Abstract
The MetaCyc database (MetaCyc.org) is a comprehensive and freely accessible resource for metabolic pathways and enzymes from all domains of life. The pathways in MetaCyc are experimentally determined, small-molecule metabolic pathways and are curated from the primary scientific literature. With more than 1400 pathways, MetaCyc is the largest collection of metabolic pathways currently available. Pathways reactions are linked to one or more well-characterized enzymes, and both pathways and enzymes are annotated with reviews, evidence codes, and literature citations. BioCyc (BioCyc.org) is a collection of more than 500 organism-specific Pathway/Genome Databases (PGDBs). Each BioCyc PGDB contains the full genome and predicted metabolic network of one organism. The network, which is predicted by the Pathway Tools software using MetaCyc as a reference, consists of metabolites, enzymes, reactions and metabolic pathways. BioCyc PGDBs also contain additional features, such as predicted operons, transport systems, and pathway hole-fillers. The BioCyc Web site offers several tools for the analysis of the PGDBs, including Omics Viewers that enable visualization of omics datasets on two different genome-scale diagrams and tools for comparative analysis. The BioCyc PGDBs generated by SRI are offered for adoption by any party interested in curation of metabolic, regulatory, and genome-related information about an organism.
INTRODUCTION
MetaCyc (MetaCyc.org) is a highly curated, non-redundant reference database of small-molecule metabolism. It contains metabolic pathway and enzyme data that have been experimentally demonstrated in the scientific literature (1) (Figure 1). Because MetaCyc contains only experimentally determined pathways and enzymes, and due to its tight integration of data and references, MetaCyc is a uniquely valuable resource in fields including genome analysis, metabolism and metabolic engineering. The metabolic pathways and enzymes in MetaCyc are derived from organisms representing all domains of life (Tables 1 and 2). In the past, microbial and plant metabolism were emphasized, but current curation also focuses on vertebrate metabolism.
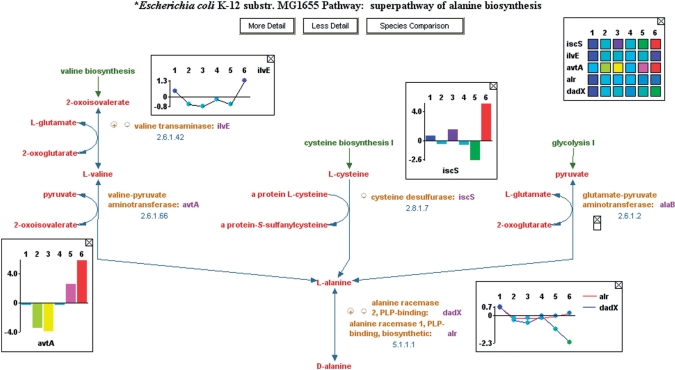
Figure 1.
An example of a pathway showing omics data pop-ups. Pathways can be displayed at varying levels of detail, and this pathway’s display depicts an intermediate level of detail including enzymes, EC numbers and genes, but no chemical structures. Notice the green arrows at the top of the pathway, which provide hyperlinks to related upstream pathways. After an omics dataset has been uploaded, the user can easily add data pop-ups to the pathway diagram displaying the omics data for genes or metabolites. The user can select from heat map, bar graph or plot representation. Multiple time points are supported.
Table 1.
List of species that have more than 15 experimentally elucidated pathways represented in MetaCyc
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Escherichia coli | 248 | Arabidopsis thaliana | 239 | Methanosarcina barkeri | 18 |
| Bacillus subtilis | 53 | Homo sapiens | 126 | Methanocaldococcus jannaschii | 17 |
| Pseudomonas aeruginosa | 47 | Saccharomyces cerevisiae | 95 | Methanosarcina thermophila | 17 |
| Pseudomonas putida | 41 | Glycine max | 62 | Methanothermobacter thermautotrophicus | 16 |
| Salmonella typhimurium | 33 | Rattus norvegicus | 60 | Sulfolobus solfataricus | 16 |
| Pseudomonas fluorescens | 26 | Pisum sativum | 42 | ||
| Mycobacterium tuberculosis | 20 | Mus musculus | 38 | ||
| Klebsiella pneumonia | 18 | Zea mays | 38 | ||
| Agrobacterium tumefaciens | 17 | Solanum lycopersicum | 34 | ||
| Cupriavidus sp. PS12 | 15 | Solanum tuberosum | 34 | ||
| Mycobacterium smegmatis | 15 | Oryza sativa | 32 |
The species are grouped by taxonomic domain and are ordered within each domain based on the number of pathways (number following species name) to which the given species was assigned. Some pathways may be labeled with a higher-level taxon, such as genus, if all the species within that genus are thought to have the given pathway. However, such higher-level taxa are not included in this table.
