Abstract
The Immuno Polymorphism Database (IPD) (http://www.ebi.ac.uk/ipd/) is a set of specialist databases related to the study of polymorphic genes in the immune system. The IPD project works with specialist groups or nomenclature committees who provide and curate individual sections before they are submitted to IPD for online publication. The IPD project stores all the data in a set of related databases. IPD currently consists of four databases: IPD-KIR, contains the allelic sequences of Killer-cell Immunoglobulin-like Receptors, IPD-MHC, is a database of sequences of the Major Histocompatibility Complex of different species; IPD-human platelet antigens, alloantigens expressed only on platelets and IPD-ESTDAB, which provides access to the European Searchable Tumour cell-line database, a cell bank of immunologically characterised melanoma cell lines. The data is currently available online from the website and ftp directory.
INTRODUCTION
The study of the immune system constitutes many different complex areas of research. The Immuno Polymorphism Database (IPD) is a set of specialist databases related to the study of polymorphic genes of the immune system. The IPD project works with specialist groups or nomenclature committees, which each curate a different section of the project. IPD currently consists of four databases: IPD-KIR, contains the allelic sequences of Killer-cell Immunoglobulin-like Receptors (KIR); IPD-MHC, is a database of sequences of the Major Histocompatibility Complex (MHC) of different species; IPD-human platelet antigens (IPD-HPA), alloantigens expressed only on platelets and IPD-ESTDAB, which provides access to the European Searchable Tumour cell-line database (ESTDAB), a cell bank of immunologically characterised melanoma cell lines. By providing a centralised resource for the work of different groups it is hoped we can bring together similar data to aid in the study and analysis of this area.
This is done by contacting various expert groups, such as the nomenclature committees for certain genes, and inviting them to contribute their data to a central resource. The individual nomenclature committees are all established within their own field, their continuing role being the identification and naming of new alleles based on the submission of new sequences to generalist databases like the European Molecular Biology Laboratory’s nucleotide sequence database (also known as EMBL-Bank) (1), the National Center for Biotechnology Information’s GenBank (2) and the DNA DataBank of Japan (DDBJ) (3). The main difference between the data held within a specialist system like IPD and the generalist databases is the further curation of the files by experts in the relevant field. This additional step allows for improvements in data quality and the addition of more specialised information. This may result in there being differences in the entries kept in the different databases; in this case the entry in IPD should be considered the most accurate. The inclusion of some nomenclature committees has meant the online publication of sequence alignments for the first time. This is particularly important when it comes to the sequences of polymorphic genes. Also because the IPD project is able to provide the work of different committees in a common format, it is easier to compare the sequences of different species.
The IPD project stores all the data in a set of related databases. Those sections with similar data, IPD-KIR and IPD-MHC share the same database structure. The sharing of a common database structure makes it easier to implement common tools for data submission and retrieval. Other unrelated sections like IPD-ESTDAB currently have their own unique structure.
IPD-KIR
The KIR are members of the immunoglobulin super family (IgSF) formerly called Killer-cell Inhibitory Receptors. KIRs have been shown to be highly polymorphic both at the allelic and haplotypic levels (4). They are composed of two or three Ig-domains, a transmembrane region and cytoplasmic tail, which can in turn be short (activatory) or long (inhibitory). The Leukocyte Receptor Complex (LRC), which encodes KIR genes, has been shown to be polymorphic, polygenic and complex in a manner similar to the MHC. Because of the complexity in the KIR region and KIR sequences a KIR Nomenclature Committee was established in 2002, to undertake the naming of KIR allele sequences. The first KIR Nomenclature report was published in 2003 (5), which coincided with the first release of the IPD-KIR database. The number of officially named KIR alleles has increased since the initial release which contained 89 alleles. As of September 2009, there are over 450 alleles, which code for over 230 unique protein sequences. This has meant that since its initial release there have been nine further releases of the IPD-KIR database, the latest being in February 2009.
