Abstract
About 15% of human colorectal cancers and, at varying degrees, other tumor entities as well as nearly all tumors related to Lynch syndrome are hallmarked by microsatellite instability (MSI) as a result of a defective mismatch repair system. The functional impact of resulting mutations depends on their genomic localization. Alterations within coding mononucleotide repeat tracts (MNRs) can lead to protein truncation and formation of neopeptides, whereas alterations within untranslated MNRs can alter transcription level or transcript stability. These mutations may provide selective advantage or disadvantage to affected cells. They may further concern the biology of microsatellite unstable cells, e.g. by generating immunogenic peptides induced by frameshifts mutations. The Selective Targets database (http://www.seltarbase.org) is a curated database of a growing number of public MNR mutation data in microsatellite unstable human tumors. Regression calculations for various MSI–H tumor entities indicating statistically deviant mutation frequencies predict TGFBR2, BAX, ACVR2A and others that are shown or highly suspected to be involved in MSI tumorigenesis. Many useful tools for further analyzing genomic DNA, derived wild-type and mutated cDNAs and peptides are integrated. A comprehensive database of all human coding, untranslated, non-coding RNA- and intronic MNRs (MNR_ensembl) is also included. Herewith, SelTarbase presents as a plenty instrument for MSI-carcinogenesis-related research, diagnostics and therapy.
INTRODUCTION
The completion of the human genome project in 2003 provided the data basis for genome-wide analyses (1). Now it became within reach to systematically investigate the whole human genome for sequence motifs or structures by computer assisted investigation to clarify the association of genome variation or mutation with certain human diseases using the human genome draft as a consensus. Currently, there are more than 22 000 known protein-coding genes annotated within the ∼3 G of base pairs within Human Ensembl (rel. 55.37, http://www.ensembl.org/Homo_sapiens/) leading to more than 100 000 transcripts. Sequence motifs of special interest comprise single nucleotide polymorphisms (SNPs), splice site recognition patterns or promoter motifs, regulatory motifs and binding sites.
The human genome sequence also facilitated the systematic search for human microsatellites that had been started earlier based on EMBL DNA and mRNA data (2). Microsatellites are especially prone to deletion and insertion mutations during DNA replication with a strong dependency of mutability from their length (3). They are distributed non-randomly throughout the whole human genome within non-coding and coding regions (4). Their function, however, is nearly unknown. Mononucleotide repeats (MNRs) seem to represent the most interesting kind of microsatellites. The length of coding MNRs (cMNRs) is conserved (5). Length alterations of cMNRs of 1 or 2 nucleotides lead to frameshift mutations. The length of non-coding MNRs however can vary highly from individual to individual. However, there are also a number of so-called quasi-monomorphic MNRs of higher length (20–40 bp) within non-coding regions that show a significantly restricted length variation within the human population which may indicate the possibility of functional relevance of these non-coding MNRs. It is well known, that alterations in polypyrimidine MNRs in the 5′ local neighborhood of splice donor sites can lead to exon skipping (6,7), which will result in a frameshift situation in two-thirds (8). In addition, shortening or elongation of MNRs within 5′ UTRs can have an impact on the transcription level, of those in the 3′ UTR on transcript stability of the respective mRNA (9).
Microsatellite alterations are corrected by the DNA mismatch repair system (MMR). The functional inactivation of the MMR system results in the manifestation of microsatellite mutations which is termed ‘microsatellite instability’ (MSI). The MSI phenotype is found in >90% of tumors developing in MMR germline mutation carriers among hereditary non-polyposis colorectal cancer (HNPCC) or Lynch syndrome patients and ∼15% of sporadic cancers (10). Colorectal MSI–H tumors are characterized by certain clinico-histopathological properties such as a better prognosis compared to tumors of the CIN phenotype (11–13). Moreover, a dense lymphocyte infiltration is a characteristic feature of MSI–H colorectal cancer (14,15). There is evidence that the obviously enhanced immunogenicity of MSI–H cancers may be caused by the generation of immunogenic peptides. Insertion/deletion mutations at coding microsatellites lead to a shift of the translational reading frame and thus may lead to the translation of frameshift peptides (neopeptides) that can be recognized as foreign neoantigens by the host’s immune system (16,17), reviewed in (18). Frameshift peptides may be generated once the MMR system is inactivated, but maybe as early as haploinsufficiency of one MMR gene becomes relevant which might be assumed by the finding of immune response against frameshift-induced neopeptides in healthy HNPCC mutation carriers without any history of tumor development (19). Notwithstanding, the MSI is assumed to be the underlying mechanism for the further malignant transformation and evolution of these tumors.