Table 2.
Distribution of pathways in MetaCyc based on the taxonomic classification of associated species. Taxonomic groups (phyla for Bacteria and Archaea, kingdoms for Eukarya) are grouped by domain and are ordered within each domain based on the number of pathways (number following taxon name) associated with the taxon
| Bacteria | Eukarya | Archaea | |||
|---|---|---|---|---|---|
| Proteobacteria | 750 | Viridiplantae | 643 | Euryarchaeota | 95 |
| Firmicutes | 190 | Fungi | 204 | Crenarchaeota | 31 |
| Actinobacteria | 171 | Metazoa | 174 | ||
| Bacteroidetes/Chlorobi | 54 | Euglenozoa | 16 | ||
| Cyanobacteria | 42 | ||||
| Deinococcus-Thermus | 22 | ||||
| Thermotogae | 12 | ||||
| Spirochaetes | 9 | ||||
| Aquificae | 9 | ||||
| Planctomycetes | 5 | ||||
| Fusobacteria | 4 | ||||
| Chloroflexi | 4 | ||||
| Chlamydiae -Verrucomicrobia | 2 | ||||
| Thermodesulfobacteria | 2 | ||||
| Nitrospirae | 2 | ||||
| Chrysiogenetes | 1 |
Euglenozoa are listed separately as this group does not belong to any of the other eukaryotic kingdoms. A pathway may be associated with multiple organisms.
In conjunction with its role as a general reference on metabolism, MetaCyc is used as a reference database for the PathoLogic component of the Pathway Tools software (2) to computationally predict the metabolic network of any organism having a sequenced and annotated genome (3). In this automated process, a predicted metabolic network is created in the form of a Pathway/Genome Database (PGDB). BioCyc (BioCyc.org) is a collection of more than 500 organism-specific PGDBs that were generated in this way both at SRI and by other groups. The editing capability of Pathway Tools enables computationally predicted PGDBs to be improved and updated by manual curation. Interested scientists may adopt and curate existing PGDBs through the BioCyc Web site (biocyc.org/intro.shtml#adoption), or create new PGDBs using MetaCyc and Pathway Tools (biocyc.org/download.shtml). More than 80 groups have used Pathway Tools and MetaCyc to create PGDBs for their organisms of interest, including important model organisms such as Saccharomyces cerevisiae (4), Arabidopsis thaliana (5), Oryza sativa (6), Mus musculus (7), Bos taurus (8), Medicago truncatula (9), Dictyostelium discoideum (10), Leishmania major (11), Chlamydomonas reinhardtii (12), several Solanaceae species (13) and many pathogenic bacteria (14) (see http://biocyc.org/otherpgdbs.shtml for a more complete list).
A web server included in Pathway Tools enables the publishing of PGDBs through either the internet or an internal network. The Navigator component of Pathway Tools allows the browsing and analysis of PGDBs either locally or over the Internet. A detailed description of Pathway Tools can be found at http://bioinformatics.ai.sri.com/ptools/ and in (15).
PGDBs generated by Pathway Tools and MetaCyc are an excellent platform for the integration of genome information with many other types of data regarding metabolism, regulation, and genetics. They provide powerful tools for analyzing omics datasets from experiments related to gene transcription, metabolomics, proteomics, ChIP-chip analysis, etc. (Figure 2). The PGDBs accelerate research in many fields including biochemistry, molecular biology, biotechnology, bioinformatics, metabolic engineering and systems biology (16–19). Both MetaCyc and organism-specific PGDBs can also be used as educational tools.
