Abstract
MINT (http://mint.bio.uniroma2.it/mint) is a public repository for molecular interactions reported in peer-reviewed journals. Since its last report, MINT has grown considerably in size and evolved in scope to meet the requirements of its users. The main changes include a more precise definition of the curation policy and the development of an enhanced and user-friendly interface to facilitate the analysis of the ever-growing interaction dataset. MINT has adopted the PSI-MI standards for the annotation and for the representation of molecular interactions and is a member of the IMEx consortium.
INTRODUCTION
Biologists and bioinformaticians have made extensive use of protein interaction information to interpret experimental results in the context of a global protein interaction network and to test new hypotheses. Protein interaction databases have played a major role in capturing this information from the literature and in presenting it in a structured format to interested users. Nevertheless no single database covers the entire interaction information reported in the literature and, to achieve the largest possible coverage, users or online resources are forced to combine data downloaded from different databases with different data models and ontologies (1–4).
To facilitate the exchange and integration of molecular interactions by data providers, databases and data users, the Molecular Interaction group of the Human Proteome Organization—Protein Standard Initiative, has proposed a data representation standard (current version PSI-MI 2.5) (5). This standard has been adopted by the major protein–protein interaction (PPI) databases and has formed the basis for the emergence of the International Molecular Exchange (IMEx) consortium (http://imex.sourceforge.net/) (6). IMEx follows the model of similar initiatives in different domains of biological data, such as the nucleotide sequence exchange between EMBL (7), Genbank (8) and DDBJ (9), and aims at distributing the curation workload between participating databases thus avoiding work duplication and increasing literature coverage. MINT, the Molecular INTeraction database, participates in this community effort. The databases adhering to this consortium have developed and adopted a common curation manual (http://imex.sourceforge.net/doc/imex-curationManual.doc), describing both the information that should be captured by the member databases and how the information should be represented. The consortium currently comprises four active members: DIP (10), Intact (11), MatrixDb (12) and MINT (13). Additional public databases have already offered to join the effort.
The PSI-MI controlled vocabulary, a major component of the PSI-MI and IMEx data representation standard, defines interactions in a broad sense. Two proteins are said to interact if they score positive in any of the many experimental procedures used to detect molecular interactions, without implying that they make direct physical contact. However, the experimental evidence is clearly defined in associated database records leaving users free to assess confidence for a given piece of experimental evidence. That is, each entry is annotated with the supporting methods, some of which are understood to provide evidence of direct physical contact between the partner proteins such as X-ray crystallography, nuclear magnetic resonance, biochemical assays carried out on purified proteins, and those for which the presence of bridging molecules cannot be excluded such as, pull-down, co-immunoprecipitation and two hybrid assays.
A recent major advance in the protein interaction field is the community definition of the Minimal Information required for reporting a Molecular Interaction eXperiment (MIMIx) (14). Authors reporting protein interaction information in their manuscripts are asked to follow this guidelines, ensuring that this ‘minimal information’ is unambiguously described with controlled vocabularies and cross-references to major public databases. The checklist contains information such as the name and organism of the proteins, their experimental role (bait, prey), or the detection method.
MINT, as a member of the IMEx consortium, is one of the major PPI repositories. It contains interactions experimentally verified and published in peer-reviewed journals. All the interactions are manually curated by professional curators. Over the past three years the database content has grown (more than 19 000 experimental evidences have been added since the last report in 2006), the curation policy has been updated to meet PSI standards, and the web interface has become more user friendly with the development of new query and graphic tools.
In addition each interaction is now annotated with a score ranging from 0 to 1 reflecting the quality and quantity of experimental information supporting the interaction.
ANNOTATION POLICY
Over the past three years in concert with other databases, mainly Intact and DIP, the annotation policy has been reviewed to meet the standards of PSI-MI and the guidelines of the IMEx consortium.
One major advance of the PSI-MI standard has been the replacement of the ‘physical interaction’ term with two new definitions that permit to discriminate between interactions that have a clear experimental evidence for direct contact between the two partners (physical association) and those where the direct contact is not demonstrated (association).
