Abstract
Much of the information on the Cytochrome P450 enzymes (CYPs) is spread across literature and the internet. Aggregating knowledge about CYPs into one database makes the search more efficient. Text mining on 57 CYPs and drugs led to a mass of papers, which were screened manually for facts about metabolism, SNPs and their effects on drug degradation. Information was put into a database, which enables the user not only to look up a particular CYP and all metabolized drugs, but also to check tolerability of drug-cocktails and to find alternative combinations, to use metabolic pathways more efficiently. The SuperCYP database contains 1170 drugs with more than 3800 interactions including references. Approximately 2000 SNPs and mutations are listed and ordered according to their effect on expression and/or activity. SuperCYP (http://bioinformatics.charite.de/supercyp) is a comprehensive resource focused on CYPs and drug metabolism. Homology-modeled structures of the CYPs can be downloaded in PDB format and related drugs are available as MOL-files. Within the resource, CYPs can be aligned with each other, drug-cocktails can be ‘mixed’, SNPs, protein point mutations, and their effects can be viewed and corresponding PubMed IDs are given. SuperCYP is meant to be a platform and a starting point for scientists and health professionals for furthering their research.
INTRODUCTION
The family of Cytochrome P450 enzymes has been the focus of pharmaceutical research for decades, as evidenced by the more than 100 000 articles in PubMed. Drug metabolism is a complex biochemical network, which consists of many different parts and reactions in the human organism. Some drugs are excreted unchanged in urine and feces without passing any metabolic treatment in the liver, but most of the drugs have a multi-step metabolism, which is mainly associated with Cytochrome P450 (CYP). These enzymes belong to the family of monooxygenases, which reach maximum of absorption at 450 nm. This work focuses on human CYPs (Table 1).
Table 1.
CYP-overview
| CYP | Tissue sites | Localization | Typical reaction |
|---|---|---|---|
| 1A1 | Lung, several extrahepatic sites, peripher blood cells | ER | Benzopyrene 3-hydroxylation |
| 1A2 | Liver | ER | Caffeine N3-demethylation |
| 1B1 | Many extrahepatic sites, incl. lung and kidney | ER | 17β-Estradiol 4-hydroxylation |
| 2A6 | Liver, lung and several extrahepatic sites | ER | Coumarin 7-hydroxylation |
| 2A7 | Only information is identification in human genome | ER | |
| 2A13 | Nasal tissue | ER | Activation of 4-(methylnitrosamino)-1- (3-pyridyl)-1-butanone |
| 2B6 | Liver, lung | ER | (S)-Mephenytoin N-demethylation |
| 2C8 | Liver | ER | Taxol 6α-hydroxylation |
| 2C9 | Liver | ER | Tobutamine methyl hydroxylation |
| 2C18 | Liver | ER | |
| 2C19 | Liver | ER | (S)-Mephenytoin 4′-hydroxylation |
| 2D6 | Liver | ER | Debrisoquine 4-hydroxylation |
| 2E1 | Liver, lung and other tissues | ER | Chlorzoxazone 6-hydroxylation |
| 2F1 | Lung | ER | 3-Methylindole activation |
| 2J2 | Lung | ER | Arachidonic acid oxidation |
| 2R1 | Only information is identification in human genome | ||
| 2S1 | Lung | ER | |
| 2U1 | Only information is identification in human genome | ||
| 2W1 | Only information is identification in human genome | ||
