Abstract
MouseIndelDB is an integrated database resource containing thousands of previously unreported mouse genomic indel (insertion and deletion) polymorphisms ranging from ∼100 nt to 10 Kb in size. The database currently includes polymorphisms identified from our alignment of 26 million whole-genome shotgun sequence traces from four laboratory mouse strains mapped against the reference C57BL/6J genome using GMAP. They can be queried on a local level by chromosomal coordinates, nearby gene names or other genomic feature identifiers, or in bulk format using categories including mouse strain(s), class of polymorphism(s) and chromosome number. The results of such queries are presented either as a custom track on the UCSC mouse genome browser or in tabular format. We anticipate that the MouseIndelDB database will be widely useful for research in mammalian genetics, genomics, and evolutionary biology. Access to the MouseIndelDB database is freely available at: http://variation.osu.edu/.
INTRODUCTION
An ultimate goal of genetics research is to link phenotypic differences with different genomic variants, and vice versa. Hundreds of distinct mouse strains are characterized by wide-ranging functional differences. This extensive phenotypic variation has helped to make the mouse a premier model organism, mimicking many aspects of human diversity and diseases. Understanding the genomic differences that distinguish different mouse strains and species will improve the usefulness of different mouse lineages as model organisms, facilitate further evolutionary analysis of ancestral relationships for mouse species and strains and shed new light on the genetic basis for variation among human individuals and in human diseases (1,2).
Recently, much attention has been given to the types of variation that exist within or between mammalian species (3–5), particularly short variations such as single nucleotide polymorphisms (SNPs) (6,7). Identification and analysis of such variants has been accomplished by many groups, as exemplified by the HapMap project compiling human data (8). These studies have helped to facilitate the recent discovery of genes associated with certain diseases by genome-wide linkage analyses. In addition to SNPs, insertion/deletion (indel) polymorphisms are another important form of variation (9–15). Indels are comprised of blocks of nucleotides that are present in one individual, strain or lineage, but absent at the orthologous locus in another. In addition to being useful in genotyping studies, indel polymorphisms can have direct functional consequences. As they are longer than SNPs, and may introduce or alter promoters, terminators, alternative splice sites and/or other determinants of transcriptional variation (16–19), indel polymorphisms could contribute significantly both to differences in gene structure and expression, and to various disease processes. In addition to indel polymorphisms, other important forms of structural variation, including copy number variants and polymorphic segmental duplications, also have been studied extensively (5,20–22).
A rich potential source of information about genomic variation exists in unassembled, conventional whole-genome shotgun (WGS) sequence traces obtained from different individuals within or between species. Recently, such traces have been used to identify human SNPs (23,24) and simple tandem repeat (STR) and short indel polymorphisms (10,11,25), as tools to identify such polymorphisms from sequence traces have been developed (26). To identify intermediate length (101–10 000 nt) indels distinguishing between mouse lineages, we recently aligned ∼26 million WGS traces from four unassembled mouse strains to the C57BL/6J reference genome assembly (19,27). Most such mouse indels of this intermediate length range are made up of repetitive elements. An overwhelming majority of such polymorphisms appears to have resulted from endogenous retrotransposon integration events (19), which is clearly distinct from human indels (12,25,28,29).
There are now several genome browsers and databases available which provide data on SNPs, STRs and other forms of variation (23,24,30–33). These browsers are mostly focused on human variants, although other species including mouse have been developed (34). Other databases tabulate forms of structural variation that distinguish human individuals or populations, including polymorphic transposon integrants and other indels in humans, but in some cases lack contextual information about neighboring genomic features (25,35,36). By contrast, MouseIndelDB is an integrated searchable database that presents high-resolution information about indel polymorphisms that distinguish inbred mouse strains. Through their presentation as a custom track on the UCSC mouse genome browser, these mouse indel data now can be visualized easily in the context of many other important and regularly updated genomic features including annotated genes and expressed sequence tags, CpG islands, other variants, including SNP polymorphisms, and conserved regions (37). These data are freely available for user-initiated queries, either focused upon local features or in categories according to mouse strain, class of indel polymorphism, and chromosome number, at: http://variation.osu.edu/. Included in this report is an example of indel polymorphism data found in MouseIndelDB that was used to screen for a nearby, linked recombinant gene trap cassette, thereby illustrating how a new genotyping assay to distinguish between inbred mouse strains can be developed using MouseIndelDB.