Further details can be found in the curation manual. According to the IMEx criteria each article is curated in its entirety, following the PSI-MI recommendations, and all the interactions and the experimental details of their supporting evidences are captured in the database entries.
There has recently been some vociferous debate between some data providers and databases regarding the role of database curators with respect to assessment of data quality or reliability (15) (Thorneycroft et al. submitted). To clarify, the MINT curation process does not involve any judgment of the accuracy of the published evidence and curators do not make any assessment or ad-hoc reliability ranking of the different interactions reported in a peer reviewed article. Their task is to faithfully represent the experimental information reported by an author. This supporting evidence can then be used by database users to filter the data according to their own reliability standards.
In order to facilitate the navigation of the results of a query, the MINT database associate to each interaction a reliability score (described below) that takes into account the experimental support.
MINT relies on the work of two professional curators and, as a member of the IMEx consortium, has been assigned the task of curating four journals: FEBS Letters, EMBO Journal, EMBO Reports and more recently the FEBS Journal. All PPI described in an article published by one of these journals are added to MINT according to the IMEx manual. Each entry is double checked by a second curator. Furthermore, as many high-throughput datasets are published by journals not covered by the IMEx consortium, these articles are curated in rotation by the three member databases.
Whereas most entries are curated after publication, MINT has begun collaborating with FEBS Letter and FEBS Journal on an editorial procedure that, as recommended by the MIMIx guidelines, involves pre-publication participation of the manuscript authors in the curation process (16). The journal editorial offices submit accepted articles to MINT curators who, in concert with authors, process the protein interaction information as database entries. The processed information is returned to the journal publisher as a structured digital abstract (SDA), where all the interactions described in the article are summarized in a short structured sentence that uses a controlled vocabulary and is appended at the end of the traditional abstract. This SDA can be easily parsed by automatic software and, in the online version of the manuscript, they are hyperlinked to relevant databases.
LIGHT CURATION
As a consequence of limited support (i.e. small curation team) and of the curation depth required by IMEx, MINT does not cover all the protein interactions reported in the literature. To increase the coverage in domains of particular interest to our experimental group, MINT also contains entries that are not fully IMEx compliant, albeit adhering to the PSI-MI model and controlled vocabularies. This type of less detailed curation has been dubbed ‘light curation’ and has allowed an increase in the number of articles curated per time unit. For example MINT has a very high coverage of the experimental evidence supporting interactions mediated by modular domains such as SH3, SH2 or 14-3-3 domains, Similarly most interactions between viral and host proteins are annotated in MINT. Many of the articles supporting these interactions were curated to a lower level of detail than recommended by IMEx, and only the information required by the MIMIx guidelines were captured by curators. The differences between IMEx curation and light (MIMIx based) curation is summarized in Table 1.
Table 1.
Differences between IMEx and light curation
| Annotation | IMEX curation | LIGHT curation | Additional information/examples | |
|---|---|---|---|---|
| Publication | Reference | √ | √ | PMID/D.O.I. |
| Interaction | Figure | √ | √ | Figure, Table |
| Interaction | Interaction type | √ | √ | Direct, physical, enzymatic reaction |
| Experiment | Detection method | √ | √ | Co-immunoprecipitation, two-hybrid |
| Experiment | Biosource | √ | Taxid, cell type, tissue | |
| Interactor | Author given name | √ | √ | |
| Interactor | Cross reference | √ | √ | Uniprotkb, refseq |
| Interactor | Organism | √ | √ | Taxid |
| Interactor | Experimental role | √ | √ | Bait/prey |
| Interactor | Biological role | √ | Enzyme, enzyme target western blot | |
| Interactor | Participant identification | √ | ||
| Interactor | Expression level | √ | Endogenous/over-expressed purification | |
| Interactor | Sample process | √ | ||
| Interactor | Tag | √ | ||
| Interactor | Binding site | √ | Range, domain | |
| Interactor | Modification | √ | Phosphorylation, resulting/required position, amino-acids | |
| Interactor | Mutation | √ |
It is important to understand that entries curated according to the light curation model are as accurate as the IMEx ones. The same controlled vocabularies, proposed by the PSI-MI consortium, are used and in both cases, for instance, the annotator makes a distinction between direct interactions and physical associations, where the experimental evidence cannot prove direct association between the partners. Most of these light curation entries are annotated by experimentalists that are specialists in the given biological domain and are reviewed by one of the professional curators.