| 3A4 | Liver, small intestine | ER | Testosterone 6β-hydroxylation |
| 3A5 | Liver, lung | ER | Testosterone 6β-hydroxylation |
| 3A7 | Ffetal liver | ER | Testosterone 6β-hydroxylation |
| 3A43 | mRNA detected in gonads | ER | |
| 4A11 | Liver | ER | Fetty acid ω-hydroxylation |
| 4A22 | Only information is identification in human genome | ER | |
| 4B1 | Lung | ER | Lauric acid ω-hydroxylation |
| 4F2 | Liver | ER | Leukotriene B4 ω-hydroxylation |
| 4F3 | Neutrophils | ER | Leukotriene B4 ω-hydroxylation |
| 4F8 | Seminal vesicles | ER | Prostaglandin ω-2 hydroxylation |
| 4F11 | Liver | ER | |
| 4F12 | Liver | ER | Arachidonic acid ω-,ω-2-hydroxylation |
| 4F22 | Only information is identification in human genome | ||
| 4V2 | Only information is identification in human genome | ||
| 4X1 | Only information is identification in human genome | ||
| 4Z1 | Only information is identification in human genome | CYP 4A20 | |
| 5A1 | Platelets | ER | Thromboxane A2 synthetase reaction |
| 7A1 | Liver | ER | Cholesterol 7α-hydroxylation |
| 7B1 | Brain | ER | Dehydroepiandrosterone 7α-hydroxylation |
| 8A1 | Aorta, others | ER | Prostacyclin synthase reaction |
| 8B1 | Liver | ER | 7α-hydroxyprogesterone 12-hydroxylation |
| 11A1 | Adrenals, other steroidogenic tissues | Mitochondrium | Cholesterol side-chain clevage |
| 11B1 | Adrenals | Mitochondrium | 11-Deoxycortisol 11-hydroxylation |
| 11B2 | Adrenals | Mitochondrium | Corticosterone 18-hydroxylation |
| 17A1 | Steroidogenic tissue | ER | Steroid 17α-hydroxylation |
| 19A1 | Steroidogenic tissue, adipose, brain | ER | Androgen aromatization |
| 20A1 | Only information is identification in human genome | ||
| 21A2 | Steroidogenic tissue | ER | 17-Hydroxyprogesterone 21-hydroxylation |
| 24A1 | Kidney | Mitochondrium | 25-Hydroxyvitamin D3 24-hydroxylation |
| 26A1 | Several | ER | Retinoic acid 4-hydroxylation |
| 26B1 | Brain | ER | Retinoic acid 4-hydroxylation |
| 26C1 | Only information is identification in human genome | ER | |
| 27A1 | Liver | Mitochondrium | Sterol 27-hydroxylation |
| 27B1 | Kidney | Mitochondrium | Vitamin D3 1-hydroxylation |
| 27C1 | Only information is identification in human genome | ||
| 39A1 | Liver | ER | 24-Hydroxycholesterol 7-hydroxylase |
| 46A1 | Brain | ER | Cholesterol 24-hydroxylation |
| 51A1 | Liver, testes | ER | Lanosterol 14α-demethylation |
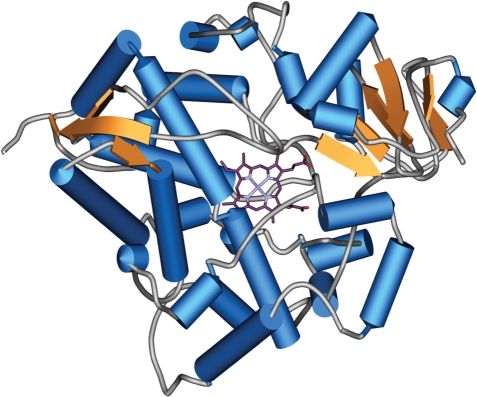
The chemical reaction is: R–H + O2 + NADPH + H+ → R–OH + H2O + NADP+ (1). CYPs catalyze a large amount of chemical reactions, such as alcohol oxidations, dehydrogenation and isomerizations. One of the most difficult tasks of medical science is to find combinations of drugs that do not affect each other’s metabolic pathways. The Human Genome Project identified 57 human CYPs (2), ordered into 18 families and 43 subfamilies by sequence similarities. In general, the human CYPs share the same fold, but because of its spacious binding site CYP 3A4 is capable of metabolizing at least 422 drugs (Figure 1).
Figure 1.
Plot of the 3D structure of CYP 3A4. Helices are shown as cylinders (light blue) and beta-sheets as orange arrows. The heme moiety is depicted in stick representation. The majority of the structure is made up of an alpha-helical arrangement, but N- and C-terminal small beta-domains can be found.