The online tools available for IPD-KIR include those for performing allele queries, sequence alignments and cell queries. As the database is based on the work of a nomenclature committee, the website includes links to a portable document format (PDF) file of recent nomenclature reports. From the data contained within these reports the database is also able to provide individual allele reports (Figure 1). These pages contain the official allele name, any previous designations, the EMBL, GenBank, or DDBJ accession number(s) and a reference linked wherever possible to the PubMed abstract. Where possible additional details on the source of sequence are also provided. This source material is normally in the form of a cell-line or DNA from which each allele in the database was isolated and characterised and it’s availability. The information contained within this dataset can be searched independently from the allele data.
Figure 1.
Allele report. The figure shows part of the report provided for each KIR allele. The report provides cross-references to an SRS entry (KIR00002), the source entries in EMBL-BANK (AY789055-U24076) and to the seminal citations in PubMed. Other information provided includes links to the source material, known ethnic origins and genomic DNA, cDNA and translated protein sequence.
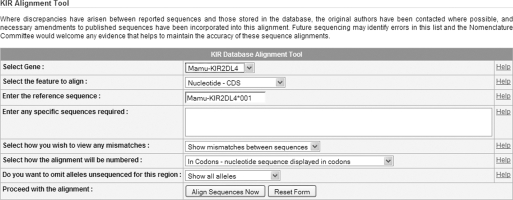
Within each IPD section alleles of a particular gene may differ from each other by as little a single nucleotide. With sometimes hundreds of alleles for a particular gene we need to be able to graphically represent where these differences lie. These polymorphic positions are also often conserved within certain groups of alleles. For this reason the sequences are displayed as multiple sequence alignments which highlight the polymorphic positions. These alignments allow a visual interpretation of sequence similarity, so that polymorphic positions can easily be identified and motifs found in multiple alleles are easily identified. The sequence alignments are available via a link from the section homepage. The sequence alignment tool uses the same basic interface for both IPD-KIR database and IPD-MHC. The interface provided (Figure 2) lets the user define a number of key variables for the alignments, before producing an online output, which can be printed or downloaded. The first step in any alignment is to select the locus of interest. The tool provides a drop-down list of all loci. The selection of a locus automatically updates the list of features, which can be aligned, as well as the default reference sequence used for the alignment. The types of feature available for alignment are the nucleotide coding sequence and individual exons, the signal peptide, mature protein and full-length protein sequence. We also include the genomic sequence, which covers some of the 5′ and 3′ untranslated regions, exons and introns into the database for over half of the alleles in the KIR system. The alignment tool options also allow the user to display a subset of alleles of a particular locus, omit alleles unsequenced for a particular region and also to align against a particular reference or consensus sequence. The alignment tool uses standard formatting for the display of sequence alignments. The alignment tool does not perform a sequence alignment each time it is used, but it extracts pre-aligned sequences, allowing for faster access. The alignments adhere to a number of conventions for displaying evolutionary events and numbering. The numbering of the alignments is based upon the sequence of the reference allele. For a nucleotide sequence, the A of the initiation Methionine codon is denoted nucleotide +1 and the nucleotide 5′ to +1 is numbered −1. There is no nucleotide zero (0). All numbering is based on the ATG of the reference sequence. If a nucleotide sequences is displayed in codons, then the protein numbering is applied. For amino acid-based alignments, the first codon of the mature protein, after cleavage of the signal sequence is labelled codon 1 and the codon 5′ to this is numbered −1. In all sequences the following conventions are used. Where identity to the reference sequence is present the base will be displayed as a hyphen (-). Non-identity to the reference sequence is shown by displaying the appropriate base at that position. Where an insertion or deletion has occurred, this is be represented by a period (.). If the sequence is unknown at any point in the alignment, this is be represented by an asterisk (*). In protein alignments for null alleles, the 'Stop' codons are represented by an X and the sequence following the termination codon, is not marked up and appears blank. The flexibility of the new alignment tool means that unlike in previous alignments you can now display a small subset of sequences against an allele of your choice, using a number of display options (Figure 3).