The differences between the two curation models do not affect most users, mainly looking for high quality data, but should be taken into consideration when the analysis requires more details about the experimental setup. The entries that are not annotated according to the IMEx manual are clearly labeled in the web-interface and in the exported files. In addition, a ‘light curator’ may choose to capture from an article only those interactions related to his topic of interest, for instance interactions involving a specific domain, and skip additional interaction information that may be present in the same article. In this case the publication is clearly labeled with a ‘caution’ annotation, visible in the web pages and exported with the interactions. This label may be used by users that only want to focus on IMEx data, and by curators who may later complete those entries. In 2009 the oldest interactions in MINT, for which these policies had not been followed, have been blocked and hidden to the users. These entries will be reviewed before re-insertion in the public dataset.
SISTER DATABASES
Once an interaction is curated and validated in MINT it is automatically imported, according to its properties, by one or more sister databases. Three sister databases are presently supported.
HomoMINT (http://mint.bio.uniroma2.it/homomint) (17) is a database of human interactions that are either experimentally verified or inferred from model organisms. Each time an interaction between human proteins is deposited into MINT, it is automatically imported into HomoMINT. The interactions between proteins of model organisms (for instance rat, mouse, yeast, worm or fly) are imported into HomoMINT after mapping to the human orthologs. Orthologs are retrieved from Ensembl compara through the Biomart webservice (http://www.ensembl.org/biomart/martview) (18). The information about the species in which the interaction was experimentally demonstrated is maintained in the HomoMINT entry.
The database of domain–peptide interactions, DOMINO (http://mint.bio.uniroma2.it/domino) (19), focuses on interactions mediated by domains (SH3, SH2, 14-3-3, PDZ, etc.). Wherever the domain mediating an interaction is specified in the MINT entry, the entry is automatically imported into DOMINO. In addition DOMINO contains interactions between domains and peptides that are not present in proteins (for instance peptides selected by phage display) and offers a different interface, including the Domino Viewer applet in which the modular composition of the proteins is displayed.
Finally, all virus–virus and virus–host interactions in MINT are automatically transferred to VirusMINT (http://mint.bio.uniroma2.it/virusmint) (20). VirusMINT has a specialized interface which focuses on virus interactomes, completed with host interactions connecting the viral networks.
A SCORING SYSTEM FOR INTERACTION CONFIDENCE
Interactions stored in MINT are not equally reliable. This is partly due to experimental false positives, especially in high throughput experiments and partly to the different sensitivity and specificity of the diverse experimental setups.
Thus, as it remains difficult for the final user to assess the quality of each binary interaction, we have developed a scoring system to facilitate the evaluation of the reliability of each single interaction, with particular focus on direct physical interactions (21).
The MINT scoring system reflects the quantity and quality of independent supporting evidence stored in the database. We arbitrarily defined the function ‘Cumulative Evidence’ as the sum of all the supporting evidence weighted by coefficients that reflects the confidence in the specific approach. This is based on:
– The size of the experiment: experiments are defined large scale if the article reporting them describes more than 50 interactions, otherwise they are defined small scale. As only the 0.01% of the stored articles report more than 50 interactions we considered this a reasonable threshold to distinguish between large and small scale experiments.
– The type of experiment supporting the interaction.
– It emphasizes evidence of direct interaction (i.e. two-hybrid) with respect to experimental support that does not provide unequivocal evidence of direct interaction (i.e. in vivo co-immunoprecipitation).
– The number of interaction partners detected in a single purification.
– The sequence similarity of ortholog proteins, for interactions mapped to the human proteome in HomoMINT.
