Experimental Factor Ontology (EFO) (11)—ensures that users retrieve all the results relevant to their query, not just those that exactly match the text of their query. More information on GXA can be found in an e-learning tutorial at http://www.ebi.ac.uk/training/elearningcentral.
PROTEOMES
UniProt (14) is the globally recognised ‘gold-standard’ data resource for information about proteins. UniProt is produced by the UniProt Consortium, a collaboration between the EMBL-EBI, the Swiss Institute of Bioinformatics (SIB) and the Protein Information Resource (PIR). The UniProt Knowledgebase, the centrepiece of the UniProt Consortium’s activities, provides an expertly and richly curated protein database consisting of two sections: UniProtKB/Swiss-Prot contains manually curated information on well-characterised proteins and UniProtKB/TrEMBL contains automatically annotated information on protein sequences mostly sourced from the ENA (3).
As completely sequenced genomes have their full complement of protein-coding genes characterised, it becomes feasible to provide richly annotated complete proteomes in UniProtKB/Swiss-Prot. A first draft of the human proteome, comprising 20 325 protein-coding sequences, was released in September 2008. This data set has now been re-annotated to improve the depth and quality of the information provided. New splice variants and polymorphisms have been added to existing records, and records have been created for newly discovered protein sequences. UniProt has joined the Consensus CDS (CCDS) project (15), a collaborative effort including the Wellcome Trust Sanger Institute, the University of California, Santa Cruz, the US National Center for Biotechnology Information and the EMBL-EBI, to identify a core set of consistently annotated and high-quality human and mouse protein-coding regions. The long-term goal is to support convergence towards a standard set of gene and protein annotations.
The complete proteome for the fission yeast Schizosaccharomyces pombe is also now available. Comparison with the proteome of the evolutionarily distant budding yeast, Saccharomyces cerevisiae, provides a powerful tool for orthologue prediction.
Another innovation in UniProt is full cross-linking with PRIDE (16), the EMBL-EBI’s standards-compliant resource for mass spectrometry based proteomics. This allows PRIDE submitters to improve the exposure of their data, and allows PRIDE data to be used to annotate UniProt protein entries.
The increasing number of publications on protein identification by mass-spectrometry provided the impetus for the launch of PRIDE in 2005, and the PRIDE team has worked closely with publishers to ensure that published proteomics data are not lost to further analysis. For instance, the submission process has now been made much easier thanks to the new tool PRIDE Converter (17). As a result, PRIDE is now the recommended submission point for proteomics data for several journals, including Nature Biotechnology (18), Nature Methods (19) and Proteomics. PRIDE is also a founding partner of the ProteomExchange consortium (http://www.proteomexchange.org) (20). The core members of this consortium (PRIDE, NCBI Peptidome, Tranche, PeptideAtlas and GPMDB) are working on a system to allow proteomics data sharing between members of the consortium, with PRIDE and NCBI Peptidome as the initial ProteomExchange submission points. In addition to the ProteomExchange initiative, PRIDE and NCBI Peptidome have agreed to replicate and share their data, to ensure that they become optimally visible to the scientific community.
Protein families and domains are invaluable pointers that help biologists to find distantly related proteins and to predict their functions. A daunting array of resources, each with different strengths and weaknesses, is available to search genomes and proteomes for ‘protein signatures’—diagnostic entities that are used to recogne a particular domain or protein family. InterPro (21) is an integrated documentation resource for protein families, domains and functional sites. The member databases of InterPro use different methods and types of biological information to derive protein signatures from well-charactered proteins. By uniting the member databases, InterPro capitalises on their individual strengths, producing a powerful integrated diagnostic tool for protein sequence classification. In 2009, a new member database joined the InterPro consortium and was integrated into the resource: HAMAP (22) provides high-quality automatic annotation of microbial proteomes and complements the existing ten databases already contained in InterPro, giving an in-depth perspective on protein families from the prokaryotic and archaeal kingdoms. Signatures from all member databases continue to be integrated and the total number of entries now stands at 19 170, an increase of just under 2500 signatures in the past year. InterPro has also launched a BioMart (23) for more advanced querying of its data, which includes web service access and links to other BioMarts. Ensembl, UniProt, PDBe, Reactome and PRIDE also have BioMarts, enabling advanced queries to be performed across many of the EMBL-EBI’s core data resources.