The identification of the relevant genes among the multitude of all genes is a key for understanding this process and may reveal target structures for both diagnostic and therapeutic concepts. Since the observed mutation frequencies of MNRs of the same type and length widely scatter (20), positive or negative selection processes in affected cell clones are held to be responsible for that finding. Growth-promoting alterations are assumed to show up with higher frequencies than coincidental mutational events without effect in so-called bystander genes. In contrast, alteration of genes of proteins with essential cellular functions is supposed to emerge with a reduced mutation frequency. A recently developed statistical model aims to identify positive and negative selected targets (PSTs and NSTs, in short) (20). A significant deviation from the average mutation rate in respect to a given MNR length will mark off genes with elevated or reduced mutation frequency, classifying them either as PSTs or NSTs.
Since closing of data collection for the original described model (20) we continued the extensive literature review on data concerning genes with coding and non-coding MNRs and their mutation rate in MSI–H colorectal, gastric or endometrial cancer samples by also parsing detail information and recruitment of other data sources, that is public databases, e.g. the Cancer Genome Project. All these data were organized in a relational database with an easily readable web interface. All MNR detail data included within SelTarbase are cross-linked with important genomic and protein information sources at EMBL, NCBI, SBI and many more, providing quick connections for sequence-related information. Especially with regard to the possible immunologic impact of cMNR mutations we developed some useful tools [prediction of frameshift transcripts and peptides, presentation of the resulting neopeptides, forecast of nonsense-mediated RNA decay (NMD) sensitivity] for the deduction of possible implications of the underlying mutation.
DATA MINING AND EVALUATION
The first extensive survey of the literature (April 2002) revealed 110 publications referring to mutation analyses of 245 coding and non-coding microsatellites in 177 genes either in MSI-H colorectal, gastric or endometrial cancer. This provided the basis for SelTarbase version 2003 (20). The PubMed (http://www.ncbi.nlm.nih.gov/sites/entrez?db=PubMed) is routinely screened once per week for search terms (microsatellite instability, MSI, MMR and HNPCC). Manuscripts, of which title or abstract contain evidence for MNR mutation data, are analyzed as full text.
Detailed information about statistical methods and parameters as well as the regression model and software used are available via the online help: Help and Documentation at http://www.seltarbase.org/?topic=help.
Core database
SelTarbase core database currently contains approximately 40 000 records (version latest, 21st release, 200908) concerning mutational data of more than 3400 MNRs from about 400 human genes derived from approximately 500 publications. With the release 201001 a larger number of more than 100 longer, mostly coding MNRs will become available (S.M. Woerner and S. Korff, unpublished data). In total, this corresponds to about 220 000 single observations combining all four entities [colon (incl. rectum), stomach, endometrium and colon culture]. The home page of SelTarbase presents information about the current database content regarding the number of references that were screened for MNR mutation data and how many of them have been included, the number of genes and MNRs each stratified by entity. Alongside the same parameters of the previous release (version last) are compared to give a short impression of the amount of new data of the current release (version latest). Most data are derived from colon tumor analysis followed by stomach, endometrium and colon culture at nearly comparable level.
SelTarbase prediction
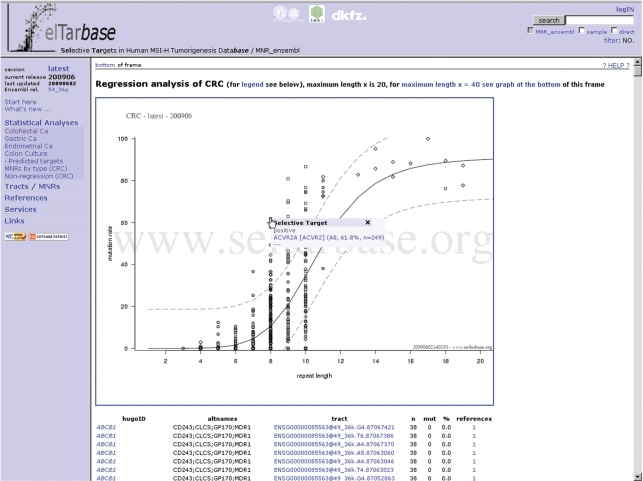
SelTarbase prediction facilitates the assigning of statistically deviant mutation frequencies based on our previously published model (20). For the four entities colon, stomach, endometrium and colon culture individual result tables are built up. Based on these, sigmoid regression calculations are performed to generate a mean mutation approximation depending on MNR length. For detailed information about the regression model see the online help: Help and Documentation at http://www.seltarbase.org/?topic=help. Figure 1 shows the graphical output of the sigmoid regression analysis for colon. Of the 1793 MNRs included in this regression analysis, 47 are significantly higher and 23 are significantly lower mutated than the average and predicted as PST or NST genes. The proportion of significantly deviating MNRs therefore is 4.0% for colon (3.9% for stomach, 3.7% for endometrium and 2.5% for colon culture).
