Abstract
TriTrypDB (http://tritrypdb.org) is an integrated database providing access to genome-scale datasets for kinetoplastid parasites, and supporting a variety of complex queries driven by research and development needs. TriTrypDB is a collaborative project, utilizing the GUS/WDK computational infrastructure developed by the Eukaryotic Pathogen Bioinformatics Resource Center (EuPathDB.org) to integrate genome annotation and analyses from GeneDB and elsewhere with a wide variety of functional genomics datasets made available by members of the global research community, often pre-publication. Currently, TriTrypDB integrates datasets from Leishmania braziliensis, L. infantum, L. major, L. tarentolae, Trypanosoma brucei and T. cruzi. Users may examine individual genes or chromosomal spans in their genomic context, including syntenic alignments with other kinetoplastid organisms. Data within TriTrypDB can be interrogated utilizing a sophisticated search strategy system that enables a user to construct complex queries combining multiple data types. All search strategies are stored, allowing future access and integrated searches. ‘User Comments’ may be added to any gene page, enhancing available annotation; such comments become immediately searchable via the text search, and are forwarded to curators for incorporation into the reference annotation when appropriate.
INTRODUCTION
The Trypanosomatidae are a group of unicellular, flagellated, obligate parasites, including many important pathogens of humans and animals. African trypanosomes (Trypanosoma brucei, T. congolense and T. vivax) are endemic in rural areas of sub-Saharan Africa, where they cause sleeping sickness in humans, and wasting disease (nagana) in cattle. These diseases are invariably fatal if left untreated (1). In 2004, 17 580 cases of human infection were reported, but due to chronic under-reporting in poor, rural areas, actual infection rates are thought to reach 300 000 new cases annually (2,3). Millions of cattle are also at risk, and trypanosomiasis severely constrains cattle grazing in endemic regions. Trypanosoma cruzi is endemic in south and central America, causing Chagas disease (American Trypanosomiasis) in approximately 8–9 million infected individuals, and ∼14 000 deaths annually (4). Leishmania parasites are found throughout the world (old world: L. major; new world: L. infantum and L. braziliensis), infecting an estimated 12 million individuals, with approximately 2 million new cases reported annually (5). These parasites exhibit a variety of spectral pathologies, including severely debilitating cutaneous disease, and visceral symptoms that may be fatal.
The genomes of multiple Trypanosomatidae have been sequenced (6–9) and are available from sources such as GeneDB (http://GeneDB.org) (10) and the primary sequence nucleotide databases (DDBJ, EMBL and GenBank); genome projects are also underway for a variety of other species of biological and evolutionary interest. Whilst GeneDB specializes in the display of highly-curated annotation, it has been difficult to integrate this information with other available ‘omics’ datasets: expression profiling data, proteomics results, etc., hindering the research on these organisms, including the development of new therapies and diagnostics. To this end, the TriTrypDB initiative was undertaken as a collaborative effort between the EuPathDB team at the Universities of Pennsylvania and Georgia (http://EuPathDB.org) (11), the GeneDB group at the Wellcome Trust Sanger Institute and researchers from the Seattle Biomedical Research Institute, culminating in the release of the first version of TriTrypDB (http://TriTrypDB.org) in early 2009. This collaboration has proved to be an effective means for providing the scientific community with up-to-date annotation and curation, and access to tools enabling sophisticated queries against genomic scale datasets.
DATA IN THE CURRENT RELEASE
TriTrypDB (release 1.1) houses the genome sequences of T. brucei TREU 927 strain [11 chromosomes, 26 megabases (Mb)] (6); T. cruzi CL Brener, Esmeraldo and non-Esmeraldo-like haplotypes (41 chromosomes, 67 Mb) (7,12); L. major Friedlin strain (36 chromosomes, 32.8 Mb) (8), L. infantum JPCM5 strain (36 chromosomes, 32 Mbs) (9); L. braziliensis Viannia strain (35 chromosomes, 32 Mb) (9); and L. tarentolae (sequence kindly provided in advance of publication, by Marc Ouellette, Université Laval, and Martin Olivier from McGill University). TriTrypDB also includes selected transcript and proteomics expression data (with more to follow over the coming months). Transcript expression information is derived from both microarray data (L. infantum differentiation time series and data provided pre-publication by Dan Zilberstein and Peter Myler) and expressed sequence tags (EST libraries from T. brucei, T. cruzi, L. braziliensis, L. infantum and L. major extracted from dbEST; http://www.ncbi.nlm.nih.gov/dbEST). Protein expression data based on tandem mass spectrometry of whole parasites and subcellular fractions is available for T. brucei (13), T. cruzi (14), L. braziliensis, L. infantum and L. major [(15), and Marc Ouellette, pre-publication].