DATABASE DEVELOPMENT
Data sources and software
Conventional WGS sequence traces from four unassembled mouse strains (A/J, DBA/2J, 129S1/SvImJ and 129X1/SvJ), generated at Celera, were downloaded from the National Center for Biotechnology Information (NCBI) trace archive database (4,19). After removing sequence traces containing <300 bases of quality >Q20, ∼26 million raw traces with an average length of 800 nt remained. Thus, a total of ∼18 billion nucleotides were available for alignment to the C57 reference genome. Genomic sequences for the C57 reference mouse genome (release 36.1/mm8, Mar. 2006) were downloaded from NCBI (27). MySql v5.05 was used for all relational tables. RepeatMasker output from the mouse reference genome was downloaded from the UCSC website, and RepeatMasker Open-3.0 was downloaded from http://www.repeatmasker.org/ (38).
Sequence trace alignments
We previously aligned inbred laboratory mouse WGS sequence traces to the mm8 mouse reference genome (Supplementary Figure S1) using GMAP (39) and in some cases Blat as described below (40). A custom Perl script was used to categorize them (Supplementary Figure S2) (19). Further details are available from the authors upon request. Weakly aligned traces were set aside, including those with shorter anchor alignment lengths or lower identities (Supplementary Figure S2).
Our analysis resulted in the identification of two distinct categories of indel polymorphisms. In the first group, the aligned sequence traces identified polymorphic insertions that are present in the reference C57 genome but absent from at least one of the unassembled strains’ genomes (Supplementary Figures S1 and S2) (19). In this category, the WGS traces aligned to the reference genome with >90% identity and >200 nt anchoring sequence at each end (both 5′ and 3′), where the inserted sequence length is of intermediate size, i.e. between ∼100 nt and 10 Kb (19).
The second group of WGS sequence traces (∼8% of the total) aligned well, but only at one end. We found that a large majority of these traces contain poor quality sequences at the unaligned end. A small number of traces that align well only at one end identify a polymorphic insertion present in the unassembled genome, but absent from the C57 reference genome. These sequence traces were filtered into strong and weak alignment groups based on their alignment scores and other criteria (‘polymorphism in strain X’, Supplementary Figures S1 and S2). Since we previously found that most polymorphic integrants present in the C57 reference genome are caused by endogenous retrotransposition by LINE (L1), SINE and ERV-K LTR retrotransposons, with L1 variants found most frequently (19), we used RepeatMasker (38) to identify mouse L1 retrotransposon sequences within such sequence traces (Supplementary Figure S2). This approach is comparable to a recently published strategy (41). Those repeat sequences contained within the traces were then masked, while the remaining, nonrepetitive sequences were re-aligned to the reference genome using Blat (40). Resulting alignments were used to categorize and map portions of polymorphic L1 integrants present in the unknown strains but absent from the reference genome at orthologous loci.
Resulting information about each trace in these two groups, including their categorization and their mapped chromosomal coordinates (mm8), was loaded into relational databases (Mysql v. 5.05). We used the UCSC ‘liftOver’ tool to map these indel traces to the mm9 mouse reference genome (42).
DATABASE CONTENT AND WEB INTERFACE
Overview of MouseIndelDB content
A total of 12 951 unique insertions between 101 nt and 10 Kb were identified in the C57 reference genome but absent from at least one of the other four mouse strains studied (Table 1). Most of these reference genome insertions are repetitive elements, particularly retrotransposon integrants (19), while the rest are simple repeats. In many cases, individual insertional polymorphisms were identified by more than one aligned WGS sequence trace, so they were clustered into unique integrants (19). An additional 9193 previously unreported L1 retrotransposon insertions, present in at least one of the four unassembled mouse strain genomes but absent from the C57 reference genome, have been incorporated into the MouseIndelDB database. These indels were identified by a total of 37 500 WGS sequence traces.
Table 1.