Figure 1.
Graph of sigmoid regression analysis. Result of colorectal cancer mutation data: ncMNRs (non-coding i.e. untranslated, intronic and pseudogene MNRs, open diamonds), cMNRs (amino acid coding MNRs, grey filled circles), and nMNRs (intergenic MNRs, open squares). Red marked MNRs changed since previous release, respectively represent new data. Regression analysis was performed combined for all MNR types. The fitted regression line is drawn as solid black line, the upper and lower prediction lines as bold dashed grey lines. If the pointing device is moved over data points that reside above the upper respectively below the lower prediction line, a popup will appear, showing the name of the gene and the mutation frequency as well as the number of analyzed tumor samples. If the pointing device is moved over the fitted regression line (solid black line), a popup will appear, showing the length × with the corresponding mean mutation frequency as well as the lower and upper prediction thresholds for the length ×.
Human MNR database: MNR_ensembl
MNR_ensembl is a comprehensive database containing coding, untranslated, non-coding RNA- and intronic MNRs within the human genome. It currently contains approximately 558 000 cMNRs, 874 000 uMNRs, 3700 ncrMNRs and 25 400 000 iMNRs of length x ≥ 4. Due to alternative splicing the complete number of MNRs within coding, untranslated, and intronic regions of the human genome is approximately 26 500 000 (Ensembl rel. 55.37). A somewhat older version of all mouse c/u/iMNRs is also available (mouse rel. 45.36f).
MNR_ensembl is fully integrated within SelTarbase including the possibility to use the transcription and translation information function, thus it is easy to have a look at all possible frameshift transcripts and peptides as well as the prediction of NMD sensitivity for any human cMNR.
Web interface
SelTarbase is based on a relational database built with MySQL. The web interface was implemented with perl cgi-scripts. The database can be searched for gene names, tract pseudonyms (e.g. BAT26), accession numbers (EMBL, Ensembl, Entrez Gene and Unigene), gene description (MNR_ensembl), author names, title, PubMed ID and DOI as well as colorectal cancer cell line names. Scrolling through MNR detail data can be customized by limiting the list to single contributing references or by coding status and MNR length (even in combination). MNR contribution of references can also be shown as alphabetically ordered MNR lists grouped by the initial of the gene name.
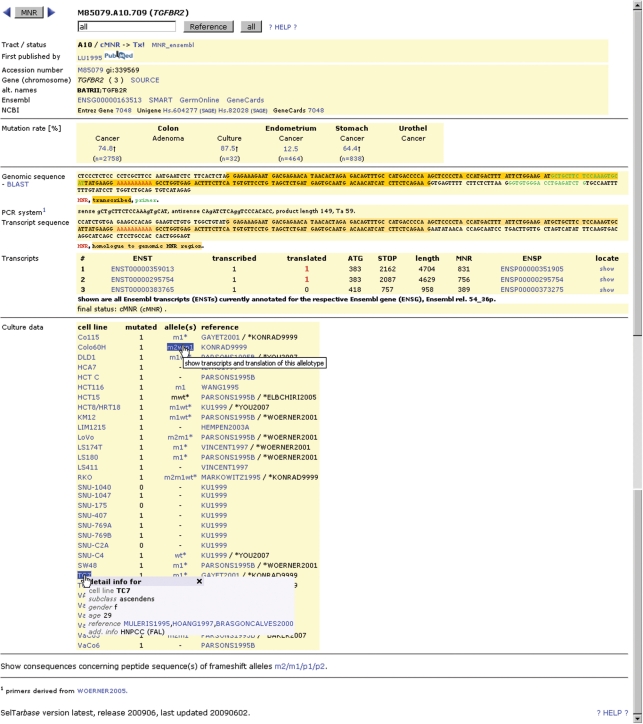
Figure 2a and b exemplarily show the MNR detail data of the A10 cMNR (BATRII) within the TGFBR2 gene. Following information is provided: MNR type (nucleotide and length, reported length if differing), coding status, the reference firstly reported data for this MNR (ordered by publication year and PubMed ID), accession number of EMBL, Ensembl, Entrez Gene and Unigene. All accession numbers are linked to the respective databases at EBI (http://www.ebi.ac.uk/embl), EMBL (http://www.ensembl.org/Homo_sapiens) and NCBI (http://www.ncbi.nlm.nih.gov/sites/entrez, http://www.ncbi.nlm.nih.gov/UniGene). Furthermore, there are links to other useful gene information databases: SOURCE (http://source.stanford.edu), SMART (http://smart.embl-heidelberg.de/smart), GermOnline (http://www.germonline.org/Homo_sapiens), GeneCards (http://www.genecards.org) and SAGE (http://cgap.nci.nih.gov/SAGE), followed by the entity related mutation frequencies and contributing sample numbers including information of the contributing references. The next paragraph presents the genomic sequence surrounding the MNR (with BLASTN option using the currently linked Ensembl genome), PCR primers for fragment or sequence analysis of this MNR—if available, as well as the transcript sequence surrounding the MNR in case of coding or untranslated MNRs. In addition, all known Ensembl transcripts of the corresponding gene are listed together with information about the transcription and translation status of the MNR, the ATG-, the stop codon- and the MNR-position. For each listed Ensembl transcript/peptide a link to the LOCATE subcellular localization database (http://locate.imb.uq.edu.au) is provided. Finally, all known mutation information of MSI–H colorectal cancer cell lines available appears in detail.
Figure 2.
The MNR detail information page. Most details are mentioned within the 2nd paragraph of section Web Interface. Using the buttons ‘MNR’ or ‘Reference’ several filters can be defined for scrolling from MNR to MNR, either restricting to MNRs of one contributing reference and/or regarding coding status and MNR length. Filters can be used in combination. If the pointing device is moved over cell line names, a popup will appear, showing additional information regarding this cell line including links to publications from which this information is drawn (Supplementary references of SelTarbase).
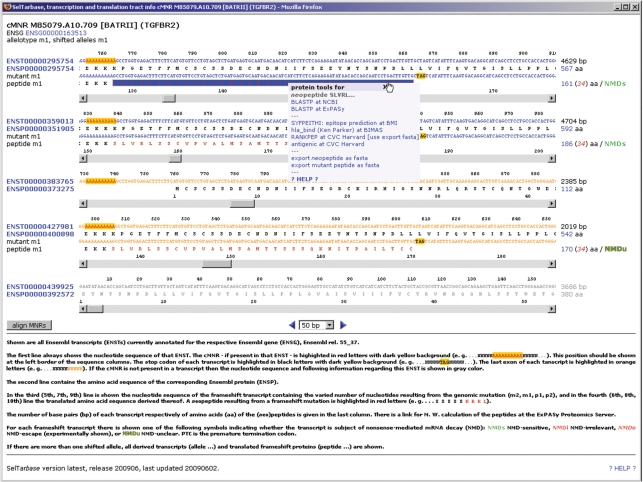
The allele status of coding MNRs—if exactly known—will link to another SelTarbase function that is shown in Figure 3: MNR transcription and translation information at sequence and amino acid level. All annotated Ensembl transcripts and the corresponding frameshift sequences (1 respectively 2-nt insertion/deletion), and the amino acid sequences derived thereof are shown. This function provides tools to perform a BLASTP of frameshift peptides via NCBI (http://www.ncbi.nlm.nih.gov/blast) and ExPASy (http://www.expasy.org), to calculate the molecular weight of wild-type and mutated peptides/proteins at ExPASy, and to export nucleotide and peptide sequences in FASTA format for further usage. Moreover, this function displays theoretical information about degradation by NMD as NMDs (sensitive), NMDu (unknown) or NMDi (insensitive) for each of the frameshift transcripts. In case of missing cell line data, this function is also available with the option of all four theoretical allele types (minus 2, minus 1, plus 1 and plus 2) as prelinked from the MNR information page, in order to be able to check predicted transcripts and frameshift peptides in the same way.
Figure 3.