USING TriTrypDB
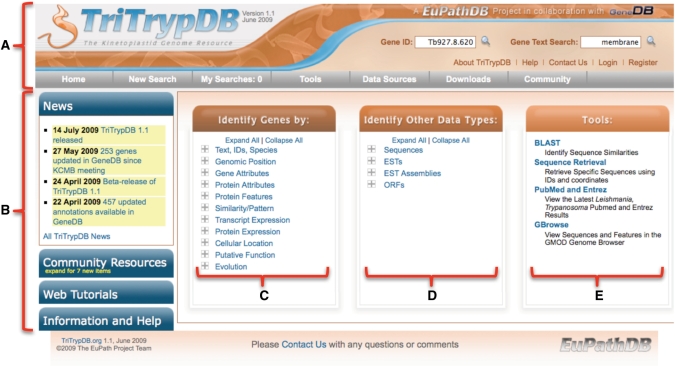
The home page of TriTrypDB is based on the recently re-designed EuPathDB web page, and includes five main sections (Figure 1). The top of the page is an interactive banner (Figure 1A), which appears on all pages and includes (i) the TriTrypDB logo, (ii) windows for ID and text searches, (iii) links providing quick access to useful pages (help and information pages, a ‘Contact Us’ link and links for registration/login) and (iv) a tool bar (grey) with links to access diverse searches (see below), the user’s personal search history, tools, downloads, data sources and other links. The left side of the home page (Figure 1B) provides a series of expandable windows presenting news items (such as release notes), tutorials (demonstrating website usage), community resources and additional information and help. New items added since the user’s last visits are indicated by yellow numbers. Three panels in the middle of the page provide access to searches and tools: the panel indicated as Figure 1C includes diverse searches pertaining to genes; Figure 1D accesses searches against other data types (assemblies, ESTs, Open Reading Frame (ORFs), etc., with more data types to follow, as already implemented for other EuPathDB component databases); Figure 1E provides links to tools such as BLAST, sequence retrieval and a genome browser.
Figure 1.
Screen shot of the TriTrypDB home page. (A) Interactive banner present on all TriTrypDB web pages, including quick search windows and a tool bar (grey). (B) Side bar components contain expandable sections for release notes, community resources, tutorials and help (new items are highlighted with a yellow alert). (C) Gene searches; clicking on ‘+’ symbols reveals a list of searches available within each category. (D) Searches of non-gene entities, such as ESTs and ORFs of genome sequence. (E) Links to available tools, including the genome browser (based on GBrowse), BLAST against TriTrypDB, the sequence retrieval tool and recent PubMed records pertaining to TriTrypDB organisms.
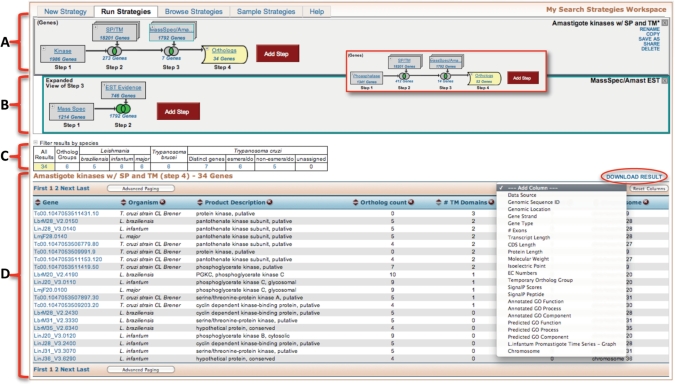
Visitors to TriTrypDB may select from approximately 80 different searches against the TriTryp genomes and datasets. Importantly, searches can be combined in an integrated and graphical manner (Figure 2A and B), and results are displayed in tabular lists below the growing search strategy (Figure 2C and D). In the example presented as Figure 2, the search strategy begins with a text search for the word kinase using either the ‘Gene Text Search’ window in the interactive banner (Figure 1A), or accessing the text search query page from ‘Identify Genes by’ section (Figure 1C). The latter provides a greater range of user options, such as defining which fields to search. All searches are also accessible by positioning the (mouse) cursor over ‘New Search’ in the banner tool bar (Figure 1A); for example, users seeking kinases may also wish to consider searching GO annotations, Interpro domains, etc. The text query presented in Figure 2 yields 1986 gene records, in any of the species supported by TriTrypDB, which contain the word kinase, and is displayed as a graphical image in the search strategy window (step 1 of Figure 2A).