Figure 2.
Alignment interface. The alignment interface provides a user-friendly method of viewing sequence alignments with output options easily selected.
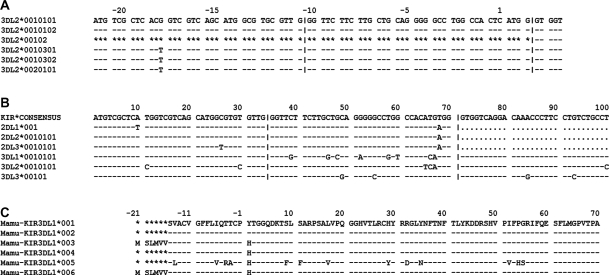
Figure 3.
Alignment formats available from IPD. In these alignments a dash (−) indicates identity to the reference sequence and an asterisk (*) denotes an unsequenced base. The first alignment shows the default output for the nucleotide sequence of KIR3DL2. The second alignment shows alleles from six different genes. The pipes indicate exon boundaries and the periods represent the non-expressed pseudoexon 3 in the 2DL genes. The final alignment shows the translated protein sequences of the Rhesus Macaque (Macaca mulatta) 3DL1 gene.
Further recent additions to the tools available from the IPD-KIR site include a KIR ligand calculator (6). The ligand calculator allows the user to define which KIR ligands are present in a transplant setting based on the HLA typing of a patient and prospective donor. This is because recent transplant strategies based on KIR-ligand mismatch to predict NK cell alloreactivity have resulted in less relapse, less GvHD and better overall survival in patients with acute myeloid leukaemia (AML) (7). The KIR-ligands are HLA molecules that can be grouped into three major categories based on the amino acid sequence determining the KIR-binding epitopes in HLA-C and HLA-B molecules. All expressed HLA-C alleles are of the C1 or C2 group (8) and most HLA-B alleles can be classified as either Bw4 or Bw6 (9). The receptors KIR2DL1, KIR2DL2 and KIR3DL1 bind KIR-ligand C2, C1 and Bw4, respectively, resulting in inhibition of NK cell mediated lysis. The output lists the ligands associated with the HLA typing entered. In the case of two digit typings the most common ligand for the allele type is displayed. A link to the full list of alleles matching the type given and their associated ligands is also provided. For two digit typings any exceptions to the list are also listed along with their motif. For example B*15 alleles are predominately Bw6 however there are a number of alleles, B*1513 for example, which contain the Bw4 motif. The output classifies alleles in the exceptions list as ‘Rare’ if they have a gene frequency of <0.001 and have been seen in less than three unrelated individuals (10). This classification allows users to make a judgement call over whether the exceptions are likely to be seen in their samples.
The typing of KIRs is dependant on up to date lists of alleles and primers, and many typing laboratories have spreadsheets detailing probe hit patterns for different alleles. Each time a new database is released it is necessary to update these ever-expanding lists. The new Probe and Primer Search Tool allows uses to enter a list of primer sequences and the tool will search the know coding sequences for these and report any matches in file format suitable for cutting and pasting into existing spreadsheets. The tool is currently limited to coding sequence but as the number of genomic sequences in the database expands then the tool will be modified to search these regions as well. A number of groups involved in developing KIR typing have proposed A Community Standard Reporting Format for KIR Genotyping Data, these guidelines are available from the IPD-KIR website.
The IPD-KIR database is also been expanded to include the KIR sequences from other species, most recently work has begun on including the sequences of KIR alleles found in Rhesus Macaques (Macaca mulatta) (11–13). These sequences have been compiled through direct submission and through data-mining existing sequences from the generalist sequence databanks. The first release of the official Mamu-KIR nomenclature will include 107 alleles covering 13 loci. Further loci will be added once haplotype studies have allowed the identification of the genes present. Sequences from other primate species like Crab eating Macaque (Macaca fascicularis) have also been submitted to the database, and these sequences will be included at a later date. The non-human KIR sequences will be included into the IPD-KIR section and be accessible using the same tools as the human KIR sequences.