Transcription and translation information page: transcripts/peptides are scrollable from 5′ to 3′ using different units of 10–500 bp (default 50 bp). When the page is loaded all MNR containing transcripts are aligned according to MNR position showing the MNR at the left border of the sequence window. Transcripts that do not contain the MNR are greyed, and no frameshift information is calculated. More information about this function is provided within the 2nd paragraph of section Web Interface.
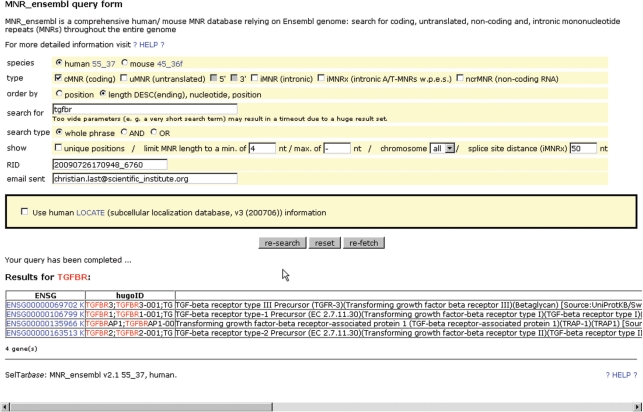
There is also the possibility to register with SelTarbase. Registration is free for all users and allows for usage of higher CPU-consuming functions e.g. model recalculation and searching the MNR database MNR_ensembl. It can be searched in whole or in part (coding, untranslated, non-coding RNA- and/or intronic MNRs) for Ensembl accession numbers (ENSG), hugoIDs, or keywords of the short gene description provided with the gene name within Ensembl (Figure 4). More detailed information is given in legend of Figure 4.
Figure 4.
MNR_ensembl query form: searching for human coding, untranslated, other non-coding and intronic MNRs: results can be reported for single categories or in combination. Ensembl accession numbers (ENSG), the hugoID or gene description key words can be searched for. Ordering is possible by genomic position or by MNR length in descending order. Results can be restricted to certain minimum and maximum MNR lengths with an absolute minimum of 4 nt. Due to alternative splicing of genes or several transcripts MNRs can be reported several times as cMNR(s), uMNR(s) and/or iMNR(s). Using the option unique positions, a complete MNR list of a gene can be generated listing each MNR at a given genomic position only once. The annotation hierarchy in this case is cMNRs > uMNRs > iMNRs, what means that an MNR once reported as cMNR will not be able to be reported as uMNR or iMNR using that option. All cMNRs will be listed as they would be transcribed within one large transcript including all annotated exons of that gene. Additionally, the search can further be limited to only those transcripts (genes) which are related to a specific subcellular localization according to the GO term(s) used in the LOCATE database (http://locate.imb.uq.edu.au). Additionally, a somewhat elder version of all mouse MNRs is also available (mouse rel. 45.36f).
An additional useful feature of SelTarbase is the possibility to upload new user mutation data in order to have the regression recalculated including these unpublished data. Data can be provided with a minimum of information not to disclose unpublished data using a self-chosen name, the nucleotide type and length and a virtual position as MNR ID. If one intends to complement data already included within Seltarbase, then it is recommended to apply the exact MNR name used in SelTarbase in order to correctly collate these data. This tool is able to process data uploaded from different platforms/operating systems and also to provide result files usable on different platforms without the need of special software, packed in the user’s suitable format (zip, rar, tar and bz2).
Documentation and help for all applications included within SelTarbase is available through the web interface.
FUTURE PROSPECTS AND UPDATES
Regression calculation is updated monthly provided that new includable data becomes available. The human MNR tables (MNR_ensembl) as well as the underlying human genome and transcripts are subsequently updated whenever a new Ensembl release will be provided (usually four times a year). The coding status of each MNR included or reported within MNR_ensembl relies on the respective Ensembl release mentioned within SelTarbase (currently rel. 55.37).
The inclusion of further entities (colon adenoma, urothelial and ovarian cancer) is primed within SelTarbase and will become available as soon as sufficient data are available for a reliable regression analysis of these entities. Additionally, the implementation of further tools for characterizing frameshift transcripts and peptides is in progress.