STRUCTURES
Structural biology has had an enormous impact on our understanding of biology and medicine—as evidenced by four Nobel Prizes awarded to workers in this field in this century alone (2002, 2003, 2006, 2009). Three-dimensional structures give us mechanistic insight into how macromolecules work, and help to explain how their functions are disrupted by mutation or interaction with small molecules. As structural genomics efforts begin to bear fruit (for example, the Midwest Center for Structural Genomics deposited its 1000th structure in the PDB in July 2009), the demand for efficient access to standard ways of viewing and describing protein structures grows.
The Protein Databank in Europe (PDBe) (24), formerly known as the Macromolecular Structure Database, is the European partner of the worldwide Protein Databank Organisation (wwPDB) (25), the other partners being the Research Collaboratory for Structural Bioinformatics (RCSB) (26) and the BioMagResBank (BMRB) (27) in the USA, and the Protein Data Bank Japan (PDBj) (28). wwPDB maintains the worldwide repository of bio-macromolecular structure data.
Year 2009 witnessed the handover of the PDBe group’s leadership to Gerard Kleywegt upon the retirement of Kim Henrick. The tireless work of Kim and his team in data remediation and automated analyses of structural data have now been complemented by newly designed PDBeView Atlas pages. These provide an overview of an individual PDB entry in a user-friendly layout and serve as a starting point to further explore the information available in the PDBe database. PDBe’s involvement with the X-ray crystallography, Nuclear Magnetic Resonance spectroscopy and cryo-Electron Microscopy communities have also resulted in improved tools for structure deposition and analysis.
SMALL MOLECULES
As researchers look beyond the genome with the aim of understanding all the processes of life, the need for a public database of biologically relevant ‘small molecules’ (not directly encoded by the genome) grows increasingly strong. ChEBI, the EMBL-EBI’s database of Chemical Entities of Biological Interest (29), has been designed with two aims in mind: first, to provide standard descriptions of molecules that enable other databases to annotate their entries consistently and second, to bridge the gap between small molecules and the macromolecules that they interact with in living systems. ChEBI is a freely available, manually annotated database of small molecular entities. It focuses on chemical nomenclature and structures, and provides a wide range of related chemical data such as formulae, links to other databases and an ontology for the chemical space.
ChEBI has grown 30-fold over the past two years. Much of this growth is due to the ChEMBL dataset—a large collection of information on the properties and activities of drugs and a large set of drug-like small molecules, which was transferred from the publicly listed company Galapagos NV into the public domain in 2008, thanks to a substantial grant from the Wellcome Trust (30). Other sources of new data in ChEBI include data from the PDBeChem database—a library of ligands, small molecules and monomers that are referenced in PDB entries; and small molecules associated with Patents, generated by the Oscar3 project in collaboration with the European Patent Office. A small number of entries have also been added through direct submissions, and a web-based submission tool has been developed for this purpose.
An important new feature of ChEBI is a chemical structure-based search function, which uses a new algorithm developed in the open source OrChem project (31). A new text-search function has also been introduced. Finally, ChEBI has greatly expanded its range of cross-links to other databases, both within and beyond the EMBL-EBI.
INTERACTIONS, PATHWAYS AND SYSTEMS
Computational systems biology today allows research to move beyond the identification of molecular ‘parts lists’ for living organisms, towards synthesising information from different omics-based approaches to generate and test new hypotheses about how biological systems work. Neither experimental nor computational biology alone will be sufficient to uncover a systems-level understanding of biology. The ability to analyse data from transcriptomics, proteomics, protein-interaction studies, pathways and network analysis, and infer how molecules function within systems, is therefore becoming a required skill for experimental biologists. Such analyses form the basis of new hypotheses.