Many drugs also inhibit or induce the activity of CYPs, which is important to health professionals trying to dose medicines. If a drug induces a CYP that is also active in another drug’s metabolism, the dosage of the first drug must be enhanced to achieve a therapeutic effect. In case of inhibition of a CYP, the dosage of the drug can be reduced, which also lowers side effects (3). Information on CYP-structures (4), binding sites (5,6), interactions and different genotypes (7,8) must be combined to allow reduce side effects and to determine correct dosages of medicine. David Nelson and colleagues listed human CYPs with alignments (9) and also address nomenclature. Flockhart created a CYP–drug-interaction table with the intention of avoiding undesired side effects when prescribing more than one drug (10), a key issue in an aging society. In 1997, Rendic and Di Carlo (11) compiled a comprehensive collection of data on CYP reactions and drug interactions. From the patients’ perspective, it is useful to know what kind of food or additional drugs they should avoid when taking their medications. Drug interactions such as these can be checked at the University of Maryland (http://www.umm.edu/adam/drug_checker.htm). Another field of interest is the diversity of human CYPs in different ethnic groups. Pharmacogenomic studies reported that African–American patients are precribed higher dosages of antipsychotic medications compared to Caucasians (12). Information on mutations in CYPs is widely spanned accross the internet. An up-to-date resource on available 3D CYP structures from the Protein Data Bank (13) is presented by CYPED (14). The Center for Molecular Design collected exhaustive information on the CYPs of many species in their Knowledgebase (http://cpd.ibmh.msk.su/), including sequences and links to the Nomenclature Committee (15). Despite the large amount of information on CYPs, optimizing multiple drug prescriptions using CYP metabolism is still complicated (16). To overcome these problems, SuperCYP aims at providing a user-friendly platform enabling health professionals to optimize drug cocktails regarding the degree of CYP capacity utilization. Furthermore, a number of scientific issues were addressed: comprehensive information about mutations influencing the metabolism of drugs (17,18), racial differences (19), comparative analysis of drug binding sites to explain their promiscuousness. To this end, multiple sequence alignments are required and will be the basis of homology-built 3D structures of all human CYPs (20,21).
MATERIALS AND METHODS
Information on CYPs was collected from scientific literature (22) and various web resources: e.g. Nelsons Homepage (23), Flockharts Interaction Table (http://www.medicine.iupui.edu/Flockhart/table.htm), University of Maryland’s Drug Checker, PubChem (24), PDB (13). Some information was gathered from FDA-files. Abstracts of PubMed were automatically filtered for relevant articles using specific keywords. The abstracts were screened for WHO-drugs and their synonyms, as was a set of human CYPs with synonyms. A team of scientists manually processed the papers found in PubMed. Each drug was attributed to those CYPs that are involved in drug metabolism as substrate, inhibitor or inducer. In addition, mutations on the protein level were retrieved utilizing the mutation/gene association text mining system described by Winneburg and Schroeder (25). The tool retrieved ∼550 distinct mutations from more than 400 scientific journal abstracts. In total, 450 new amino acid substitutions were retrieved in addition to those 500 already obtained from PubMed. The approach was adapted to gather information on alleles, the change of enzyme/transcription activity and populations. As with the first literature screening, the automated predictions were manually verified before being included in the database, and in all cases PubMed-IDs as references are provided to track details regarding the given information.
SuperCYP is designed as a relational database on a MySQL server. For chemical functionality, the MyChem package is included, which aims to provide a complete set of functions for handling chemical data within MySQL. Most of the functions used by MyChem depend on Open Babel (26). For displaying 3D structures, Jmol—an open-source Java viewer for chemical structures in 3D—is used. For visual inspection of the alignments, JALVIEW is installed. ChemSketch was applied as a built-in molecule editor, which allows users to screen using self-edited molecules. The website is built with PHP and javascript, web access is enabled via Apache Webserver 2.2.
DATABASE
The SuperCYP website was developed as a user-friendly platform for researchers and health professionals. The navigation bar on the left side offers ‘FAQs’ or Frequently Asked Questions, for first-time users.
‘Drug search’ enables the user to search for a drug and find information on its metabolism. ‘Get Information’ leads to a table listing CYPs involved in the metabolism of the drug. Here there is also a description of possible consequences and after clicking on the drug name on the results page ‘Drug search’ enables the user to get information for compounds by means of the CAS-number or name.
The ‘ATC tree’ is the WHO classification system that classifies drugs into different groups according to anatomic site of action, their therapeutical effect and chemical structure. It is the basis for drug alternative drug recommendations. In a Java applet, the user finds a drop-down tree with major and minor branches of classification. All drugs affiliated with a minor branch are listed in a table and information on the CYP metabolism is provided.