DISCUSSION
Since human tumors showing MSI considerably differ in a number of clinical as well pathologic parameters from tumors showing chromosomal instability, the MSI phenotype is of very high concern regarding tumorigenesis (11–13) and immunology (18). The functional loss of the mismatch repair capacity—acquired or inherited—is the initial step; a huge number of mutation manifestations spread all over the genome is the consequence (21–23). A defined number of such mutations providing a selective advantage for malignant transforming/transformed cells is believed to be the driving force of MSI tumorigenesis (20,24). However, due to the mentioned differences between CIN and MSI tumors certain affected genes may share. But, the pivotal players of MSI tumorigenesis can be investigated using a different strategy. As an additional point MSI tumors represent a certain variation of human tumors by the generation of tumor-specific neoantigens. Frameshift peptides (neopeptides) resulting from numerous coding MNR mutations are providing a special immunologic situation maybe explaining the better prognosis of MSI tumors compared to CIN tumors. Taken together, systematic and profound information about human microsatellites would be of great value for scientific, diagnostic and therapeutic purposes.
Since 1995 a number of genes suspected to be involved in MSI tumorigenesis by MNR mutations [e.g. TGFBR2, BAX and others (25,26)] were described, and more or less intensely characterized. As functional evidence for the consequence of MNR mutations within those genes is difficult to show, the mutation frequency often was used as a clue for the transformation impact. However, mutation rates of MNRs widely scatter. Hence, the question was how to differentiate suspicious from average-mutated MNRs. Therefore, different attempts were undertaken to identify the key players of MSI tumorigenesis. We developed a statistical model in order to accomplish such a discrimination (20). Since 2003 the basic data set tremendously increased and mathematics were adapted accordingly. As a result of the regression analysis, from the very first beginning of the model up to now, genes such as TGFBR2, BAX and ACVR2A with the highest evidence for a MSI-tumorigenesis driving force show a significantly elevated mutation frequency in MSI–H gastrointestinal cancer. Meanwhile, there are some other genes showing significantly increased mutation frequencies by the regression analysis included within SelTarbase, for which more and more evidence for an involvement in MSI tumorigenesis is emerging, such as EPHB2, MRE11A, MYH11 and TCF-4 (27–30).
SelTarbase is a comprehensive curated mutation data collection of human MSI-H tumor and colorectal cell line data. This information is systematically organized and straightforward accessible. Being routinely revised and extended it always provides an up-to-date resource of known (public) data for the community as simple information, for data or sample controlling, or helping to prevent from multiple analysis. The superimposed regression analyses may help researchers to focus functional investigations of MSI-related genes by superseding additional time- and money-consuming analyses. Within SelTarbase integrated tools, e.g. the prediction of frameshift transcripts and peptides or the prediction of transcripts regarding NMD may further assist the decision process or the investigation outline of forthcoming immunology related experiments. The systematic information of and the prediction tools implemented may also facilitate the selection progression of appropriate candidates for vaccination and other diagnostic or therapy-related options. For new, unpublished MNR mutation data submission and recalculation of the regression analysis may be supportive to classify the candidate(s) within the number of already known MNR data. As further part of SelTarbase, MNR_ensembl provides a current, complete database of all human coding, untranslated, other non-coding and intronic MNRs as potential targets of MSI in MSI-H tumors and cell lines based on the current human genome included within the Ensembl database (Homo sapiens, rel. 55.37) and may lead to direct mutation analyses of new highly promising candidate genes without the need of own bioinformatic utilities and capabilities. Taken together, SelTarbase opens a broad variety of possibilities to researchers committed in the field of MSI tumorigenesis, diagnostics and therapy.
Availability
SelTarbase is available at http://www.seltarbase.org. Data can be retrieved as html as well as downloaded as plain text files.
FUNDING
The Deutsche Krebshilfe; and the Klaus Tschira Foundation (to S.M.W.). Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the expert help and support of T. Woerner. Further, we thank J. Lammarsch and R. Bogus (Rechenzentrum der Universität Heidelberg, University of Heidelberg) for the initial setup and maintenance of the hardware running SelTarbase. Finally, we would like to express our gratitude to J. Lacroix, J. Kopitz, and M. Kloor for critical reading the manuscript and helpful discussion.