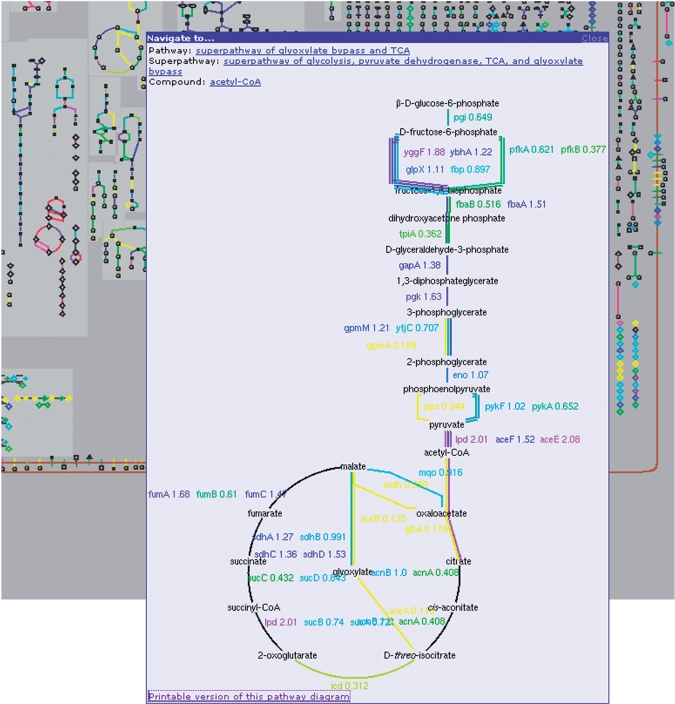
Figure 2.
The omics viewers enable visualization of omics datasets on genome-scale diagrams. The background of this figure shows part of the cellular overview, with gene transcription data superimposed over the enzymatic reactions that are catalyzed by the enzymes encoded by the assayed genes. By clicking on a single pathway within the diagram, the user can open a pop-up window that shows the pathway along with the omics data in full scale. The numbers next to gene names indicate the value of the omics data. The colors provide another quantitative indication (for more information, see http://biocyc.org/ov-expr.shtml). When multiple enzymes are known to catalyze the same reaction, multiple, color-coded lines are displayed for that reaction, each describing one gene/enzyme.
During the past 2 years, we again significantly expanded the data content of MetaCyc and BioCyc. We also added supporting enhancements to the Pathway Tools software. The expanded and enhanced databases and software are described in the following sections.
METACYC ENHANCEMENTS
Expansion of MetaCyc
All pathways in MetaCyc are curated from the experimental literature. Since the last Nucleic Acids Research publication (2 years ago) (20), we added 507 new base pathways (pathways comprised of reactions only, where no portion of the pathway is designated as a subpathway) and 129 superpathways (pathways composed of at least one base pathway plus additional reactions or pathways), and updated 104 existing pathways, for a total of 740 new and revised pathways. The total number of base pathways grew by 43%, from 977 (version 11.5) to 1399 (version 13.5) (the total increase is less than 507 pathways because some existing pathways were deleted from the database during this period); while the total number of superpathways grew by 120%, from 106 (version 11.5) to 235 (version 13.5).
Along with the increase in pathway number, the number of enzymes, reactions, chemical compounds, and citations in the database grew by 35%, 25%, 29% and 37%, respectively; and the number of referenced organisms increased by 75% (currently at 1795).
MetaCyc pathway distribution
The pathways in MetaCyc are classified by an ontology developed at SRI that is constantly updated to reflect curation needs (Table 3). The four top-level categories (or classes) of this ontology are biosynthesis, degradation/utilization/assimilation, generation of precursor metabolites and energy and detoxification.
Table 3.
