Abstract
Most of the proteins in a cell assemble into complexes to carry out their function. In this work, we have created a new database (named ComSin) of protein structures in bound (complex) and unbound (single) states to provide a researcher with exhaustive information on structures of the same or homologous proteins in bound and unbound states. From the complete Protein Data Bank (PDB), we selected 24 910 pairs of protein structures in bound and unbound states, and identified regions of intrinsic disorder. For 2448 pairs, the proteins in bound and unbound states are identical, while 7129 pairs have sequence identity 90% or larger. The developed server enables one to search for proteins in bound and unbound states with several options including sequence similarity between the corresponding proteins in bound and unbound states, and validation of interaction interfaces of protein complexes. Besides that, through our web server, one can obtain necessary information for studying disorder-to-order and order-to-disorder transitions upon complex formation, and analyze structural differences between proteins in bound and unbound states. The database is available at http://antares.protres.ru/comsin/.
INTRODUCTION
It has been found that many proteins have no unique tertiary structure under physiological conditions although they are functional (1–4). Such proteins are called intrinsically unstructured (or intrinsically disordered or natively unfolded) proteins. The size of unstructured region(s) in these proteins can vary from a few amino acid residues to dozens or even hundreds of amino acid residues. Moreover, an entire protein can be completely unstructured. Since disordered regions of the protein chain often play an important role in the functioning of the protein, the prediction and examination of the disordered regions have been the subject of much recent attention (5,6). It has been shown that disordered proteins (and disordered regions in globular proteins) have certain properties that distinguish them from globular proteins with well-defined, ordered spatial structures. Typically, disordered regions have lower aromatic content and higher net charge as well as lower sequence complexity and higher flexibility (2,3,7).
The Database of Protein Disorder [named DisProt (8)] provides information about proteins that lack fixed 3D structure in their putative native states, either in their entirety or in part (http://www.disprot.org/index.php). The latest version (Release 4.9) of this database contains 523 proteins. DisProt emphasizes mainly proteins with large disordered regions. Consequently, proteins from PDB (mostly globular proteins usually possessing short disordered regions) are underrepresented in this database. Recently, protein segments (shorter than 70 residues) assumed to be disordered when unbound and also observed to be bound to other proteins/chains were collected from PDB (9, www.pdb.org) to obtain the 372 chains that make up the MoRF (molecular recognition features) dataset (10). MoRFs represent a class of disordered regions that exhibit molecular recognition and binding functions (10). It should be noted that MoRFs are predicted to be disordered in unbound form and ordered in the complex.
While individual cases of disorder in complexes are known and have been discussed in the literature, the first large-scale analysis of this phenomenon was done in our recent paper on a carefully assembled dataset (11). The set of protein structures selected in this work forms the seed of the database that we are presenting now. To gain a clear insight into the abundance of disordered regions in structures of unbound proteins and protein complexes (bound states) as well as disorder-to-order and order-to-disorder transitions upon complex formation, we create an exhaustive database (named ComSin) of protein structures in bound (‘Complex’) and unbound (‘Single’) states. The usage of this database is not restricted only to the tasks connected with investigations of disordered regions in proteins and their complexes. ComSin can be used to analyze any structural differences between proteins in bound and unbound states and to explore changes induced by protein binding, among other purposes.
DESCRIPTION OF THE DATABASE
Composing the database
The ComSin database was created based on the Conserved Domain Database (CDD) (12). CDD represents a collection of well-annotated multiple sequence alignment models for protein domains. CDD includes curated domains, which use 3D-structure information to explicitly define domain boundaries and provide accurate alignments and functional annotation, as well as domain models imported from a number of external source databases [Pfam (13), SMART (14) and COG (15)]. Conserved domains were mapped onto sequences from PDB following the protocol described in (16). Protein chains from the MMDB database (17) were used as query sequences to RPS-BLAST (18) with default parameters (E = 0.01) against domains in CDD v.2.08 (12). We selected only those MMDB X-ray structures that had resolution better than 3 Å.