REFERENCES
- 1.International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 2.Woerner SM, Gebert J, Yuan YP, Sutter C, Ridder R, Bork P, von Knebel Doeberitz M. Systematic identification of genes with coding microsatellites mutated in DNA mismatch repair-deficient cancer cells. Int. J. Cancer. 2001;93:12–19. doi: 10.1002/ijc.1299. [DOI] [PubMed] [Google Scholar]
- 3.Sagher D, Hsu A, Strauss B. Stabilization of the intermediate in frameshift mutation. Mutat. Res. 1999;423:73–77. doi: 10.1016/s0027-5107(98)00227-9. [DOI] [PubMed] [Google Scholar]
- 4.Ellegren H. Microsatellites: simple sequences with complex evolution. Nat. Rev. Genet. 2004;5:435–445. doi: 10.1038/nrg1348. [DOI] [PubMed] [Google Scholar]
- 5.Metzgar D, Bytof J, Wills C. Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res. 2000;10:72–80. [PMC free article] [PubMed] [Google Scholar]
- 6.Ejima Y, Yang L, Sasaki MS. Aberrant splicing of the ATM gene associated with shortening of the intronic mononucleotide tract in human colon tumor cell lines: a novel mutation target of microsatellite instability. Int. J. Cancer. 2000;86:262–268. doi: 10.1002/(sici)1097-0215(20000415)86:2<262::aid-ijc17>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 7.Giannini G, Rinaldi C, Ristori E, Ambrosini MI, Cerignoli F, Viel A, Bidoli E, Berni S, D'A;mati G, Scambia G, et al. Mutations of an intronic repeat induce impaired MRE11 expression in primary human cancer with microsatellite instability. Oncogene. 2004;23:2640–2647. doi: 10.1038/sj.onc.1207409. [DOI] [PubMed] [Google Scholar]
- 8.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 9.Suraweera N, Iacopetta B, Duval A, Compoint A, Tubacher E, Hamelin R. Conservation of mononucleotide repeats within 3′ and 5′ untranslated regions and their instability in MSI-H colorectal cancer. Oncogene. 2001;20:7472–7477. doi: 10.1038/sj.onc.1204952. [DOI] [PubMed] [Google Scholar]
- 10.Imai K, Yamamoto H. Carcinogenesis and microsatellite instability: the interrelationship between genetics and epigenetics. Carcinogenesis. 2008;29:673–680. doi: 10.1093/carcin/bgm228. [DOI] [PubMed] [Google Scholar]
- 11.Samowitz WS, Curtin K, Ma KN, Schaffer D, Coleman LW, Leppert M, Slattery ML. Microsatellite instability in sporadic colon cancer is associated with an improved prognosis at the population level. Cancer Epidemiol. Biomarkers Prev. 2001;10:917–923. [PubMed] [Google Scholar]
- 12.Hemminki A, Mecklin JP, Jarvinen H, Aaltonen LA, Joensuu H. Microsatellite instability is a favorable prognostic indicator in patients with colorectal cancer receiving chemotherapy. Gastroenterology. 2000;119:921–928. doi: 10.1053/gast.2000.18161. [DOI] [PubMed] [Google Scholar]
- 13.Gryfe R, Kim H, Hsieh ET, Aronson MD, Holowaty EJ, Bull SB, Redston M, Gallinger S. Tumor microsatellite instability and clinical outcome in young patients with colorectal cancer. N. Engl. J. Med. 2000;342:69–77. doi: 10.1056/NEJM200001133420201. [DOI] [PubMed] [Google Scholar]
- 14.Dolcetti R, Viel A, Doglioni C, Russo A, Guidoboni M, Capozzi E, Vecchiato N, Macri E, Fornasarig M, Boiocchi M. High prevalence of activated intraepithelial cytotoxic T lymphocytes and increased neoplastic cell apoptosis in colorectal carcinomas with microsatellite instability. Am. J. Pathol. 1999;154:1805–1813. doi: 10.1016/S0002-9440(10)65436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Smyrk TC, Watson P, Kaul K, Lynch HT. Tumor-infiltrating lymphocytes are a marker for microsatellite instability in colorectal carcinoma. Cancer. 2001;91:2417–2422. [PubMed] [Google Scholar]
- 16.Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan YP, Bork P, von Knebel Doeberitz M. Frameshift peptide-derived T-cell epitopes: a source of novel tumor- specific antigens. Int. J. Cancer. 2001;93:6–11. doi: 10.1002/ijc.1298. [DOI] [PubMed] [Google Scholar]