– The number of different publications supporting the interaction.
The resulting score ranges between 0 and 1 and only well supported interactions obtain a value close to 1. More details and updates about the score are available at http://mint.bio.uniroma2.it/mint/doc/MINT-confidence-score.html.
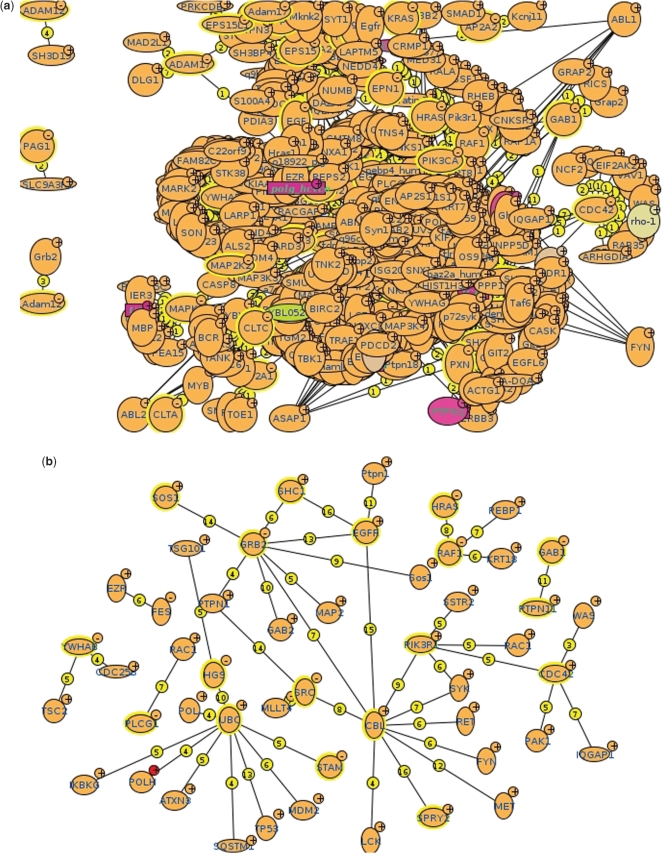
The scoring system, as illustrated in Figure 1, is an effective tool for filtering interactions. In panel (a) we have displayed all the proteins that, according to the MINT database interact with the proteins participating in the EGFR pathway as defined by the Reactome database (22). Panel (b) shows the network that is obtained after removing interactions that are below a certain confidence threshold. Interestingly, the remaining interactions can be easily recognized by any biologist familiar with this pathway (e.g. EFGR-GRB2, EFGR-SHC1).
Figure 1.
Exploring the EGFR pathway in MINT. In panel (a) the EFGR pathway interactome is shown. The network in panel (b) is obtained by removing interactions below a certain confidence threshold (0.72).
WEB INTERFACE
MINT can be queried online using the web interface available at http://mint.bio.uniroma2.it/mint/. The list of molecules that have been shown to interact with a chosen query protein may be displayed either as text on an HTML page or as a graph in the Viewer applet.
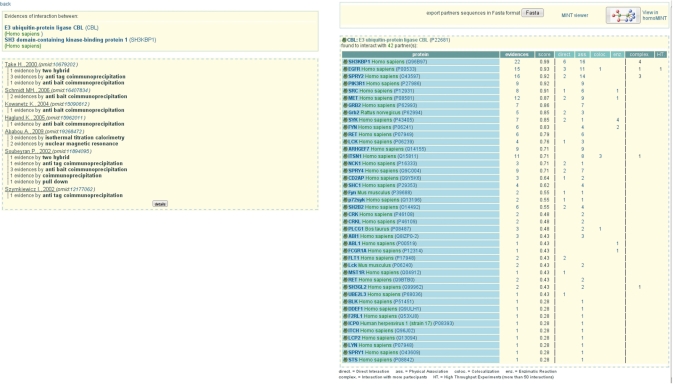
In the HTML output page (Figure 2), the list of interactors is associated with MINT reliability scores. We have recently added new columns to provide an overview of the type of evidence supporting the interaction. We distinguish between experimental evidence for direct interactions, associations (we group here both PSI-MI terms association and physical association), enzymatic reactions and co-localizations. MINT does not capture evidence of genetic interactions. The number of experiments supporting the association of the two proteins in a larger complex and the number of high throughput experiments are indicated in the last two columns.