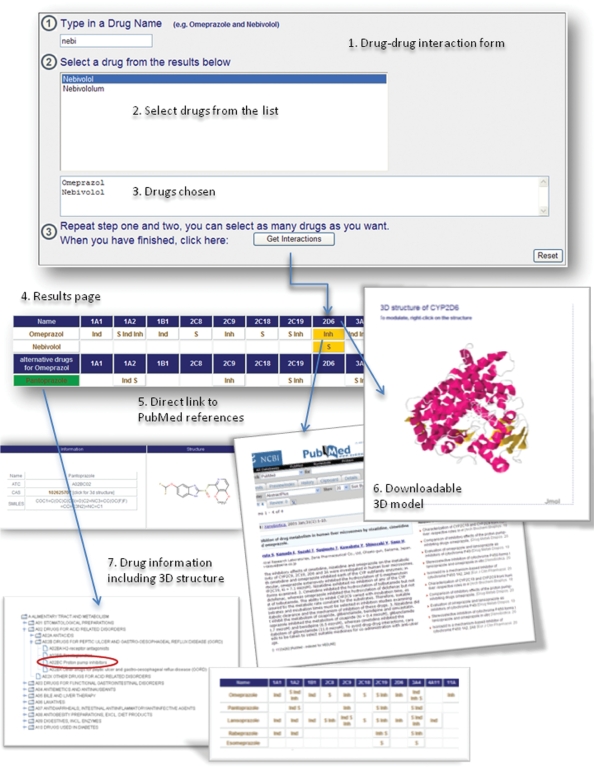
‘Drug–drug interaction’ is the main feature of the database. It allows users to enter names of several different drugs and to check interactions between these drugs, but they also receive alternative drug options.
As an example, Omeprazol, a proton pump inhibitor, and Nebivolol, a beta-blocker, interact on the CYP level. After selecting the drugs, the database provides detailed information on drug structures and ATC group plus CAS numbers (Figure 2).
Figure 2.
Queries and results of the SuperCyp web-interface explaining the various possibilities of the ‘Drug–drug-interaction’ option with the help of two example drugs: Omeprazole and Nebivolol.
The successive ‘results’ page warns that Omeprazol has an inhibitory effect on CYP 2D6, whereas Nebivolol is a substrate. The colored background of the table illustrates this dual use of the CYP metabolism pathway. To avoid this and to optimize the drug composition, Omeprazol can be substituted with other drugs from the same ATC-group, for example Pantoprazol, achieving a comparable effect, but using another pathway.
The proposal of Pantoprazol is derived from the assumption that it does not interact with CYP 2D6. All data on the proposed drugs are provided, and the reference to related publications is given.
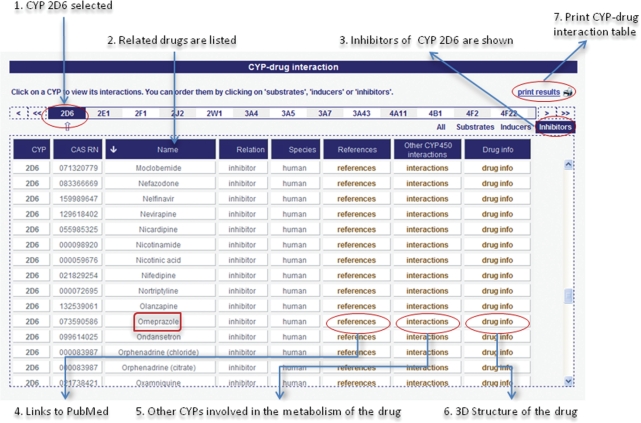
The ‘CYP–Drug-interaction’ allows users to browse substrates, inducers and inhibitors of a certain CYP (Figure 3).
Figure 3.
Results of the SuperCyp web-interface for CYP 2D6 explaining the functionality of the ‘CYP–drug interaction’ table.
After the user has selected a CYP from the task menu, all known relations with drugs are listed in a table. Then users can specify the relation and focus on substrates, inducers or inhibitors. Respective drugs are given in a table and combined with further information on the particular drug and all CYP interactions. References are linked to PubMed and other scientific websites or articles. The ‘Drug Info’ button is linked to the SuperDrug Database (27), which provides a large number of more specific information on the particular drug.