We ensured that each chain has only one CDD domain that covers at least 70% of the full chain sequence. Later on, we will consider complete protein chains (rather than separate domains) in the form of pairs (chain in the bound state versus the same chain in the unbound state). Once CDD families are assigned, we identify all interacting chains within a PDB entry. Two chains qualify as interacting if they have at least five residue–residue contacts. A contact takes place between a residue from one chain and a residue from the other when any non-hydrogen atom of one residue is within 6 Å of any non-hydrogen atom of the other residue. The set of residues that make contacts between the chains form the interface. To ensure that interactions are biological and not spurious, such as from crystal packing, we verify interactions using the Conserved Binding Modes (CBM) database (19) and the PISA algorithm (20). In ComSin, we compare disorder content in an unbound (‘single’) and bound states (‘complex’). To do so, chains from ‘single’ and ‘complex’ states were aligned to ensure 90–100% sequence identity in the non-gapped alignment (different levels of sequence similarity of 90, 95 and 100% are used).
Sequence identity is determined as follows:
where N_identical is the number of amino acid residues that are identical in the unbound protein and its close homolog in the bound state according to their BLAST (21) alignment, L_bound is the size (total number of residues) of the unbound protein and L_bound is the size of the bound homologous protein.
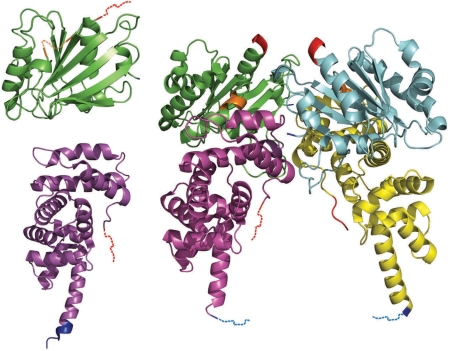
Figure 1 shows an example of two proteins in bound and unbound forms from the composed database. In the left part of the figure, ADP-ribosylation factor (top) and exchange factor ARNO (bottom) are shown in unbound forms (PDB entries 1RRF and 1R8M, respectively). In the right part of the figure, a complex of these two proteins is shown (PDB entry 1R8Q). Colored dashed lines indicate disordered residues.
Figure 1.
ADP-ribosylation factor (green and blue chains) and exchange factor ARNO (magenta and yellow chains) in unbound (left; PDB entries 1RRF and 1R8M, correspondingly) and bound (right; PDB entry 1R8Q) forms. Blue, red and orange dashed lines correspond to disordered regions at N-terminus, at C-terminus and in the central part of protein chains, correspondingly.
Statistics of the database
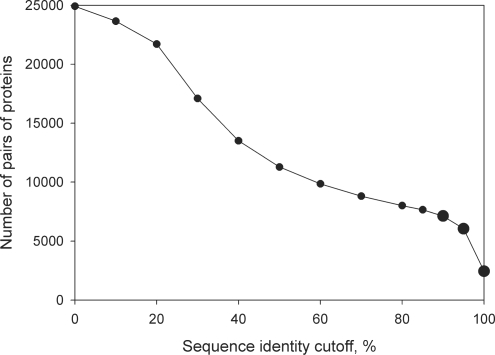
In total, we obtained 24 910 pairs of homologous proteins observed in unbound and bound states, with a wide range of sequence identity between the homologs. There are 2448, 6051 and 7129 single–complex pairs at 100, 95 and 90% identity level cutoffs, respectively (Figure 2). The number of CDD families is 352, 521 and 576 at 100, 95 and 90% identity level cutoffs, respectively. The further average values are calculated as follows. At first, the average value for each family is calculated. Then, the average between these average values is calculated. Thus, we consider each family with an equal weight. At the 100% identity level, the bound chains (‘complexes’) contain 3.7 protein chains per complex on average. The average size of the protein chains is 220 residues. Of 2448 single–complex pairs, 1975 are homo-oligomers (that is, all chains in the complex have identical sequences) and 473 are hetero-oligomers (there are chains with non-identical sequences in the complex). For the 90% identity level cutoff, the complexes contain 3.9 protein chains per file on average. The average size of protein chains is 223 residues. Of 7129 chains in complexes, 6060 are homo-oligomers and 1069 are hetero-oligomers.