Figure 2.
Proteins partners of the E3 ubiquitin-protein ligase CBL. In the right frame interactors are ordered according to the MINT score. The total number of evidences is provided as well as the number of direct interactions, physical associations (the two PSI-MI terms physical association and association are grouped here), colocalizations and enzymatic reactions. Even if enzymatic reactions are considered according to PSI-MI standard as a subtype of direct interactions, there are counted here only as enzymatic reaction. The number of evidences by high throughput experiment and the number of evidences in which the two proteins are part of a larger complex are indicated in the last two columns. Each figure in the different columns is linked to the description of the evidences supporting the interaction. By clicking the ‘22’ link on the first row one obtains the information in the the left frame where the evidences for SH3KBP1 as a partner of CBL are grouped by publication and detection method.
In the Viewer applet, the proteins are represented as nodes. Edges are drawn between interacting proteins. The number of evidences that support the interaction is displayed on each edge. The network can be extended by clicking on the “+” symbol in the small circles (it is possible to undo this operation by right clicking on the same symbol). A protein can be removed from the displayed graph by right-clicking on it. The size of the nodes and the distance between them can be controlled through the slide bars in the upper part of the graphic frame. A third slide bar allows the user to hide interactions whose scores are below a chosen threshold. In the graphic display algorithm, the node repulsion force is proportional to the number of partners. As a consequence, two proteins with many interactors (hubs) will tend to lie further away in the graph display when compared with proteins with fewer partners. This graph display rule facilitates the identification of hubs in the graph. As an additional feature, to help identify interaction partners in complex graphs, the action of clicking on a node brings all its partners forward and all the other nodes in the network decrease in size. Only bait–prey partners are represented in the viewer (spoke model), but all components of the same complex are also brought forward during this operation.
Any network displayed by the viewer can be exported as a list of protein pairs along with their confidence score (button score), or in either PSI-MITAB and PSI-MI XML format, both described later in this document. The network exported in a standard PSI-MI format may be easily imported and analyzed in visualization software such as Cytoscape (23).
Both the HTML page and the Viewer applet are connected to relevant information in other pages of the MINT website. Hyperlinks are provided to the source of the descriptions [this information is imported from the Uniprot knowledge base (24)] or the full description of the interactions and the experiments by which they are supported.
A visit to the MINT web-site typically starts by searching for a gene name, a protein name or a cross-reference [to UniprotKB, RefSeq (25), etc.]. If the protein is present in the MINT database, the query returns an HTML page describing the protein on the left frame and listing the interactors in the right frame as described in the previous paragraph. In addition to the information about the protein imported from Uniprot, a list of ortholog proteins available in MINT is appended. If the protein is human or has a human ortholog, it is possible to switch to HomoMINT, where the network of experimentally verified human protein interactions is extended with the interactions transferred from model organisms.
An additional page describes all the experimental details and provides a link to sister databases whenever the interaction is relevant for the specific topic. Sister databases offer several advantages, including additional data (for instance inferred interactions, or interactions between ‘non-natural’ peptides and proteins) and an interface adapted to the specific needs of the specialized database (display of the modular structure of the proteins, visualization of viral-host networks).
Finally, a new tool. ‘Connect’, has been added to the search page. This tool permits interrogation of the database with a list of proteins and returns the entire network of interactions connecting them. The search is performed on a list of proteins cross-references (for instance a list of Uniprot accession numbers), and the user can choose whether to include in the network additional proteins that connect the query proteins (Figure 3). Since the algorithm underlying this tool is demanding up to ∼100 proteins can be submitted.
Figure 3.
The ‘connect’ tool. The network is obtained by searching for 13 SH3 containing proteins. The node size has been reduced to the minimum with the scrollbar to provide a lighter view of the network. The proteins in the query are identified by a yellow border.