The distribution of pathways in MetaCyc based on pathway ontology
| Biosynthesis (902) | Secondary Metabolites Biosynthesis (351) |
| Cofactors, Prosthetic Groups, Electron Carriers Biosynthesis (160) | |
| Amino acids Biosynthesis (105) | |
| Fatty Acids and Lipids Biosynthesis (101) | |
| Carbohydrates Biosynthesis (70) | |
| Hormones Biosynthesis (40) | |
| Amines and Polyamines Biosynthesis (32) | |
| Cell structures Biosynthesis (31) | |
| Nucleosides and Nucleotides Biosynthesis (20) | |
| Other Biosynthesis (16) | |
| Aromatic Compounds Biosynthesis (13) | |
| Siderophore Biosynthesis (7) | |
| Metabolic Regulators Biosynthesis (4) | |
| Aminoacyl-tRNA Charging (4) | |
| Degradation/Utilization/ Assimilation (639) | Aromatic Compounds Degradation (152) |
| Amino Acids Degradation (113) | |
| Inorganic Nutrients Metabolism (72) | |
| Secondary Metabolites Degradation (58) | |
| Carbohydrates Degradation (52) | |
| Amines and Polyamines Degradation (40) | |
| Chlorinated Compounds Degradation (39) | |
| Carboxylates Degradation (30) | |
| Degradation/Utilization/Assimilation -Other (26) | |
| C1 Compounds Utilization and Assimilation (24) | |
| Fatty Acid and Lipids Degradation (18) | |
| Alcohols Degradation (14) | |
| Hormones Degradation (15) | |
| Aldehyde Degradation (12) | |
| Nucleosides and Nucleotides Degradation and Recycling (9) | |
| Cofactors, Prosthetic Groups, Electron Carriers Degradation (2) | |
| Generation of precursor metabolites and energy (124) | Fermentation (34) |
| Respiration (25) | |
| Chemoautotrophic Energy Metabolism (14) | |
| Methanogenesis (12) | |
| Electron Transfer (11) | |
| TCA cycle (9) | |
| Glycolysis (6) | |
| Photosynthesis (6) | |
| Pentose Phosphate Pathways (4) | |
| Other (3) | |
| Detoxification (16) | Methylglyoxal Detoxification (8) |
| Arsenate Detoxification (3) | |
| Acid Resistance (2) | |
| Mercury Detoxification (1) |
Pathways classified directly under the top-level classes are not shown. The number of pathways within each class is presented in parentheses. Some pathways may be classified under more than one class.
In version 13.5, the largest top-level class is Biosynthesis, with 902 base pathways. Its main subclasses are secondary metabolites biosynthesis (351); cofactors, prosthetic groups, and electron carriers biosynthesis (160); amino acids biosynthesis (105); and fatty acids and lipids biosynthesis (101).
The second-largest top-level class is degradation/utilization/assimilation, with 639 base pathways. Within this group, the largest subclasses are aromatic compounds degradation (152), amino acids degradation (113), inorganic nutrients metabolism (72), secondary metabolites degradation (58), and carbohydrates degradation (52).
The third-largest top-level class, generation of precursor metabolites and energy, contains 124 base pathways. its largest subclasses are fermentation (34), respiration (25), chemoautotrophic energy metabolism (14) and methanogenesis (12).
The final top-level class, detoxification, is much smaller, with only 16 base pathways.
During the previous 2 years, the number of metazoan pathways in MetaCyc increased by 67%, from 104 to 174 pathways.
The list of pathways added to MetaCyc since the last NAR publication is too long to give here. For a complete report, please see the MetaCyc release notes history at http://metacyc.org/release-notes.shtml.
Electron transfer pathways
Following the introduction of support for electron transfer reactions into the database schema, we added a total of 11 electron transfer pathways to the database. This type of pathways utilizes a different display algorithm that conveys features such as the direction of the electron flow, the cell-compartment locations where the substrates are transformed, and the optional translocation of protons across membranes. For an example of such pathways, see the pathway ‘succinate to cytochrome bd oxidase electron transfer’.
Interactions with other databases
IUBMB
MetaCyc is regularly updated with data from the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology (NC-IUBMB), which includes new and modified EC numbers. The last supplement incorporated is supplement 14.
NCBI Taxonomy
Starting with version 12.0, the full NCBI Taxonomy database (21) is integrated into Pathway Tools, enabling specification of the taxa in which MetaCyc pathways occur using NCBI Taxonomy, and allowing taxonomic querying of MetaCyc pathways and enzymes.
Gene Ontology
We continue to update the mapping between MetaCyc and Gene Ontology (GO) process and function terms (22). In May 2009, we submitted to GO updated mappings between MetaCyc pathways and reactions and GO biological process and molecular function terms. An updated file is found at http://www.geneontology.org/external2go/metacyc2go.