IPD-MHC
The MHC sequences of many different species have been reported (14–17), along with different nomenclature systems used in the naming and identification of new genes and alleles in each species. The sequences of the MHC from a number of different species are highly conserved between species (18). The nomenclature for MHC genes and alleles in species other than humans and mice has historically been overseen either informally by groups generating sequences, or by formal nomenclature committees set up by the International Society for Animal Genetics (ISAG). This work is now overseen by the Comparative MHC Nomenclature Committee (19) and is supported by ISAG and the Veterinary Immunology Committee (VIC) of the International Union of Immunological Societies (IUIS). By bringing the work of different nomenclature committees and the sequences of different species together it is hoped to provide a central resource that will facilitate further research on the MHC of each species and on their comparison.
The first version of the IPD-MHC database involved the work of groups specialising in non-human primates (NHP), canines (DLA) (15,20) and felines (FLA) and incorporated all data previously available in the IMGT/MHC database (21). This version included data from five species of ape, 16 species of new world monkey and 17 species of old world monkey. Since the first version we have been able to add sequences from cattle (BoLA), teleost fish, rats (RT1), sheep (OLA) and swine (SLA) (22–24). Future versions are planned to include chicken, horse, prosimians and pinipeds (seal and sea lion) sequences. As well as adding new species to the database, the individual sections are also updated to include new sequences, depending on the number of sequences to be added updates are performed approximately twice a year.
For each species, there are some differences in the spectrum of data covered but all sections provide the core nomenclature pages and sequence alignments. The nomenclature and alignments follow a similar structure to that of the IPD-KIR section, and the same basic tools are used in both sections. Currently the IPD-MHC sequence alignments are limited to species-specific alignments; however, we are working to allow cross species alignments and the inclusion of human sequences from the IMGT/HLA database (25) for comparative purposes.
The initial release of the database was limited to access plain text alignments and simple HTML tables detailing the nomenclature. We are in the process of migrating each section to use tools similar to IPD-KIR, this means user-defined alignments, online search tools and more detailed allele reports. Currently only the NHP section has been moved to this new version, this was undertaken when the nomenclature used for some species was updated. The NHP section now contains information for over 40 species of primate, with nearly 3000 alleles.
IPD-HPA
Human platelet antigens (HPA) are alloantigens expressed only on platelets, specifically on platelet membrane glycoproteins. These platelet-specific antigens are immunogenic and can result in pathological reactions to transfusion therapy. The HPA nomenclature system was adopted in 1990 (26,27) to overcome problems with the previous nomenclature. Since then more antigens have been described and the molecular basis of many has been resolved. As a result the nomenclature was revised in 2003 (28) and included in the IPD project. The IPD-HPA section contains nomenclature information and additional background material. The different genes in the HPA system have not been sequenced to the same level as some of the other projects and so currently only single nucleotide polymorphisms (SNP) are used to determine alleles. This information is presented in a grid of SNP for each gene The IPD and HPA nomenclature committee hope to expand this to provide full sequence alignments when possible.
IPD-ESTDAB
IPD-ESTDAB is a database of immunologically characterised melanoma cell lines. The database works in conjunction with ESTDAB cell bank (29,30), which is housed in Tübingen, Germany and provides immunologically characterised tumour cells. The IPD-ESTDAB section of the website provides an online search facility for cells stored in this cell bank. This enables investigators to identify cells possessing specific parameters important for studies of immunity, immunogenetics, gene expression, metastasis, response to chemotherapy, and other tumour biological experimentation. The search tool allows for searches based on a single parameter, or clusters of parameters on over 250 different markers for each cell. The detailed reports produced can be then be used to identify cells of interest, which can then be obtained from the cell bank.