Figure 2.
Screen shot of the search strategy and results summary page. (A) The expanding search strategy—a search strategy is built by adding steps, which constitute a search combined with the previous step using Boolean operators (intersect, union and minus). Any step in a strategy may be revised, deleted or expanded—the insert in the red box shows the effect of revising the first step in the strategy in A. (B) An example of a nested strategy—the search feeding into Step 3 in (A) was expanded to include other searches without the need to re-run the entire strategy. (C) The filter table, which represents a summary of results in all species represented in TriTrypDB, provides a bird’s-eye view of all results and allows quick access to those results by simply clicking on the cells in the table. (D) Tabular representation of results [highlighted in yellow in the strategy in (A) and table in (C)—this table is interactive allowing the addition, deletion and reordering of columns]. Results of searches may be downloaded by clicking on the download results link (red circle).
The user may be satisfied with this result, or may wish to revise or combine it with other searches: for example, how many of these kinases are predicted to be secreted? Clicking on the ‘add step’ button opens a pop-up window containing all available searches, from which the user can select the ‘cellular location’ query ‘predicted signal peptide’ (data not shown), specify appropriate parameters (or accept the defaults) and select how to combine this search with the previous one in the strategy (i.e. which Boolean operator to use: intersection, union or minus). Intersecting genes predicted to contain a signal peptide with those containing the word kinase are displayed, along with a Venn diagram representing how these searches were combined, in step 2 (Figure 2A). This strategy can be further expanded by asking which of these results is supported by proteomics evidence in general or by proteomics evidence from specific parasite life cycle stages as shown in Figure 2 (Step 3).
This search yields a limited number of hits, for several reasons, including the limited amount of functional genomics evidence available. The search can readily be expanded, however, as indicated in the main body of Figure 2. For example, the signal peptide search can be expanded to consider genes that contain a signal peptide and/or a transmembrane domain (as it is not clear that all secreted proteins are properly annotated and accurately recognized by SignalP). Similarly, expression data can be expanded by considering genes with either proteomics evidence or EST support (this information appears as a nested ‘sub-strategy’ 3 in Figure 2B). The ability to apply an ortholog transform on any search result (i.e. identify orthologs for a set of genes), based on orthologs identified by OrthoMCL (http://orthomcl.org) (16), provides another powerful method for expanding searches. In the strategy shown, an ortholog transform identifies any secreted kinetoplastid parasite gene for which expression evidence is available for an ortholog in any other member of the kinetoplastida (Step 3, Figure 2A).
All steps, in all searches, may be revised, renamed, transformed, deleted or expanded as nested sub-strategies. Nested strategies allow a user to expand a specific step as a separate branch of the strategy. For example, revising the first step in Figure 1 to substitute phosphatase for kinase results in changes which are propagated through all subsequent steps (red inset).
The results of a search strategy are summarized by species (filter table) (Figure 2C), and displayed as an interactive gene list (Figure 2D). The filter table provides a bird’s-eye view of results of a search across all species in TriTypDB, and allows the user to click on any results in this table to display a particular species in the gene list shown below. Similarly, clicking on the results shown in any of the graphical icons in Figure 2A and B changes the table and gene list (Figure 2C and D). The gene list may also be modified, by adding, deleting or moving columns (by dragging them to the preferred position). Finally, all results can also be downloaded by clicking on the ‘download results’ link.
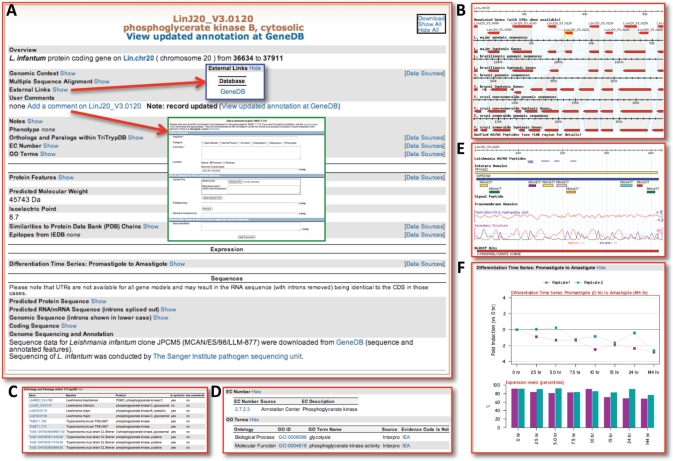
Viewing a gene page can be achieved by either entering a specific gene ID in the ID search window in the interactive banner (Figure 1A), or the ID search query (Figure 1C) or by clicking on the gene ID in the results table of a search strategy (Figure 2D). The gene page (Figure 3A) contains all available information for a gene displayed on a single page, including synteny maps (Figure 3B), information on orthologs and paralogs (Figure 3C), EC (enzyme commission) numbers and genome ontology associations (Figure 3D), proteomics (Figure 3E), microarray and EST data (Figure 3F) and the actual sequence (amino acid and nucleotide) of the displayed gene (data not shown). In addition, links to User Comments (and access to the gene-specific comment form; green insert in Figure 3A) and linkouts to the gene record on GeneDB (blue insert in Figure 3A) are available through the gene page. Similarly, GeneDB provides links to appropriate records in TriTrypDB.