‘Polymorphism’ shows single nucleotide polymorphisms for a particular CYP. All known alleles (15,28) are shown and if there is a decrease or increase in activity or expression, this information is provided. Nucleotide changes and their effects, as well as enzyme activity and assay type are given with corresponding references. Some mutation entries address the protein level directly, in which cases information on SNPs may be missing. However, it is desirable to include protein data, as they provide valuable insights into structure-function relationships.
Example: For CYP11B2, which encodes the enzyme aldosterone synthase (P450aldo), no SNPs were retrieved through keyword searches. However, our mutation/gene association text mining system found 54 protein mutations in 41 PubMed abstracts, which were then added to the database. Among those, the substitution of the highly conserved arginine at position 384 by proline reportedly led to a complete loss of function of this enzyme as part of the autosomal recessively inherited disorder CMO-I deficiency in male Caucasians.
‘Alignments’ uses a structure-based alignment program to match the amino acid sequence of all CYPs. It is possible to create a multiple sequence alignment from any number of sequences or to align them with external sequences by uploading a file or entering a sequence in FASTA format. Users may also draft a convenient output with Jalview.
‘Three-dimensional structures’ displays protein structures of human CYPs. Existing structures were extracted from the PDB. Theoretical models were generated with Swiss-Model (29) or built manually. All structures are downloadable as PDB-files and more information on the CYP is given in the box on the right side.
Clicking the ‘Browse’-button leads to a Java applet, where all CYPs are listed in a drop-down tree, ordered by main families and subfamilies. Each CYP is viewable as a model and further information on its interactions is provided.
RESULTS
Comprehensive data on the 57 human CYPs are stored in the SuperCYP database. For all CYPs, the sequences can be viewed and aligned. Around 1000 SNPs and more than 1200 protein mutations are listed and ordered by their effect on expression and/or activity.
The Protein Data Bank (13,30) provides several crystal structures of Cytochome P450 enzymes. Table 2 lists those nine CYPs that are represented in PDB, while families 4, 5, 7, 11, 17, 19, 20, 21, 24, 26, 27, 39, 46 and 51 are not. Based on the known structures, the SuperCYP database provides theoretical 3D models of the 48 missing human CYPs (Table 2).
Table 2.
Overview of the coverage of the CYP-classes with experimentally determined structures
| CYP | PDB-ID |
|---|---|
| 1A2 | 2hi4 |
| 2A6 | 1z10, 1z11, 1fdu, 2fdv, 2fdw, 2fdy, 2pg5, 2pg6, 2pg7 |
| 2A13 | 2p85 |
| 2C8 | 1pq2, 2nni, 2vn0 |
| 2C9 | 1og2, 1og5, 1r9o |
| 2D6 | 2f9q |
| 2R1 | 3c6g |
| 3A4 | 1w0e, 1w0f, 1w0g, 1tqn, 2j0d, 2v0m |
| 8A1 | 2iag |
With 1170 drugs and ∼3800 interactions, SuperCYP provides the largest number of CYP relations and corresponding information available online. Additionally, checking the tolerability of drug-cocktails and finding alternative uses of metabolic pathways has been made more efficient with the database.
DISCUSSION
The degradation of compounds by CYPs plays an important role in drug–drug interactions that are responsible for harmful adverse effects, e.g. deadly acute renal failure (31). Detailed knowledge about CYP–drug relations is therefore essential for recognizing incompatible drug combinations and to allow individualized therapies. Besides all-inclusive information about drugs, CYPs and their relations, descriptive data such as known CYP-mutations, their phenotypic effects, or the structural information about the CYPs and drugs will enable systematic approaches to quantitative structure–activity relationships. SuperCYP indicates potentially perilous drug–drug interactions and proposes alternative drugs improving mixture or dosage. In conclusion, the server provides a comprehensive resource on CYPs as well as a discovery tool for analysis of drug degradation and drug-cocktail optimization.
AVAILABILITY AND REQUIREMENTS
SuperCYP is available at http://bioinformatics.charite.de/supercyp and can be obtained via a Creative Commons Attribution-Noncommercial-Share Alike 3.0 License. To access all features of the website the latest version of Java should be installed. The database will be updated every 6 months.