Figure 2.
Dependence of the number of complex–single pairs on the sequence identity cutoff. There are 2448, 6051 and 7129 single–complex pairs at 100, 95 and 90% identity level cutoffs, respectively.
An overview of the disordered regions in Protein Data Bank
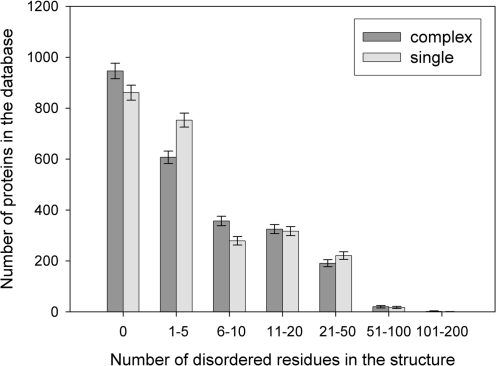
Using the ComSin database, we have analyzed the abundance of disorder in the structures of the same proteins (i.e. at 100% identity level) in bound and unbound states. Figure 3 shows the number of structures with a given number of disordered residues for complex and single states. As can be seen from this figure, although there are somewhat larger number of structures without disordered residues at all in complexes compared to the single structures, there is no definite tendency to have more disordered residues in unbound structures compared to the bound ones (if disordered residues are present). Moreover, the average numbers of disordered residues per protein are not significantly different for single or complex states (the average over complexes is 8.9 ± 0.6 disordered residues per structure and the average over single structures is 9.3 ± 0.7 disordered residues per structure). Among the 2448 pairs of identical proteins in our database, there was an equal number of disordered residues in bound and unbound states in 1091 cases. Of these, in 702 proteins disordered residues were absent both in bound and in unbound states. In 728 proteins, the number of disordered residues in the unbound state was greater than that in the bound state (implying possible disorder-to-order transition). On the other hand, order-to-disorder transition is observed in 629 cases (the number of disordered residues in the bound state was greater than that in the unbound state). Thus, the numbers of cases of disorder-to-order and order-to-disorder transitions are comparable.
Figure 3.
A histogram of the number of proteins in the database by the number of disordered residues in the structure, for 2448 pairs of homologous proteins with 100% identity level.
The role of disordered regions in complexes has been analyzed previously in several studies (11,22,23). It has been proposed that disordered regions can be energetically beneficial in proteins and their complexes due to a number of reasons: they can provide an increase in backbone conformational entropy upon ligand binding, can accommodate sites for post-translational modifications and can provide interfaces for binding other partners (22,24–29). Thus, it is not surprising that disorder is widespread in protein complexes.
Description of the server
The ComSin server is designed as follows. The main page contains a general description of the information a user may obtain through this database. On the ComSin search page, there are several filters for selecting a subset of single–complex pairs of structures. One can search for pairs of structures with a given cutoff level of sequence identity between proteins in single and complex states. Presently, one of the three cutoffs of sequence identity can be selected: 100 (completely the same protein in bound and unbound form), 95 or 90%. For the bound state, one can also choose to view only homo- or only hetero-oligomers. In addition, the database may be searched using a PDB code or a CDD family identifier as an input. One can also select only those pairs that are considered valid by PISA (20) and/or by CBM analysis (19). The PISA and CBM algorithms were used for validation of oligomeric states and the biological relevance of each interaction. PISA validation is based on calculation of the stability of multimeric states inferred from the crystalline state. To ensure that the interactions observed in the complex (bound state) are biological and not spurious, such as from crystal packing, we applied the conserved binding mode (CBM) analysis that confirms interactions by finding several instances of the same domain family pair interacting in the same orientation. We also give a link to a new NCBI server, IBIS (Inferred Biomolecular Interaction Server, http://www.ncbi.nlm.nih.gov/Structure/ibis/ibis.cgi) that provides a tool to investigate biomolecular interactions observed in a given protein structure together with the complex set of interactions inferred from its close homologs (30). To emphasize biologically relevant binding sites, several algorithms are used in IBIS for verification in terms of evolutionary conservation, biological importance of binding partners, size and stability of interfaces as well as evidence from the published literature.