DATA AVAILABILITY
All the entries in MINT can be freely downloaded in several formats from a public FTP site, and are programmatically accessible through a web-service.
The official and more complete format is PSI-MI XML 2.5. The former version of the PSI-MI XML format (1.0) has been deprecated and it is no longer available for downloading. The model is normalized, meaning that the experiments and interactors are not repeated in the interaction descriptions but are first listed and then referenced by the interactions. The normalized model results in lighter files. The PSI-MI XML format, in its complex structure, is not human readable but allows a complete description of molecular interactions and their experimental evidences. One of the most relevant advantages is the possibility to associate more than two proteins with an interaction, allowing the correct representation of purified complexes, without misleading binary extensions. Additionally, features such as mutations, modifications or binding sites can be annotated with their sequence.
The FTP site for PSI-MI XML export contains two directories:
PMIDs: a single file is generated for each publication in MINT.
Datasets: containing xml files with the complete dataset or separate files describing interaction entries for each of the main model organisms (human, yeast, fly and worm) or a group of phylogenetically related organisms (e.g. mammals).
PSI-MI XML files for large datasets containing more than 1000 interactions, such as those derived from manuscripts reporting high throughput experiments or datasets for intensively studied organisms, are split in smaller files and zipped together. There is no overlap between XML files contained in a single archive.
Files in the PSI-MITAB format contain the same information, but in a tab-delimited format that can be opened in a spreadsheet software and it is easier to parse. The current format is 2.6. The columns of version 2.6 are the 15 columns of version 2.5 (see http://code.google.com/p/psimi/wiki/PsimiTabFormat) extended with 16 new columns (read discussion at http://code.google.com/p/psimi/issues/detail?id=2). The ‘expansion’ column is probably the most relevant feature of the new format. An expansion strategy is applied in order to represent a complex of three or more proteins in a binary file format. There are two models that can be adopted to achieve this goal. On one hand, matrix model, one can represent the complex as the ensemble of all the possible binary interactions between the protein members. Alternatively, spoke model, one member of the complex (bait) can be associated to all the remaining members (prey). None of those models faithfully describes the topology of a complex, and the resulting protein pairs are not meant to represent the interaction between the protein in the complex. If the interaction on a row results from the expansion of a complex, the expansion mode (spoke, matrix) is specified in this field. The field is empty if the binary interaction is not the result of an expansion procedure. This allows the user to filter rows where the interaction is not supported by an experiment that is evidence for a binary interaction, or to reconstruct the complexes (for instance by grouping rows by interaction identifier and looking at the experimental role of the components).
PPI databases, adhering to PSI MI, have commonly agreed to use the spoke model as a method to represent n-ary interaction data in a binary format during the PSI-MI 2009 spring meeting. This means that if the experiment consists of the identification of many preys with a single bait (i.e. as in TAP tag technology), all possible bait–prey interactions are represented. If, on the other hand, the role of the proteins in the experiment supporting the existence of the complex is ‘neutral’ (i.e. in the case of a complex identified by purification by co-sedimentation), one protein is chosen as an ‘arbitrary’ bait. Since we feel that this policy may be misleading for some user cases, we provide two files. One containing only binary interactions and the second only complex interactions ‘exploded’ according to the spoke model. There is no overlap between those two files, which can be appended one to the other to obtain the full dataset. This issue is not relevant for XML files in which the representation of complexes is possible.
In the last additional column, ‘Caution Interaction’, we export the caution annotations described in the previous paragraphs—for instance if an interaction is curated with MIMIx standards rather than fully adhering to the IMEx curation manual, or if a publication has been only partially curated.
Finally, we provide a tab-delimited format, similar to MITAB, where all the proteins forming a complex are described in a single line (complexes are not exploded). In this format we list the baits or enzymes (for enzymatic reactions) in the first column, and the preys or enzyme targets in the second.