- 17.Saeterdal I, Bjorheim J, Lislerud K, Gjertsen MK, Bukholm IK, Olsen OC, Nesland JM, Eriksen JA, Moller M, Lindblom A, et al. Frameshift-mutation-derived peptides as tumor-specific antigens in inherited and spontaneous colorectal cancer. Proc. Natl Acad. Sci. USA. 2001;98:13255–13260. doi: 10.1073/pnas.231326898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Banerjea A, Bustin SA, Dorudi S. The immunogenicity of colorectal cancers with high-degree microsatellite instability. World J. Surg. Oncol. 2005;3:26. doi: 10.1186/1477-7819-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schwitalle Y, Kloor M, Eiermann S, Linnebacher M, Kienle P, Knaebel HP, Tariverdian M, Benner A, von Knebel Doeberitz M. Immune response against frameshift-induced neopeptides in HNPCC patients and healthy HNPCC mutation carriers. Gastroenterology. 2008;134:988–997. doi: 10.1053/j.gastro.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 20.Woerner SM, Benner A, Sutter C, Schiller M, Yuan YP, Keller G, Bork P, von Knebel Doeberitz M, Gebert JF. Pathogenesis of DNA repair-deficient cancers: a statistical meta-analysis of putative Real Common Target genes. Oncogene. 2003;22:2226–2235. doi: 10.1038/sj.onc.1206421. [DOI] [PubMed] [Google Scholar]
- 21.Aaltonen LA, Peltomaki P, Leach FS, Sistonen P, Pylkkanen L, Mecklin JP, Jarvinen H, Powell SM, Jen J, Hamilton SR. Clues to the pathogenesis of familial colorectal cancer. Science. 1993;260:812–816. doi: 10.1126/science.8484121. [DOI] [PubMed] [Google Scholar]
- 22.Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M. Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature. 1993;363:558–561. doi: 10.1038/363558a0. [DOI] [PubMed] [Google Scholar]
- 23.Thibodeau SN, Bren G, Schaid D. Microsatellite instability in cancer of the proximal colon. Science. 1993;260:816–819. doi: 10.1126/science.8484122. [DOI] [PubMed] [Google Scholar]
- 24.Duval A, Hamelin R. Mutations at coding repeat sequences in mismatch repair-deficient human cancers: toward a new concept of target genes for instability. Cancer Res. 2002;62:2447–2454. [PubMed] [Google Scholar]
- 25.Markowitz S, Wang J, Myeroff L, Parsons R, Sun L, Lutterbaugh J, Fan RS, Zborowska E, Kinzler KW, Vogelstein B, et al. Inactivation of the type II TGF-beta receptor in colon cancer cells with microsatellite instability. Science. 1995;268:1336–1338. doi: 10.1126/science.7761852. [DOI] [PubMed] [Google Scholar]
- 26.Rampino N, Yamamoto H, Ionov Y, Li Y, Sawai H, Reed JC, Perucho M. Somatic frameshift mutations in the BAX gene in colon cancers of the microsatellite mutator phenotype. Science. 1997;275:967–969. doi: 10.1126/science.275.5302.967. [DOI] [PubMed] [Google Scholar]
- 27.Alazzouzi H, Davalos V, Kokko A, Domingo E, Woerner SM, Wilson AJ, Konrad L, Laiho P, Espin E, Armengol M, et al. Mechanisms of inactivation of the receptor tyrosine kinase EPHB2 in colorectal tumors. Cancer Res. 2005;65:10170–10173. doi: 10.1158/0008-5472.CAN-05-2580. [DOI] [PubMed] [Google Scholar]
- 28.Wen Q, Scorah J, Phear G, Rodgers G, Rodgers S, Meuth M. A mutant allele of MRE11 found in mismatch repair-deficient tumor cells suppresses the cellular response to DNA replication fork stress in a dominant negative manner. Mol. Biol. Cell. 2008;19:1693–1705. doi: 10.1091/mbc.E07-09-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alhopuro P, Phichith D, Tuupanen S, Sammalkorpi H, Nybondas M, Saharinen J, Robinson JP, Yang Z, Chen LQ, Orntoft T, et al. Unregulated smooth-muscle myosin in human intestinal neoplasia. Proc. Natl Acad. Sci. USA. 2008;105:5513–5518. doi: 10.1073/pnas.0801213105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuilliere-Dartigues P, El-Bchiri J, Krimi A, Buhard O, Fontanges P, Flejou JF, Hamelin R, Duval A. TCF-4 isoforms absent in TCF-4 mutated MSI-H colorectal cancer cells colocalize with nuclear CtBP and repress TCF-4-mediated transcription. Oncogene. 2006;25:4441–4448. doi: 10.1038/sj.onc.1209471. [DOI] [PubMed] [Google Scholar]