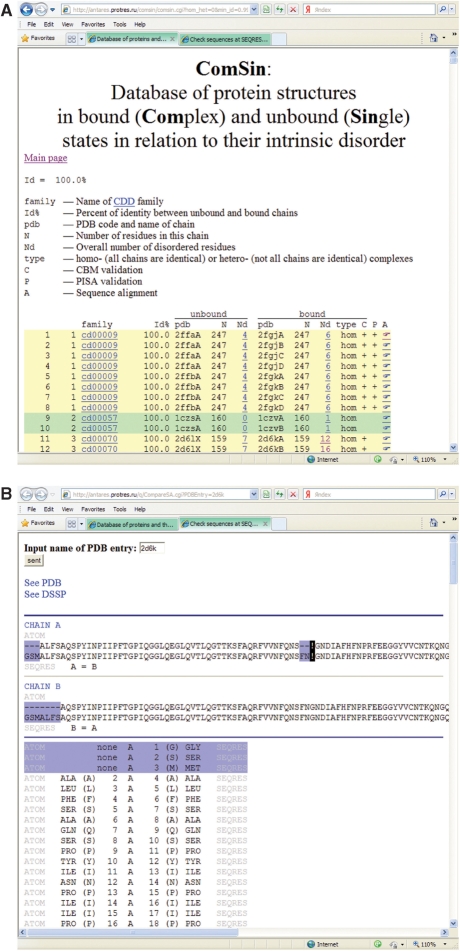
By clicking the button ‘Search ComSin’, the user obtains a list of protein pairs that satisfy the selected filtering criteria. For example, for 100% identity between bound and unbound states (the default value), one will obtain a list of 2448 pairs of protein structures. Each line corresponds to the same protein (if 100% identity is selected) or close homologs (if 95% or 90% identity is selected) observed in unbound and bound states (Figure 4A). The first column shows the numbering of the pair in the current list. The next two columns contain information on the CDD domain family to which both unbound protein and its bound homolog belong. By clicking on the CDD family name, one can see the description of the family at the NCBI web site. In the next column, sequence identity between the unbound protein and its bound homolog is shown. Further, there are three columns related to the structure in the unbound form: the PDB code including the name of the chain, size of the chain and number of disordered residues in the chain. Similarly, the next three columns correspond to the structure of the bound homolog (PDB code, size of the chain and the number of disordered residues). The next two columns correspond to the whole complex. The first column describes the type of the complex: homo- or hetero-oligomers. The column C indicates CBM validation of the complex, and the column P indicates PISA validation of the bound and unbound states. In the last column (A), references to sequence alignments of each pair are given.
Figure 4.
(A) A screenshot of ComSin results filtered for sequence identity 100%. (B) A screenshot of an individual page with information on disordered regions for PDB entry 2D6K.
Disordered regions are defined as regions with missing coordinates in X-ray-resolved structures. For each chain, we searched for amino acid residues with missing coordinates for Cα atoms in the corresponding PDB entry by comparing the ATOM and SEQRES records (in an X-ray-resolved structure, disordered regions are supposed to be present in SEQRES record but absent in ATOM record). By clicking on the number of disordered residues for a single or a complex structure, one can view a file showing the positions of disordered regions (one file per PDB structure, see Figure 4B). The sequence of the corresponding protein (according to SEQRES and according to ATOM fields) is given in horizontal (short) and vertical (long) view; disordered residues are marked in blue (Figure 4B). In addition, one can open the corresponding PDB and DSSP (31) files for this protein that will provide additional structural information.