DISCUSSION
The IPD project provides a resource for those interested in the study of polymorphic sequences in the immune system. By accommodating related systems in a single database, data can be made available in common formats aiding use and interpretation. As the projects grow and more sections are added, the benefit of having expertly curated sequences from related areas stored in a single location is becoming more apparent. This is particularly true of the IPD-MHC project, where cross-species studies are able to utilise the high quality sequences provided by the different nomenclature committees in a common standardised format, ready for use. The initial release of the IPD Database contained only four sections and a small number of tools; however, as the database has grown and more sections and species have been added, more tools have been added to the website. We plan to use the existing database structures to house data for new sections of the IPD project as they become available. The files will also be made available in different formats to download from the website, ftp server and included into SRS, BLAST and FASTA search engines at the European Bioinformatics Institute (31).
FUNDING
The IPD projects have received funding from the European Commission within the Fifth Framework Infrastructures program (contract no. QLRI-CT-200!-01325) for IPD-ESTDAB and by a National Institutes of Health grant (NIH/NCI P01 111412) for IPD-KIR. The work of the IPD databases is recognised and supported by the International Union of Immunological Societies (IUIS) for both KIR nomenclature through the IUIS KIR Nomenclature Committee and MHC Nomenclature by the International Society for Animal Genetics (ISAG) and the Veterinary Immunology Committee (VIC). Funding for open access charge: The Anthony Nolan Trust, a Charitable organisation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge the work of all the individual nomenclature committees for both the MHC and HPA sections, as well as our ESTDAB collaborators. The authors would also like to acknowledge the support provided by the External Services Group at the European Bioinformatics Institute which allows the IPD project to be hosted within the EBI infrastructure.
Appendix 1: Access and contact
| IPD homepage: | http://www.ebi.ac.uk/ipd/ |
| IPD-KIR homepage: | http://www.ebi.ac.uk/ipd/kir/ |
| IPD-MHC homepage: | http://www.ebi.ac.uk/ipd/mhc/ |
| IPD-HPA homepage: | http://www.ebi.ac.uk/ipd/hpa/ |
| IPD-ESTDAB homepage: | http://www.ebi.ac.uk/ipd/estdab/ |
| Contact: | ipd@ebi.ac.uk |
If you are interested in contributing to the project, there are specific guidelines for the inclusion of new sections, and interested parties should contact Prof. SGE Marsh, steven.marsh@ucl.ac.uk for further information.
REFERENCES
- 1.Cochrane G, Akhtar R, Bonfield J, Bower L, Demiralp F, Faruque N, Gibson R, Hoad G, Hubbard T, Hunter C, et al. Petabyte-scale innovations at the European Nucleotide Archive. Nucleic Acids Res. 2009;37:D19–D25. doi: 10.1093/nar/gkn765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sugawara H, Ikeo K, Fukuchi S, Gojobori T, Tateno Y. DDBJ dealing with mass data produced by the second generation sequencer. Nucleic Acids Res. 2009;37:D16–D18. doi: 10.1093/nar/gkn724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia CA, Robinson J, Guethlein LA, Parham P, Madrigal JA, Marsh SGE. Human KIR sequences 2003. Immunogenetics. 2003;55:227–239. doi: 10.1007/s00251-003-0572-y. [DOI] [PubMed] [Google Scholar]