Figure 3.
Screen shot of a gene record page in TriTrypDB. (A) The gene page, with all data ‘hidden’; any available data type can be viewed by clicking on ‘show’. Display preferences (and prior queries) are saved for registered users. (B) Genomic context view, showing SynView (19) synteny map between organisms supported in TriTrypDB. (C) Orthologs and paralogs table. (D) Tables representing EC (enzyme commission) numbers and GO (Genome Ontology) associations. (E) Protein features, including mapped peptides from proteomics experiments, InterPro domain predictions, hydropathy plots and BlastP results. (F) Evidence of transcript expression, from microarray experiments.
CURATION
We have implemented a synergistic approach to annotation, integrating the staff and expertise available as part of the GeneDB and EuPathDB projects with invaluable help from the broader scientific community. Annotators and curators at three sites [Seattle Biomedical Research Institute (USA), University of Georgia, Athens (USA) and the Wellcome Trust Sanger Institute (UK)] are all able to remotely curate, using Virtual Private Network connections to the Gene Builder interface of the Artemis annotation tool (17), which reads and writes directly to a Chado relational database at GeneDB (18). Curation currently focuses on preparing new sequence releases, updates to gene structure and function annotation and mutant phenotype annotations. This process is aided by providing members of the trypanosomatid research community the ability to directly add annotations to genes in TriTrypDB in the form of User Comments (green insert in Figure 3A). Comments made by scientific community members are forwarded to the annotators, in addition to immediately appearing on the gene record page in TriTrypDB. The comment form is designed to allow the user to input information into structured fields helpful to an annotator (in essence an expert opinion to guide the annotator), such as synonyms, experimentally-validated gene coordinates, gene product functional characterization, PubMed IDs, GenBank accession numbers and related genes (whereby the comment is replicated on related gene pages). We have found this to be a valuable forum for community input into TriTrypDB, of considerable use to curators working to improve gene annotations. Updates, committed to the GeneDB Chado database, set a flag on the relevant page in TriTrypDB, alerting users to updated annotations, a process mediated by web-services. These changes are propagated to TriTrypDB as part of the subsequent data update and release. To date, this collaborative effort has yielded modifications of 1159 gene records, including changes to the gene product name, gene structure, addition of untranslated regions and the addition of functional information and PubMed citations.
FUTURE DIRECTIONS
TriTrypDB will continue to expand both in functionality and data content over the coming years. Data types for which we anticipate storing and providing a query interface include expression data from proteomics experiments and transcriptome analysis (by microarrays and RNA-Seq); DNA-binding data (ChIP-chip and ChIP-seq); metabolomic data and metabolic pathway reconstructions; new genome sequences and annotation, including genome variation data; and re-assemblies of current genome sequences.
FUNDING
Grant from the Bill & Melinda Gates Foundation to develop a TriTrypDB component of the EuPathDB project (Grant Number 50097 to D.S.R., J.C.K., C.J.S. and P.J.M.); Wellcome Trust (Grant Number WT085822MA to M.C., D.F.S., M.B. and D.S.R.); Core funding from of the Wellcome Trust Sanger Institute by the Wellcome Trust (Grant Number WT085775/Z/08/Z). EuPathDB, which provides the infrastructure upon which TriTrypDB was constructed, is funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN266200400037C (to D.S.R., C.J.S. and J.C.K.). Funding for open access charge: Bill and Melinda Foundation (Grant Number 50096) and The Wellcome Trust (Grant Number WT085775).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to acknowledge the contribution of numerous members of the trypanosomatid research community, in the form of advice, suggestions and/or data—often made available to the community via TriTrypDB in advance to publication.