CONCLUSIONS AND FUTURE DIRECTIONS
We have collected an exhaustive database of proteins in unbound and bound states, and examined disordered regions of the same protein in unbound state and in a protein complex. The evidence pointing to the tremendous importance of intrinsic disorder in a large variety of cellular processes is accumulating and merits further study. In future work, we are planning to include into the database data on (experimentally shown or predicted) function of disordered regions in proteins in unbound and bound states. We will be grateful for any contribution to the database from the community.
FUNDING
S.H.Programs ‘Molecular and Cellular Biology’ and ‘Fundamental Sciences—medicine’ by the Russian Foundation for Basic Research (08-04-00561); Russian Science Support Foundation; Federal Agency for Science and Innovation (grant #02.740.11.0295); Intramural Research Program of the NIH, National Library of Medicine. Funding for open access charge: Intramural Research Program of the NIH, National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: reassembling the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN, Gillspie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 4.Tompa P. Intrinsically unstructured proteins. Trends Biochem. Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 5.Linding R. PhD Thesis. University of Heidelberg; 2004. Linear functional modules. Implication for protein function. [Google Scholar]
- 6.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 7.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J. Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 11.Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS Comput. Biol. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, et al. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meereis F, Kaufmann M. Extension of the COG and arCOG databases by amino acid and nucleotide sequences. BMC Bioinform. 2008;9:479. doi: 10.1186/1471-2105-9-479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci. 2006;15:352–361. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, et al. MMDB: annotating protein sequences with Entrez's; 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci. 2006;15:352–361. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meszaros B, Tompa P, Simon I, Dosztanyi Z. Molecular principles of the interactions of disordered proteins. J. Mol. Biol. 2007;372:549–561. doi: 10.1016/j.jmb.2007.07.004. [DOI] [PubMed] [Google Scholar]
- 23.Tompa P, Fuxreiter M. Fuzzy complexes: polymorphism and structural disorder in protein-protein interactions. Trends Biochem. Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 24.Huber R, Bennett WS., Jr Functional significance of flexibility in proteins. Biopolymers. 1983;22:261–279. doi: 10.1002/bip.360220136. [DOI] [PubMed] [Google Scholar]
- 25.Stivers JT, Abeygunawardana C, Mildvan AS. 15N NMR relaxation studies of free and inhibitor-bound 4-oxalocrotonate tautomerase: backbone dynamics and entropy changes of an enzyme upon inhibitor binding. Biochemistry. 1996;35:16036–16047. doi: 10.1021/bi961834q. [DOI] [PubMed] [Google Scholar]
- 26.Olejniczak ET, Zhou MM, Fesik SW. Changes in the NMR-derived motional parameters of the insulin receptor substrate 1 phosphotyrosine binding domain upon binding to an interleukin 4 receptor phosphopeptide. Biochemistry. 1997;36:4118–4124. doi: 10.1021/bi963050i. [DOI] [PubMed] [Google Scholar]
- 27.Zidek L, Novotny MV, Stone MJ. Increased protein backbone conformational entropy upon hydrophobic ligand binding. Nat. Struct. Biol. 1999;6:1118–1121. doi: 10.1038/70057. [DOI] [PubMed] [Google Scholar]
- 28.Loh AP, Pawley N, Nicholson LK, Oswald RE. An increase in side chain entropy facilitates effector binding: NMR characterization of the side chain methyl group dynamics in Cdc42Hs. Biochemistry. 2001;40:4590–4600. doi: 10.1021/bi002418f. [DOI] [PubMed] [Google Scholar]
- 29.Sigalov AB, Zhuravleva AV, Orekhov VY. Binding of intrinsically disordered proteins is not necessarily accompanied by a structural transition to a folded form. Biochimie. 2007;89:419–421. doi: 10.1016/j.biochi.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred Biomolecular Interaction Server (IBIS)—a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]