Mapped sequence traces and unique polymorphic loci currently in MouseIndelDB
| Genotype | RepeatMasker | No. of loci | No. of traces | |
|---|---|---|---|---|
| Insert in alt-strain | LINE | 9193 | 14 025 | |
| Insert in C57 ref. | LINE | 5564 | 9394 | |
| SINE | 2912 | 6193 | ||
| LTR | 3314 | 6363 | ||
| Simple repeat | 1161 | 1525 |
User queries
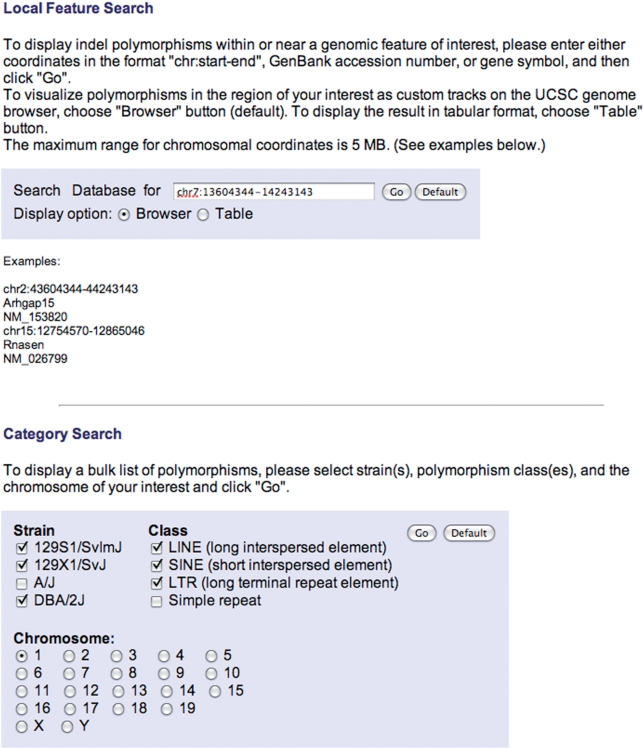
Users can initiate queries of the mouse indel polymorphisms presented in MouseIndelDB, using two query modes available at the home page at: http://variation.osu.edu/ (Figure 1). Users can alternatively focus upon local features of interest, or search the database in categories according to mouse strain(s), class of indel polymorphism(s) and chromosome number. For local feature queries, users can optionally enter a GenBank accession number, gene symbol or chromosomal coordinates in the format ‘chr:start-end’. The maximal range for chromosomal coordinates is 5 MB. Examples of these inputs are provided on the home page (Figure 1). Users can choose to display outputs via a custom track at the UCSC mouse genome browser, or in a table (see below). A choice is provided to search for polymorphisms mapped to the mm8 mouse genome assembly or to the more recent mm9 assembly (July 2007). For category queries, users can choose one or more of the mouse strains 129S1/SvImJ, 129X1/SvJ, A/J and DBA/2J, one or more of the polymorphic elements, including L1 retrotransposons, SINEs, LTR retrotransposons and simple repeats, and a chromosome number. These category searches result in tabular output.
Figure 1.
MouseIndelDB user interface. Two query modes are available at the home page of the MouseIndelDB web interface. Users can alternatively focus upon local features of interest, or search the database in categories according to mouse strain(s), class of indel polymorphism(s) and chromosome number. Default entries and other examples of optional user inputs are provided.
Custom track at UCSC mouse genome browser
We implemented a custom track at the UCSC mouse genome browser (43) to display content of the MouseIndelDB database in the context of other annotated genomic features presented alternatively according to the mm8 and mm9 reference mouse genome assemblies. In each case, a temporary Browser Extensible Data (BED) file containing indel polymorphisms up to 500 Kb upstream and 500 Kb downstream of a specified chromosomal locus is uploaded to the UCSC genome browser website.
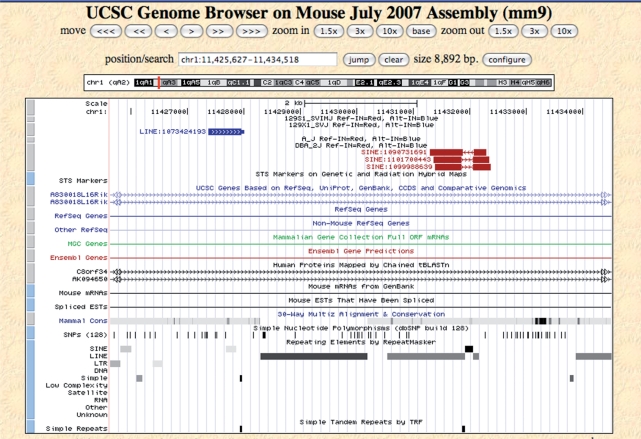
A screen-shot of the MouseIndelDB custom track on the UCSC browser is presented in Figure 2. Examples of intermediate-sized indel variants (100 nt–10 Kb) are presented here, including three WGS sequence traces from DBA/2J mice that skip over a single polymorphic SINE retrotransposon present in the reference genome but absent from DBA, while a nearby mapped sequence trace from the 129X1 strain indicates an insertional polymorphism present in that strain but absent from the reference C57 genome. Polymorphisms are color-coded, as red indels indicate integrants present in the reference genome (Ref-IN), while blue indels indicate those present in an alternative strain (Alt-IN). In cases where a polymorphic integrant is present in an alternative strain (Alt-IN, Figure 2), we also added a 50-nt thin projection on one side or the other of anchored sequence traces to indicate its genomic junction and relative position. Following conventions established on the host browser, users can also click on each feature for additional information including primary sequences and WGS trace alignment information, and can scale the chromosomal region displayed up to a limit of 500 Kb upstream and 500 Kb downstream of the original locus while depicting all indels in this region.
Figure 2.