- 5.Marsh SGE, Parham P, Dupont B, Geraghty DE, Trowsdale J, Middleton D, Vilches C, Carrington M, Witt C, Guethlein LA, et al. Killer-cell immunoglobulin-like receptor (KIR) nomenclature report, 2002. Immunogenetics. 2003;55:220–226. doi: 10.1007/s00251-003-0571-z. [DOI] [PubMed] [Google Scholar]
- 6.Yun G, Tolar J, Yerich AK, Marsh SGE, Robinson J, Noreen H, Blazar BR, Miller JS. A novel method for KIR-ligand typing by pyrosequencing to predict NK cell alloreactivity. Clin. Immunol. 2007;123:272–280. doi: 10.1016/j.clim.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruggeri L, Capanni M, Casucci M, Volpi I, Tosti A, Perruccio K, Urbani E, Negrin RS, Martelli MF, Velardi A. Role of natural killer cell alloreactivity in HLA-mismatched hematopoietic stem cell transplantation. Blood. 1999;94:333–339. [PubMed] [Google Scholar]
- 8.Khakoo SI, Thio CL, Martin MP, Brooks CR, Gao X, Astemborski J, Cheng J, Goedert JJ, Vlahov D, Hilgartner M, et al. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science. 2004;305:872–874. doi: 10.1126/science.1097670. [DOI] [PubMed] [Google Scholar]
- 9.Gumperz JE, Barber LD, Valiante NM, Percival L, Phillips JH, Lanier LL, Parham P. Conserved and variable residues within the Bw4 motif of HLA-B make separable contributions to recognition by the NKB1 killer cell-inhibitory receptor. J. Immunol. 1997;158:5237–5241. [PubMed] [Google Scholar]
- 10.Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, Noreen H, Reed EF, Senitzer D, Setterholm M, Smith A, et al. Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum. Immunol. 2007;68:392–417. doi: 10.1016/j.humimm.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 11.Bimber BN, Moreland AJ, Wiseman RW, Hughes AL, O'C;onnor DH. Complete characterization of killer Ig-like receptor (KIR) haplotypes in Mauritian cynomolgus macaques: novel insights into nonhuman primate KIR gene content and organization. J. Immunol. 2008;181:6301–6308. doi: 10.4049/jimmunol.181.9.6301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blokhuis JH, Doxiadis GG, Bontrop RE. A splice site mutation converts an inhibitory killer cell Ig-like receptor into an activating one. Mol. Immunol. 2009;46:640–648. doi: 10.1016/j.molimm.2008.08.270. [DOI] [PubMed] [Google Scholar]
- 13.Blokhuis JH, van der Wiel MK, Doxiadis GG, Bontrop RE. Evidence for balancing selection acting on KIR2DL4 genotypes in rhesus macaques of Indian origin. Immunogenetics. 2009;61:503–512. doi: 10.1007/s00251-009-0379-6. [DOI] [PubMed] [Google Scholar]
- 14.Kennedy LJ, Angles JM, Barnes A, Carter SD, Francino O, Gerlach JA, Happ GM, Ollier WE, Thomson W, Wagner JL. Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: Second report of the ISAG DLA Nomenclature Committee. Tissue Antigens. 2001;58:55–70. doi: 10.1034/j.1399-0039.2001.580111.x. [DOI] [PubMed] [Google Scholar]
- 15.Kennedy LJ, Altet L, Angles JM, Barnes A, Carter SD, Francino O, Gerlach JA, Happ GM, Ollier WE, Polvi A, et al. Nomenclature for factors of the dog major histocompatibility system (DLA), 1998. First report of the ISAG DLA Nomenclature Committee. International Society for Animals Genetics. Tissue Antigens. 1999;54:312–321. doi: 10.1034/j.1399-0039.1999.540319.x. [DOI] [PubMed] [Google Scholar]
- 16.Davies CJ, Joosten I, Bernoco D, Arriens MA, Bester J, Ceriotti G, Ellis S, Hensen EJ, Hines HC, Horin P, et al. Polymorphism of bovine MHC class I genes. Joint Report of the Fifth International Bovine Lymphocyte Antigen (BoLA) Workshop, Interlaken, Switzerland, 1 August 1992. Eur. J. Immunogenet. 1994;21:239–258. doi: 10.1111/j.1744-313x.1994.tb00197.x. [DOI] [PubMed] [Google Scholar]