FUNDING
Deutsche Forschungsgemeinschaft (SFB 449), Investitionsbank Berlin (IBB), International Research Training Group (IRTG) Berlin–Boston–Kyoto and Deutsche Krebshilfe, Bundesministerium für Bildung und Forschung (BMBF) and European Union (EU). Funding for open access charge: DFG SFB 449, BMBF.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Sarah Preissner for her assistance in completing the structural models of the CYPs.
REFERENCES
- 1.Lepesheva GI, Hargrove TY, Kleshchenko Y, Nes WD, Villalta F, Waterman MR. CYP51: a major drug target in the Cytochrome P450 superfamily. Lipids. 2008;43:1117–1125. doi: 10.1007/s11745-008-3225-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ingelman-Sundberg M. The human genome project and novel aspects of cytochrome P450 research. Toxicol. Appl. Pharmacol. 2005;207:52–56. doi: 10.1016/j.taap.2005.01.030. [DOI] [PubMed] [Google Scholar]
- 3.Lu C, Hatsis P, Berg C, Lee FW, Balani SK. Prediction of pharmacokinetic drug-drug interactions using human hepatocyte suspension in plasma and cytochrome P450 phenotypic data. II. In vitro-in vivo correlation with ketoconazole. Drug Metab. Dispos. 2008;36:1255–1260. doi: 10.1124/dmd.107.018796. [DOI] [PubMed] [Google Scholar]
- 4.Keseru GM. Application of theoretical and synthetic models to cytochrome P450 catalysed metabolic reactions. Acta Pharm. Hung. 1998;68:65–69. [PubMed] [Google Scholar]
- 5.Fernando H, Halpert JR, Davydov DR. Resolution of multiple substrate binding sites in cytochrome P450 3A4: the stoichiometry of the enzyme-substrate complexes probed by FRET and Job’s titration. Biochemistry. 2006;45:4199–4209. doi: 10.1021/bi052491b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dell H, Wong SC, Phillips IR, Shephard EA. Identification of regulatory protein-binding sites in cytochrome P450 genes by means of the gel-retardation assay. Methods Mol. Biol. 1998;107:381–394. doi: 10.1385/0-89603-519-0:381. [DOI] [PubMed] [Google Scholar]
- 7.Bertilsson L. Geographical/interracial differences in polymorphic drug oxidation. Current state of knowledge of cytochromes P450 (CYP) 2D6 and 2C19. Clin. Pharmacokinet. 1995;29:192–209. doi: 10.2165/00003088-199529030-00005. [DOI] [PubMed] [Google Scholar]
- 8.Bathum L, Andersen-Ranberg K, Boldsen J, Brosen K, Jeune B. Genotypes for the cytochrome P450 enzymes CYP2D6 and CYP2C19 in human longevitY. Role of CYP2D6 and CYP2C19 in longevity. Eur. J. Clin. Pharmacol. 1998;54:427–430. doi: 10.1007/s002280050487. [DOI] [PubMed] [Google Scholar]
- 9.Nelson DR. Mining databases for cytochrome P450 genes. Methods Enzymol. 2002;357:3–15. doi: 10.1016/s0076-6879(02)57660-6. [DOI] [PubMed] [Google Scholar]
- 10.Flockhart DA, Oesterheld JR. Cytochrome P450-mediated drug interactions. Child Adolesc. Psychiatr. Clin. N. Am. 2000;9:43–76. [PubMed] [Google Scholar]
- 11.Rendic S, Di Carlo FJ. Human cytochrome P450 enzymes: a status report summarizing their reactions, substrates, inducers, and inhibitors. Drug Metab. Rev. 1997;29:413–580. doi: 10.3109/03602539709037591. [DOI] [PubMed] [Google Scholar]
- 12.Bakare MO. Effective therapeutic dosage of antipsychotic medications in patients with psychotic symptoms: Is there a racial difference? BMC Res. Notes. 2008;1:25. doi: 10.1186/1756-0500-1-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fischer M, Knoll M, Sirim D, Wagner F, Funke S, Pleiss J. The Cytochrome P450 Engineering Database: a navigation and prediction tool for the cytochrome P450 protein family. Bioinformatics. 2007;23:2015–2017. doi: 10.1093/bioinformatics/btm268. [DOI] [PubMed] [Google Scholar]