REFERENCES
- 1.Cox FE. History of sleeping sickness (African trypanosomiasis) Infect. Dis. Clin. North Am. 2004;18:231–245. doi: 10.1016/j.idc.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 2.The World Health Organization. Human African trypanosomiasis (sleeping sickness): epidemiological update. Wkly. Epidemiol. Rec. 2006;81:71–80. [PubMed] [Google Scholar]
- 3.Simarro PP, Jannin J, Cattand P. Eliminating human African trypanosomiasis: where do we stand and what comes next? PLoS Med. 2008;5:e55. doi: 10.1371/journal.pmed.0050055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hotez PJ, Bottazzi ME, Franco-Paredes C, Ault SK, Periago MR. The neglected tropical diseases of Latin America and the Caribbean: a review of disease burden and distribution and a roadmap for control and elimination. PLoS Negl. Trop. Dis. 2008;2:e300. doi: 10.1371/journal.pntd.0000300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Geneva: World Health Organization; 2009. [(July 2009, last date accessed)]. Leishmaniasis: magnitude of the problem. http://www.who.int/leishmaniasis/burden/magnitude/burden_magnitude/en/index.html. [Google Scholar]
- 6.Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, Renauld H, Bartholomeu DC, Lennard NJ, Caler E, Hamlin NE, Haas B, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 7.El-Sayed NM, Myler PJ, Bartholomeu DC, Nilsson D, Aggarwal G, Tran AN, Ghedin E, Worthey EA, Delcher AL, Blandin G, et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science. 2005;309:409–415. doi: 10.1126/science.1112631. [DOI] [PubMed] [Google Scholar]
- 8.Ivens AC, Peacock CS, Worthey EA, Murphy L, Aggarwal G, Berriman M, Sisk E, Rajandream MA, Adlem E, Aert R, et al. The genome of the kinetoplastid parasite, Leishmania major. Science. 2005;309:436–442. doi: 10.1126/science.1112680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peacock CS, Seeger K, Harris D, Murphy L, Ruiz JC, Quail MA, Peters N, Adlem E, Tivey A, Aslett M, et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat. Genet. 2007;39:839–847. doi: 10.1038/ng2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertz-Fowler C, Peacock CS, Wood V, Aslett M, Kerhornou A, Mooney P, Tivey A, Berriman M, Hall N, Rutherford K, et al. GeneDB: a resource for prokaryotic and eukaryotic organisms. Nucleic Acids Res. 2004;32:D339–D343. doi: 10.1093/nar/gkh007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aurrecoechea C, Heiges M, Wang H, Wang Z, Fischer S, Rhodes P, Miller J, Kraemer E, Stoeckert CJ, Jr, Roos DS, et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35:D427–D430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weatherly DB, Boehlke C, Tarleton RL. Chromosome level assembly of the hybrid Trypanosoma cruzi genome. BMC Genomics. 2009;10:255. doi: 10.1186/1471-2164-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Panigrahi AK, Ogata Y, Zikova A, Anupama A, Dalley RA, Acestor N, Myler PJ, Stuart KD. A comprehensive analysis of Trypanosoma brucei mitochondrial proteome. Proteomics. 2009;9:434–450. doi: 10.1002/pmic.200800477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Atwood JA, 3rd, Weatherly DB, Minning TA, Bundy B, Cavola C, Opperdoes FR, Orlando R, Tarleton RL. The Trypanosoma cruzi proteome. Science. 2005;309:473–476. doi: 10.1126/science.1110289. [DOI] [PubMed] [Google Scholar]
- 15.Rosenzweig D, Smith D, Myler PJ, Olafson RW, Zilberstein D. Post-translational modification of cellular proteins during Leishmania donovani differentiation. Proteomics. 2008;8:1843–1850. doi: 10.1002/pmic.200701043. [DOI] [PubMed] [Google Scholar]
- 16.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carver T, Berriman M, Tivey A, Patel C, Bohme U, Barrell BG, Parkhill J, Rajandream MA. Artemis and ACT: viewing, annotating and comparing sequences stored in a relational database. Bioinformatics. 2008;24:2672–2676. doi: 10.1093/bioinformatics/btn529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mungall CJ, Emmert DB. A Chado case study: an ontology-based modular schema for representing genome-associated biological information. Bioinformatics. 2007;23:i337–i346. doi: 10.1093/bioinformatics/btm189. [DOI] [PubMed] [Google Scholar]
- 19.Wang H, Su Y, Mackey AJ, Kraemer ET, Kissinger JC. SynView: a GBrowse-compatible approach to visualizing comparative genome data. Bioinformatics. 2006;22:2308–2309. doi: 10.1093/bioinformatics/btl389. [DOI] [PubMed] [Google Scholar]