- 17.Klein J, Bontrop RE, Dawkins RL, Erlich HA, Gyllensten UB, Heise ER, Jones PP, Parham P, Wakeland EK, Watkins DI. Nomenclature for the major histocompatibility complexes of different species: a proposal. Immunogenetics. 1990;31:217–219. doi: 10.1007/BF00204890. [DOI] [PubMed] [Google Scholar]
- 18.Parham P. Virtual reality in the MHC. Immunol. Rev. 1999;167:5–15. doi: 10.1111/j.1600-065x.1999.tb01378.x. [DOI] [PubMed] [Google Scholar]
- 19.Ellis SA, Bontrop RE, Antczak DF, Ballingall K, Davies CJ, Kaufman J, Kennedy LJ, Robinson J, Smith DM, Stear MJ, et al. ISAG/IUIS-VIC Comparative MHC Nomenclature Committee report, 2005. Immunogenetics. 2006;57:953–958. doi: 10.1007/s00251-005-0071-4. [DOI] [PubMed] [Google Scholar]
- 20.Kennedy LJ, Angles JM, Barnes A, Carter SD, Francino O, Gerlach JA, Happ GM, Ollier WE, Thomson W, Wagner JL. Nomenclature for factors of the dog major histocompatibility system (DLA), 2000: second report of the ISAG DLA Nomenclature Committee. Anim. Genet. 2001;32:193–199. doi: 10.1046/j.1365-2052.2001.00762.x. [DOI] [PubMed] [Google Scholar]
- 21.Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, Stoehr P, Marsh SGE. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho CS, Lunney JK, Ando A, Rogel-Gaillard C, Lee JH, Schook LB, Smith DM. Nomenclature for factors of the SLA system, update 2008. Tissue Antigens. 2009;73:307–315. doi: 10.1111/j.1399-0039.2009.01213.x. [DOI] [PubMed] [Google Scholar]
- 23.Smith DM, Lunney JK, Ho CS, Martens GW, Ando A, Lee JH, Schook L, Renard C, Chardon P. Nomenclature for factors of the swine leukocyte antigen class II system, 2005. Tissue Antigens. 2005;66:623–639. doi: 10.1111/j.1399-0039.2005.00492.x. [DOI] [PubMed] [Google Scholar]
- 24.Smith DM, Lunney JK, Martens GW, Ando A, Lee JH, Ho CS, Schook L, Renard C, Chardon P. Nomenclature for factors of the SLA class-I system, 2004. Tissue Antigens. 2005;65:136–149. doi: 10.1111/j.1399-0039.2005.00337.x. [DOI] [PubMed] [Google Scholar]
- 25.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database—a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 26.von dem Borne AE, Decary F. ICSH/ISBT Working Party on platelet serology. Nomenclature of platelet-specific antigens. Vox Sang. 1990;58:176. doi: 10.1111/j.1423-0410.1990.tb02085.x. [DOI] [PubMed] [Google Scholar]
- 27.von dem Borne AE, Decary F. Nomenclature of platelet-specific antigens. Hum. Immunol. 1990;29:1–2. doi: 10.1016/0198-8859(90)90063-u. [DOI] [PubMed] [Google Scholar]
- 28.Metcalfe P, Watkins NA, Ouwehand WH, Kaplan C, Newman P, Kekomaki R, De Haas M, Aster R, Shibata Y, Smith J, et al. Nomenclature of human platelet antigens. Vox Sang. 2003;85:240–245. doi: 10.1046/j.1423-0410.2003.00331.x. [DOI] [PubMed] [Google Scholar]
- 29.Pawelec G, Marsh SGE. ESTDAB: a collection of immunologically characterised melanoma cell lines and searchable databank. Cancer Immunol. Immunother. 2006;55:623–627. doi: 10.1007/s00262-005-0117-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson J, Roberts Ch, Dodi AI, Pawelec G, Marsh SGE. The European Searchable Tumour Line Database (ESTDAB) Cancer Immunol. Immunother. 2009;58:1501–1506. doi: 10.1007/s00262-008-0656-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Harte N, Silventoinen V, Quevillon E, Robinson S, Kallio K, Fustero X, Patel P, Jokinen P, Lopez R. Public web-based services from the European Bioinformatics Institute. Nucleic Acids Res. 2004;32:W3–W9. doi: 10.1093/nar/gkh405. [DOI] [PMC free article] [PubMed] [Google Scholar]