PubChem
To improve the mapping between MetaCyc compounds and other compound databases, all MetaCyc compounds were incorporated into the PubChem database (23), and are linked to their PubChem entries.
KEGG
MetaCyc was updated to better link objects to corresponding entries in KEGG. 3269 MetaCyc reactions were mapped to KEGG reactions, and an additional 920 MetaCyc compounds now have links to KEGG compounds. MetaCyc pathways contain more than twice as many reactions (4950) than does KEGG (2463), although KEGG contains more total reactions (9194 in version 50) than MetaCyc (8387). MetaCyc contains 1399 pathways compared to the 155 pathways in KEGG.
BioCyc ENHANCEMENTS
Expansion of BioCyc
The BioCyc databases are organized into three tiers.
Tier 1 PGDBs were created using intensive manual curation and receive continuous updating.
Tier 2 PGDBs have received moderate amounts of review, but may not be updated on an ongoing basis.
Tier 3 PGDBs were created computationally, and received no subsequent manual review or updating.
During the past two years, the number of BioCyc PGDBs increased from 371 (version 11.5) to 508 (version 13.1), out of which two are in Tier 1 (EcoCyc and MetaCyc), 24 are in Tier 2, and the rest belong to Tier 3. Some Tier 2 PGDBs were provided by groups outside SRI [examples include MouseCyc (7) CattleCyc (8) and YeastCyc (4)]. Database authors are identified on the database summary page (Tools → Reports → Summary Statistics).
Innovations in database sharing
The extended family of Pathway Tools-based Pathway/Genome Databases includes both the SRI-created BioCyc collection and many PGDBs created outside SRI by other Pathway Tools users. This DB family exhibits a number of innovations in scientific database sharing. We believe that no one group can curate all the world’s genomes; therefore, we strongly emphasize the notion of widely distributing the workload of curating genome databases. All the Tier 3 PGDBs and some of the Tier 2 PGDBs are offered for adoption to interested parties under an open license agreement. Some groups adopt existing PGDBs within BioCyc, assuming responsibility for their ongoing curation. Other groups create their own PGDBs using Pathway Tools. We offer free technical support to facilitate this task for our academic users.
All Pathway Tools-based PGDBs share the same schema, thus facilitating comparative analyses and data exchange. An encouraged form of data exchange is the submission of experimentally determined metabolic pathways curated by curators of other PGDBs for inclusion in MetaCyc, broadening the pathways available for pathway prediction, and easing the bottleneck of data entry into MetaCyc. Conversely, Pathway Tools now includes the ability to propagate updates made to MetaCyc to other PGDBs derived from earlier versions of MetaCyc. For example, corrections made to MetaCyc chemical structures or reaction equations can be propagated to other PGDBs. Further, Pathway Tools can perform incremental pathway prediction, thus propagating newly curated pathways present in the latest version of MetaCyc to older organism-specific PGDBs.
Another means for facilitating data exchange is the PGDB registry, operated by SRI. Groups that curate PGDBs can register their databases in our PGDB registry (http://biocyc.org/registry.shtml) in a process that includes deposition of the PGDB in a downloadable format on the author’s FTP or HTTP site. With a few mouse clicks, any Pathway Tools user can download a PGDB listed in the Registry and install it into their working copy of Pathway Tools, making it available for comparative analysis, omics data analysis, etc. Thus, users can share PGDBs as easily as they exchange music files on the Internet.
Compound protonation and reaction balancing
Starting with version 13.0, all MetaCyc compounds have been adjusted to a consistent protonation state for a reference pH of 7.3, common in the cellular cytosol. This adjustment was performed using the Marvin computational chemistry software (ChemAxon Kft, Budapest Hungary). In addition, all reactions that had a mass-imbalance due only to hydrogen atoms were computationally balanced by adding or removing protons from the appropriate side of the reaction. These updated compounds and reactions eventually will be propagated into all BioCyc PGDBs, and to other PGDBs created using Pathway Tools, making it easier to apply flux-balance analysis techniques to these databases.