- 15.Sim SC, Ingelman-Sundberg M. The human cytochrome P450 Allele Nomenclature Committee Web site: submission criteria, procedures, and objectives. Methods Mol. Biol. 2006;320:183–191. doi: 10.1385/1-59259-998-2:183. [DOI] [PubMed] [Google Scholar]
- 16.Radhakrishnan ML, Tidor B. Optimal drug cocktail design: methods for targeting molecular ensembles and insights from theoretical model systems. J. Chem. Inf. Model. 2008;48:1055–1073. doi: 10.1021/ci700452r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van der Weide J, Steijns LS. Cytochrome P450 enzyme system: genetic polymorphisms and impact on clinical pharmacology. Ann. Clin. Biochem. 1999;36(Pt 6):722–729. doi: 10.1177/000456329903600604. [DOI] [PubMed] [Google Scholar]
- 18.Giacomini KM, Brett CM, Altman RB, Benowitz NL, Dolan ME, Flockhart DA, Johnson JA, Hayes DF, Klein T, Krauss RM, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin. Pharmacol. Ther. 2007;81:328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jaja C, Burke W, Thummel K, Edwards K, Veenstra DL. Cytochrome p450 enzyme polymorphism frequency in indigenous and native American populations: a systematic review. Community Genet. 2008;11:141–149. doi: 10.1159/000113876. [DOI] [PubMed] [Google Scholar]
- 20.Lewis DF, Ito Y, Goldfarb PS. Structural modelling of the human drug-metabolizing cytochromes P450. Curr. Med. Chem. 2006;13:2645–2652. doi: 10.2174/092986706778201567. [DOI] [PubMed] [Google Scholar]
- 21.Michalsky E, Goede A, Preissner R. Loops In Proteins (LIP)—a comprehensive loop database for homology modelling. Protein Eng. 2003;16:979–985. doi: 10.1093/protein/gzg119. [DOI] [PubMed] [Google Scholar]
- 22.Montellano PROd. Cytochrome P450: Structure, Mechanism, and Biochemistry. 3rd edn. Netherlands: Springer; 2004. [Google Scholar]
- 23.Nelson DR. Cytochrome P450 nomenclature, 2004. Methods Mol. Biol. 2006;320:1–10. doi: 10.1385/1-59259-998-2:1. [DOI] [PubMed] [Google Scholar]
- 24.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winnenburg R, Plake C, Schroeder M. Proceedings of the ECCB 2008 Workshop: Annotation, Interpretation and Management of Mutations (AIMM) 2008. Mutation tagging with gene identifiers applied to membrane protein stability prediction. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guha R, Howard MT, Hutchison GR, Murray-Rust P, Rzepa H, Steinbeck C, Wegner J, Willighagen EL. The Blue Obelisk-interoperability in chemical informatics. J. Chem. Inf. Model. 2006;46:991–998. doi: 10.1021/ci050400b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 28.Lee SJ, Perera L, Coulter SJ, Mohrenweiser HW, Jetten A, Goldstein JA. The discovery of new coding alleles of human CYP26A1 that are potentially defective in the metabolism of all-trans retinoic acid and their assessment in a recombinant cDNA expression system. Pharmacogenet. Genomics. 2007;17:169–180. doi: 10.1097/FPC.0b013e32801152d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 30.Sussman JL, Lin D, Jiang J, Manning NO, Prilusky J, Ritter O, Abola EE. Protein Data Bank (PDB): database of three-dimensional structural information of biological macromolecules. Acta Crystallogr. D Biol. Crystallogr. 1998;54:1078–1084. doi: 10.1107/s0907444998009378. [DOI] [PubMed] [Google Scholar]
- 31.Kusus M, Stapleton DD, Lertora JJ, Simon EE, Dreisbach AW. Rhabdomyolysis and acute renal failure in a cardiac transplant recipient due to multiple drug interactions. Am. J. Med. Sci. 2000;320:394–397. doi: 10.1097/00000441-200012000-00007. [DOI] [PubMed] [Google Scholar]