Web services allow the access to the data computationally and have become increasingly more popular in the bioinformatics community. The HUPO-PSI workgroup has defined a standard web service to access molecular databases: the PSICQUIC interface. MINT uses the PSICQUIC reference implementation, implemented at the European Bioinformatics Institute. The web-service is available using either SOAP or REST protocols at http://mint.bio.uniroma2.it/mint/psicquic. Documentation about the PSICQUIC interface, the PSICQUIC providers and the reference implementation are available at http://code.google.com/p/psicquic/.
FUNDING
Italian association for Cancer Research (AIRC), Telethon and the ENFIN FP7 network of excellence. Funding for open access charge: AIRC.
Conflict of interest statement. None declared.
REFERENCES
- 1.Jayapandian M, Chapman A, Tarcea VG, Yu C, Elkiss A, Ianni A, Liu B, Nandi A, Santos C, Andrews P, et al. Michigan Molecular Interactions (MiMI): putting the jigsaw puzzle together. Nucleic Acids Res. 2007;35:D566–D571. doi: 10.1093/nar/gkl859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aragues R, Jaeggi D, Oliva B. PIANA: protein interactions and network analysis. Bioinformatics. 2006;22:1015–1057. doi: 10.1093/bioinformatics/btl072. [DOI] [PubMed] [Google Scholar]
- 3.Cerami EG, Bader GD, Gross BE, Sander C. cPath: open source software for collecting, storing, and querying biological pathways. BMC Bioinformatics. 2006;7:497. doi: 10.1186/1471-2105-7-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. Proteomics. 2007;7(Suppl. 1):28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 7.Kulikova T, Akhtar R, Aldebert P, Althorpe N, Andersson M, Baldwin A, Bates K, Bhattacharyya S, Bower L, Browne P, et al. EMBL Nucleotide Sequence Database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2008;36:D25–D30. doi: 10.1093/nar/gkm929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugawara H, Ogasawara O, Okubo K, Gojobori T, Tateno Y. DDBJ with new system and face. Nucleic Acids Res. 2008;36:D22–D24. doi: 10.1093/nar/gkm889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct—open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;25:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 15.Cusick ME, Yu H, Smolyar A, Venkatesan K, Carvunis AR, Simonis N, Rual JF, Borick H, Braun P, Dreze M, et al. Literature-curated protein interaction datasets. Nat. Methods. 2009;6:39–46. doi: 10.1038/nmeth.1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ceol A, Chatr-Aryamontri A, Licata L, Cesareni G. Linking entries in protein interaction database to structured text: the FEBS Letters experiment. FEBS Lett. 2008;582:1171–1177. doi: 10.1016/j.febslet.2008.02.071. [DOI] [PubMed] [Google Scholar]
- 17.Persico M, Ceol A, Gavrila C, Hoffmann R, Florio A, Cesareni G. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6(Suppl. 4):S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ceol A, Chatr-aryamontri A, Santonico E, Sacco R, Castagnoli L, Cesareni G. DOMINO: a database of domain-peptide interactions. Nucleic Acids Res. 2007;35:D557–D560. doi: 10.1093/nar/gkl961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M, et al. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669–D673. doi: 10.1093/nar/gkn739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chatr-Aryamontri A, Ceol A, Licata L, Cesareni G. Protein interactions: integration leads to belief. Trends Biochem. Sci. 2008;33:241–242. doi: 10.1016/j.tibs.2008.04.002. author reply 242–243. [DOI] [PubMed] [Google Scholar]
- 22.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Killcoyne S, Carter GW, Smith J, Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol. Biol. 2009;563:219–239. doi: 10.1007/978-1-60761-175-2_12. [DOI] [PubMed] [Google Scholar]
- 24.The Universal Protein Resource (UniProt) (2009) Nucleic Acids Res. 37:D169–D74. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the entries in MINT can be freely downloaded in several formats from a public FTP site, and are programmatically accessible through a web-service.