This change resulted in certain differences between some MetaCyc reactions and the comparable reactions in other databases, such as the ENZYME Database (24). However, we believe that the representation of reactions in MetaCyc is more consistent and, within the limits of the cytosolic pH of 7.3, more accurate.
SOFTWARE IMPROVEMENTS
The following paragraphs list a number of the most salient improvements to Pathway Tools during the past 2 years.
Web site redesign
The BioCyc Web site has undergone a significant overhaul that includes a new toolbar, a new organism selector widget, and new search commands. The new object-specific search commands provide an intermediate level of search complexity that lies between the very easy-to-use Quick Search box and the sophisticated Advanced Query Page. Search pages customized for finding genes/proteins/RNAs, chemical compounds, and pathways are relatively easy to use, yet enable the user to define multi-criteria searches (e.g. find proteins that satisfy specified constraints on their pI, molecular weight, cellular location, small-molecule ligand and chromosomal location). As part of the site redesign, the Regulatory Overview tool that depicts the complete regulatory network stored within a PGDB is now available through the Web site (currently only the desktop version of Pathway Tools supports painting of omics data on the Regulatory Overview).
Web accounts
Users of the BioCyc Web site can now create accounts in which they can store Web site display preferences, specify a default organism for queries, and define and save organism lists for comparative genomics operations.
New omics display and analysis functions
The desktop version of Pathway Tools enables users to graph omics data for selected genes or metabolites, and also provides over-representation analysis for determining whether certain ontology classes (including GO MetaCyc Pathway Ontology, etc.) are over-represented in gene lists and metabolite lists. A new X–Y plot style of tracks for the Pathway Tools genome browser allows the user to visualize ChIP-chip datasets against the genome. ChIP-chip intensity measurements can be visually correlated with promoters, gene positions and operon boundaries.
Customize pathway diagrams for PowerPoint or publications
The appearance of pathway pages can now be customized in many respects (Pathway → Customize Diagram). Options include setting the font size, determining which elements are included in the drawing (such as enzyme names and gene names), and deciding whether chemical structures are displayed. The pathway diagrams can be downloaded as screen-resolution GIF images (for import into PowerPoint presentations), or as high-resolution PostScript or PDF files for import into documents.
Apple port
Pathway Tools now runs on Apple computers.
HOW TO LEARN MORE ABOUT MetaCyc AND BioCyc
The BioCyc.org and MetaCyc.org Web sites provide several informational resources, including an online BioCyc guided tour (25), a MetaCyc user guide (26) and many Webinar videos that combine narration with online demonstration of different topics (27). We routinely host workshops and tutorials (on site and at conferences) that provide training and in-depth discussion of our software for beginning and advanced users. To stay informed about recent changes and enhancements to our software, join the BioCyc mailing list at http://biocyc.org/subscribe.shtml. A list of our publications is available online (28).
DATABASE AVAILABILITY
The MetaCyc and BioCyc databases are freely and openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and of the BioCyc and MetaCyc Web sites are released four times per year.
FUNDING
National Institutes of Health, National Institute of General Medical Sciences (GM080746, GM077678, and GM75742); National Science Foundation, Division of Biological Infrastructure (grant number 0640769 to P.Z., A.K., K.D.). Funding for open access charge: National Institutes of Health, National Institute of General Medical Sciences.
Conflict of interest statement. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the preceding agencies.
ACKNOWLEDGEMENTS
We thank Dr Carol Bult and Dr Alexei Evsikov from the Jackson Laboratory for their contribution of pathways from the MouseCyc database. We thank Dr Lindsay Eltis and Dr Hao-Ping Chen from the University of British Columbia for their contribution of pathways from the Rhodococcus jostii RHA1 database. We also thank Dr Malabika Sarker from SRI International for her contribution of the Mycobacterium tuberculosis mycolate biosynthesis pathway. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the preceding agencies.