Molecular interactions provide a valuable resource for the elucidation of cellular function. IntAct provides a central, public repository of such interactions, including protein–protein, protein–small-molecule and protein–nucleic-acid interations (32). IntAct is a member of The International Molecular Exchange (IMEx) Consortium (http://imex.sf.net) (33)—a group of public interaction data providers that share curation effort and exchange molecular interaction data, similarly to successful global collaborations for protein and DNA sequences and macromolecular structures. IntAct is also MIMIx compliant, allowing researchers to submit their molecular interaction experiments in a format that complies with agreed community standards (34).
Growth of the data in IntAct has necessitated a redesign of its website, which now enables users to view pair-wise interactions as a list before narrowing down their selection criteria (for example, by choosing only those entries for which there is the strongest experimental evidence) and finally creating a graphical view of the selected interactions. Chemical structure-based searching, using the above-mentioned OrChem structure-based searching algorithm (31), has also been introduced.
Life on the molecular level is an intricate network of biochemical reactions and pathways. Biologists have been elucidating fragments of this network for a century, but a vast amount of the knowledge is scattered and largely inaccessible to computational investigation. Straightforward computational access to this information is a prerequisite for systems biology. Reactome (35) goes some way to satisfying this need as a free, online, open-source, curated pathway database encompassing many areas of human biology.
Reactome is a collaboration between the EMBL-EBI, the Ontario Institute for Cancer Research, New York University Medical Center and Cold Spring Harbor Laboratory. Information is authored by expert biological researchers, maintained by the Reactome editorial staff and cross-referenced to a wide range of other bioinformatics databases. The curated human data are used to infer orthologous events in non-human species including mouse, rat, chicken, puffer fish, worm, fly, yeast, plants and Escherichia coli. Additions in 2009 have included a large number of cell signalling and cell-adhesion pathways, and RNA metabolism; cross-links are now provided to NCBI BioSystems, and several species-centric research communities are using the Reactome system to build their own species-specific versions of Reactome, including gallus-Reactome and fly-Reactome.
PUTTING IT ALL TOGETHER
Data in, knowledge out
Integrating biological data from different sources is the holy grail of bioinformatics, and is made all the more challenging by the fact that different levels of integration are required for different types of task. The sheer volume of data demands that they are structured for analysis by computational pipelines, which, combined with the complexity of the information, poses a substantial challenge. Presenting the data and analyses to scientists in a comprehensible form is equally challenging.
The wealth of information available from the EMBL-EBI website can be especially daunting for users unfamiliar with the core data resources. This is confounded by the fact that many of our data resources are collaborative efforts, with their own websites (e.g. http://www.ensembl.org, http://www.uniprot.org, http://www.reactome.org). The EB-eye search engine, available from every page of the EMBL-EBI website, is designed to allow users to perform text-based searches across the most commonly used fields of all the EMBL-EBI’s core data resources, without any prior knowledge of the underlying data resources. The results are presented as an expandable list of ‘knowledge domains’ covering different data types (nucleotide sequences, protein sequences, macromolecules, etc.). Each knowledge domain can then be expanded, allowing the user to drill down into an individual database, or even a single field in a specific database. Extensive cross-linking between related objects in different databases then allows navigation from one data resource to another. More information about EB-eye can be found at http://www.ebi.ac.uk/inc/help/search_help.html, and there is an e-learning course on the EBeye at http://www.ebi.ac.uk/training/elearningcentral/.
The EB-eye is complemented by BioMart (23). Several of the EMBL-EBI’s core data resources now have their own BioMarts, and it’s possible to perform complex, bespoke searches across several data resources using this technology.
Databases and literature have been tightly connected ever since the first biomolecular databases appeared. Data records appearing in the databases cite the relevant literature, and, for many kinds of data, the literature quotes accession numbers or other identifiers in the databases. The scientific literature provides a natural entry point to the biomolecular databases, and we are now beginning to exploit these connections through CiteXplore, the EMBL-EBI’s portal to the literature. CiteXplore uses text-mining tools developed by researchers both within and beyond the EMBL-EBI to mark up search results with links to many of the core data resources.