The official and more complete format is PSI-MI XML 2.5. The former version of the PSI-MI XML format (1.0) has been deprecated and it is no longer available for downloading. The model is normalized, meaning that the experiments and interactors are not repeated in the interaction descriptions but are first listed and then referenced by the interactions. The normalized model results in lighter files. The PSI-MI XML format, in its complex structure, is not human readable but allows a complete description of molecular interactions and their experimental evidences. One of the most relevant advantages is the possibility to associate more than two proteins with an interaction, allowing the correct representation of purified complexes, without misleading binary extensions. Additionally, features such as mutations, modifications or binding sites can be annotated with their sequence.
The FTP site for PSI-MI XML export contains two directories:
PMIDs: a single file is generated for each publication in MINT.
Datasets: containing xml files with the complete dataset or separate files describing interaction entries for each of the main model organisms (human, yeast, fly and worm) or a group of phylogenetically related organisms (e.g. mammals).
PSI-MI XML files for large datasets containing more than 1000 interactions, such as those derived from manuscripts reporting high throughput experiments or datasets for intensively studied organisms, are split in smaller files and zipped together. There is no overlap between XML files contained in a single archive.
Files in the PSI-MITAB format contain the same information, but in a tab-delimited format that can be opened in a spreadsheet software and it is easier to parse. The current format is 2.6. The columns of version 2.6 are the 15 columns of version 2.5 (see http://code.google.com/p/psimi/wiki/PsimiTabFormat) extended with 16 new columns (read discussion at http://code.google.com/p/psimi/issues/detail?id=2). The ‘expansion’ column is probably the most relevant feature of the new format. An expansion strategy is applied in order to represent a complex of three or more proteins in a binary file format. There are two models that can be adopted to achieve this goal. On one hand, matrix model, one can represent the complex as the ensemble of all the possible binary interactions between the protein members. Alternatively, spoke model, one member of the complex (bait) can be associated to all the remaining members (prey). None of those models faithfully describes the topology of a complex, and the resulting protein pairs are not meant to represent the interaction between the protein in the complex. If the interaction on a row results from the expansion of a complex, the expansion mode (spoke, matrix) is specified in this field. The field is empty if the binary interaction is not the result of an expansion procedure. This allows the user to filter rows where the interaction is not supported by an experiment that is evidence for a binary interaction, or to reconstruct the complexes (for instance by grouping rows by interaction identifier and looking at the experimental role of the components).
PPI databases, adhering to PSI MI, have commonly agreed to use the spoke model as a method to represent n-ary interaction data in a binary format during the PSI-MI 2009 spring meeting. This means that if the experiment consists of the identification of many preys with a single bait (i.e. as in TAP tag technology), all possible bait–prey interactions are represented. If, on the other hand, the role of the proteins in the experiment supporting the existence of the complex is ‘neutral’ (i.e. in the case of a complex identified by purification by co-sedimentation), one protein is chosen as an ‘arbitrary’ bait. Since we feel that this policy may be misleading for some user cases, we provide two files. One containing only binary interactions and the second only complex interactions ‘exploded’ according to the spoke model. There is no overlap between those two files, which can be appended one to the other to obtain the full dataset. This issue is not relevant for XML files in which the representation of complexes is possible.
In the last additional column, ‘Caution Interaction’, we export the caution annotations described in the previous paragraphs—for instance if an interaction is curated with MIMIx standards rather than fully adhering to the IMEx curation manual, or if a publication has been only partially curated.
Finally, we provide a tab-delimited format, similar to MITAB, where all the proteins forming a complex are described in a single line (complexes are not exploded). In this format we list the baits or enzymes (for enzymatic reactions) in the first column, and the preys or enzyme targets in the second.
Web services allow the access to the data computationally and have become increasingly more popular in the bioinformatics community. The HUPO-PSI workgroup has defined a standard web service to access molecular databases: the PSICQUIC interface. MINT uses the PSICQUIC reference implementation, implemented at the European Bioinformatics Institute. The web-service is available using either SOAP or REST protocols at http://mint.bio.uniroma2.it/mint/psicquic. Documentation about the PSICQUIC interface, the PSICQUIC providers and the reference implementation are available at http://code.google.com/p/psicquic/.