REFERENCES
- 1.Krieger CJ, Zhang P, Mueller LA, Wang A, Paley S, Arnaud M, Pick J, Rhee SY, Karp PD. MetaCyc: a multiorganism database of metabolic pathways and enzymes. Nucleic Acids Res. 2004;32:D438–D442. doi: 10.1093/nar/gkh100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee T, Kaipa P, Gilham F, Spaulding A, Popescu L, et al. Pathway Tools version 13.0: Integrated Software for Pathway/Genome Informatics and Systems Biology. Brief. Bioinformatics. 2010 doi: 10.1093/bib/bbp043. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004;6:R2.1–R2.17. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mueller LA, Zhang P, Rhee SY. AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol. 2003;132:453–460. doi: 10.1104/pp.102.017236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36:D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evsikov AV, et al. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Seo S, Lewin HA. Reconstruction of metabolic pathways for the cattle genome. BMC Syst. Biol. 2009;3:33. doi: 10.1186/1752-0509-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urbanczyk-Wochniak E, Sumner LW. MedicCyc: a biochemical pathway database for Medicago truncatula. Bioinformatics. 2007;23:1418–1423. doi: 10.1093/bioinformatics/btm040. [DOI] [PubMed] [Google Scholar]
- 10.Fey P, Gaudet P, Curk T, Zupan B, Just EM, Basu S, Merchant SN, Bushmanova YA, Shaulsky G, Kibbe WA, et al. dictyBase—a Dictyostelium bioinformatics resource update. Nucleic Acids Res. 2009;37:D515–D519. doi: 10.1093/nar/gkn844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doyle MA, MacRae JI, De Souza DP, Saunders EC, McConville MJ, Likic VA. LeishCyc: a biochemical pathways database for Leishmania major. BMC Syst. Biol. 2009;3:57. doi: 10.1186/1752-0509-3-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.May P, Christian JO, Kempa S, Walther D. ChlamyCyc: an integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazourek M, Pujar A, Borovsky Y, Paran I, Mueller L, Jahn MM. A dynamic interface for capsaicinoid systems biology. Plant Physiol. 2009;150:1806–1821. doi: 10.1104/pp.109.136549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snyder EE, Kampanya N, Lu J, Nordberg EK, Karur HR, Shukla M, Soneja J, Tian Y, Xue T, Yoo H, et al. PATRIC: the VBI PathoSystems Resource Integration Center. Nucleic Acids Res. 2007;35:D401–D406. doi: 10.1093/nar/gkl858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 16.Valdes J, Veloso F, Jedlicki E, Holmes D. Metabolic reconstruction of sulfur assimilation in the extremophile Acidithiobacillus ferrooxidans based on genome analysis. BMC Genomics. 2003;4:51. doi: 10.1186/1471-2164-4-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim TY, Kim HU, Park JM, Song H, Kim JS, Lee SY. Genome-scale analysis of Mannheimia succiniciproducens metabolism. Biotechnol. Bioeng. 2007;97:657–671. doi: 10.1002/bit.21433. [DOI] [PubMed] [Google Scholar]
- 18.Aanensen DM, Mavroidi A, Bentley SD, Reeves PR, Spratt BG. Predicted functions and linkage specificities of the products of the Streptococcus pneumoniae capsular biosynthetic loci. J. Bacteriol. 2007;189:7856–7876. doi: 10.1128/JB.00837-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernal V, Carinhas N, Yokomizo AY, Carrondo MJ, Alves PM. Cell density effect in the baculovirus-insect cells system: a quantitative analysis of energetic metabolism. Biotechnol. Bioeng. 2009;104:162–180. doi: 10.1002/bit.22364. [DOI] [PubMed] [Google Scholar]
- 20.Caspi R, Foerster H, Fulcher CA, Kaipa P, Krummenacker M, Latendresse M, Paley S, Rhee SY, Shearer AG, Tissier C, et al. The MetaCyc Database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases. Nucleic Acids Res. 2008;36:D623–D631. doi: 10.1093/nar/gkm900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, et al. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W33. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. [(1 October 2009, date last accessed)]. SRI International, BioCyc online guided tour, http://biocyc.org/samples.shtml.
- 26. SRI International, MetaCyc user guide, http://www.metacyc.org/MetaCycUserGuide.shtml (1 October 2009, date last accessed)
- 27. SRI International, BioCyc webinars, http://biocyc.org/webinar.shtml (1 October 2009, date last accessed)
- 28. SRI International, BioCyc publication list, http://biocyc.org/publications.shtml (1 October 2009, date last accessed)