Those who need to define their own analysis methods or pipelines are supported by web services technology (36), which allows users’ own programmes to interact with the databases and tools at the EMBL-EBI. These web-services interfaces can be used to retrieve and analyse large amounts of data, or perform complex analyses that involve several nested searches spanning a range of different data resources. This provides an easy and flexible way of dealing with repetitive tasks and large queries. Another strength of web services is that they allow programmers to build complex applications without having to install and maintain the databases and analysis tools and without having to take on the financial overheads that accompany these. Moreover, web services provide easier integration and interoperability between bioinformatics applications and the data they require. A lightweight programme (a client) on the user’s computer communicates with the servers running at the EMBL-EBI. Users can create their own clients or use the perl- and java-based clients that we provide for each of our web services. Instructions on how to build clients in a variety of programming languages can be found in the tutorials at www.ebi.ac.uk/Tools/webservices/tutorial.html.
European context: technical, scientific and political challenges
The genomic era has changed research, by catalysing a shift towards asking questions on a genome-wide scale rather than one gene at a time. But perhaps even more importantly, the genomic era heralded a social change for the life-sciences: the scale of genome-sequencing projects necessitated a completely open attitude towards sharing data, both within and beyond the collaborative groups involved in generating the sequence. Biological experiments are now generating data at rates comparable to astrophysics or particle physics experiments, and it all has to be placed in the public domain and made amenable to analysis by hundreds of thousands of researchers. This requires an upgrade to the information infrastructure of a scale and nature beyond the remit or capability of conventional research funding mechanisms, both nationally and internationally.
The European Strategy Forum on Research Infrastructures (ESFRI, http://cordis.europa.eu/esfri/), which advises the European Commission on Europe’s future needs for research infrastructure, included a major upgrade to Europe’s biological data infrastructure in its 2005 Roadmap. The EMBL-EBI is coordinating ELIXIR—a preparatory phase project that is preparing the ground for building a new infrastructure for biological data. ELIXIR will provide: data resources; bio-compute centres; infrastructure for data integration, software tools and services; support for other European infrastructures in biomedical and environmental research; training and standards development. This will enable ELIXIR’s users to meet the European Grand Challenges, the most important of which are biological, namely: healthcare for an aging population, a sustainable food supply, competitive pharmaceutical and biotechnology industries, and protection of the environment.
ELIXIR will require financial support from all the European Member states. Two countries—Sweden and the UK—have already committed funds but there is still a long way to go. Over the past 18 months, with significant stakeholder input, ELIXIR’s workpackage committees have written their recommendations for ELIXIR. These are available at http://www.elixir-europe.org/page.php?page=reports, and we continue to welcome feedback on them from our users, who are also vitally important stakeholders. During the next part of ELIXIR’s preparatory phase, these recommendations will be incorporated into a business case which will lead to the construction of ELIXIR beginning perhaps as early as 2011.
The huge quantities of data that are now being generated by life-science research provide unforeseen opportunities and challenges. Already the new DNA sequencing methods are providing the technology to sequence individual genomes, to quantify expression, to measure biodiversity and its response to the environment, to study cancer differentiation and to measure a patient’s responses to therapy to name but a few. Realising the benefits of this knowledge to health and human wellbeing will depend crucially on applying computational methods to the vast repositories of data. Computational biology will necessarily move to centre stage. Biological data resources will lie at the heart of new discoveries and their applications, and we must build the infrastructure to support this endeavour. We firmly believe that this infrastructure must remain rooted in the principles of open access and international collaboration that have enabled post-genomic research to progress at such an impressive pace.
FUNDING
Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The EMBL-EBI is indebted to the support of its funders: EMBL’s Member States, the European Commission, the Wellcome Trust, the UK Research Councils, the US National Institutes of Health and our industry partners. We are also indebted to hundreds of thousands of scientists who have submitted data and annotation to the shared data collections. The authors would like to thank Guy Cochrane, Paul Flicek, Sarah Hunter, Paul Kersey, Gerard Kleywegt, Claire O’Donovan and Juan Vizcaíno for their constructive feedback on this manuscript.
REFERENCES
- 1.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bevan M, Waugh R. Applying plant genomics to crop improvement. Genome Biol. 2009;8:302. doi: 10.1186/gb-2007-8-2-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinonen R, Akhtar R, Bonfield J, Bower L, Corbett M, Cheng Y, Demiralp F, Faruque N, Goodgame N, Gibson R, et al. Improvements to services at the European Nucleotide Archive. Nucleic Acids Res. 2010;38:D39–D45. doi: 10.1093/nar/gkp998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilkinson P, Sengerova J, Matteoni R, Chen C-K, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, et al. EMMA – mouse mutant resources for the international scientific community. Nucleic Acids Res. 2010;38:D570–D576. doi: 10.1093/nar/gkp799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, McWilliam H, Richart de la Torre A, Grodowski A, Benediktovich I, Goujon M, Nauche S, Lopez R. Non-redundant patent sequence databases with value-added annotations at two-levels. Nucleic Acids Res. 2010;38:D52–D56. doi: 10.1093/nar/gkp960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.International Chicken Genome Sequencing Consortium. Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- 8.Weigel D, Mott R. The 1001 genomes project for Arabidopsis thaliana. Genome Biol. 2009;10:107. doi: 10.1186/gb-2009-10-5-107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kähäri A, et al. Ensembl Genomes: Extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brazma A, Hingamp P, Quackenbush J, Sherlock G, Spellman P, Stoeckert C, Aach J, Ansorge W, Ball CA, Causton HC, et al. Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat. Genet. 2001;29:365–371. doi: 10.1038/ng1201-365. [DOI] [PubMed] [Google Scholar]
- 11.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update—from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2008;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A. Gene Expression Atlas at the European Bioinformatics Institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: Identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barsnes H, Vizcaíno JA, Eidhammer I, Martens L. PRIDE Converter: making data sharing easy. Nat. Biotechnol. 2009;27:598–599. doi: 10.1038/nbt0709-598. [DOI] [PubMed] [Google Scholar]
- 18.Anonymous. 2007. Democratizing proteomics data. Nat. Biotechnol., 25, 262. [DOI] [PubMed] [Google Scholar]
- 19.Anonymous. 2008. Thou shalt share your data. Nat. Methods, 5, 209. [Google Scholar]
- 20.Hermjakob H, Apweiler R. The Proteomics Identifications Database (PRIDE) and the ProteomExchange Consortium: making proteomics data accessible. Expert Rev. Proteom. 2006;3:1–3. doi: 10.1586/14789450.3.1.1. [DOI] [PubMed] [Google Scholar]
- 21.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lima T, Auchincloss AH, Coudert E, Keller G, Michoud K, Rivoire C, Bulliard V, de Castro E, Lachaize C, Baratin D, et al. HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res. 2009;37:D471–D478. doi: 10.1093/nar/gkn661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart—biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Velankar S, Best C, Beuth B, Boutselakis HC, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Databank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Standley DM, Kinjo AR, Kinoshita K, Nakamura H. Protein Structure Database with new web services for structural biology and biomedical research. Brief. Bioinform. 2008;9:276–285. doi: 10.1093/bib/bbn015. [DOI] [PubMed] [Google Scholar]
- 29.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical Entities of Biological Interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Houlton S. Wellcome boost for open access chemistry. Nat. Rev. Drug Discov. 2008;7:789–790. [Google Scholar]
- 31.Rijnbeek M, Steinbeck C. An open source chemistry search engine for Oracle. J. Cheminf. 2009;1:17. doi: 10.1186/1758-2946-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. Proteomics. 2007;7(Suppl 1):28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 34.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stümpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The Minimum Information required for reporting a Molecular Interaction Experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 35.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McWilliam H, Valentin F, Goujon M, Li W, Narayanasamy M, Martin J, Miyar T, Lopez R. Web services at the European Bioinformatics Institute-2009. Nucleic Acids Res. 2009;37:W6–W10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]