MouseIndelDB custom track at UCSC mouse genome browser. Polymorphic SINE and L1 retrotransposons at a region of mouse chromosome 1 are shown on the MouseIndelDB custom track displayed at the UCSC mouse genome browser (mm9 assembly). Three WGS sequence traces from DBA/2J mice each skip over a single polymorphic SINE retrotransposon, present in the reference (designated as REF-IN) genome but absent from DBA (top, labeled as REF-IN, red rectangles, trace IDs 1090731691, 1101700443 and 109988639). The reference SINE element can be seen on the conventional RepeatMasker track (bottom). In addition, a nearby mapped sequence trace from the 129X1 strain indicates an insertional polymorphism present in that alternative (ALT-IN) strain but absent from the reference C57 genome (blue rectangle, labeled as ALT-IN, trace ID 1073424193). In the latter case, a 50-nt thin projection (thin blue rectangle, right) indicates the genomic junction and relative position of the previously unreported L1 retrotransposon integrant in the 129X1 genome. Superimposed arrows indicate the direction of sequence trace alignments compared with the reference genome.
Tabular display
Based on chromosomal coordinates entered by the user, a list of polymorphisms can be retrieved from the MouseIndelDB database. The range of chromosomal coordinates that can be queried is limited to 5 MB. Each aligned sequence trace is linked to the sequence trace ID, providing additional alignment data and a link to the indel polymorphism custom track at UCSC.
DISCUSSION AND FUTURE PLANS
Our goal in developing the MouseIndelDB database and web interface has been to identify and provide detailed access to tens of thousands of indel polymorphisms that distinguish mouse strains. The data can be queried either according to local features or in a bulk, category mode. The resulting data have been linked to a custom track at the UCSC mouse genome browser, facilitating the visualization of previously unreported indel polymorphisms in the context of other annotated features available with the mm8 and mm9 reference mouse genome assemblies. Resulting data can be downloaded in tabular format, and large data sets will be made available to users upon request.
In developing this database, we focused on mouse strains and subspecies, since to our knowledge no integrated indel polymorphism database has been described previously for mouse strains, and since millions of high-quality WGS sequence traces are available for alignment to the reference C57 genome. As several hundred distinct mouse strains have many distinct phenotypes including behavioral differences, predisposition to many different diseases and cancers, and other quantifiable characteristics (1), we expect that MouseIndelDB will prove useful in genetic and evolutionary studies addressing various forms of variation (including but not merely limited to SNPs) within and between the strains.
To highlight how the MouseIndelDB database can be queried for indel polymorphisms near a local feature of interest, we studied variants closely linked to a transgene insertion in the Sumo1 locus (Supplementary Data and Supplementary Figure S3).
We and others currently are generating additional mouse genome sequence data from other currently unsequenced strains and murine species. We plan to update MouseIndelDB frequently to include more information about various forms of polymorphisms as they become available. In particular, we plan to add more information about STR polymorphisms distinguishing between mouse lineages as it becomes available. In addition, we now are studying how some classes of indel polymorphisms are related to transcriptional variation in different strains, tissues, developmental time points, etc. Resulting novel fusion transcript data also will be incorporated together with these genomic variants in additional tracks and data available via future versions of MouseIndelDB. Using information from the Mouse Genome Database and related databases with phenotypic information (31), we plan to identify those genes to which strain-specific phenotypes have been mapped, to facilitate correlations between the various types of genomic polymorphisms available in MouseIndelDB and such variable phenotypes. Through a merging method similar to that used to consolidate overlapping indel traces (Supplementary Figures S1 and S2), we also plan to flag those short indels, SNPs and other genomic variants represented in multiple WGS sequence traces to add an evidence statistic to them. We also plan to make our polymorphism data collection available directly through on-demand tracks at the UCSC mouse genome browser (http://genome.ucsc.edu/) and through the Mouse Genome Database website at the Jackson Laboratory (http://www.informatics.jax.org/) (34).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Cancer Institute, National Institutes of Health (under contract N01-CO-12400); the Intramural Research Program, Center for Cancer Research, National Cancer Institute, National Institutes of Health; and The Ohio State University Comprehensive Cancer Center. Funding for open access charge: The Ohio State University Comprehensive Cancer Center.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Thomas Wu (Genentech) for help applying GMAP to trace analysis and program modifications, Richard Frederickson (SAIC Frederick) for preparing figures, and David Bryant (ABCC, SAIC Frederick) and Michael Koluder (Ohio State University Comprehensive Cancer Center) for help setting up websites. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
REFERENCES
- 1.Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat. Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- 2.Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat. Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- 3.Varki A, Altheide TK. Comparing the human and chimpanzee genomes: searching for needles in a haystack. Genome Res. 2005;15:1746–1758. doi: 10.1101/gr.3737405. [DOI] [PubMed] [Google Scholar]
- 4.Wade CM, Daly MJ. Genetic variation in laboratory mice. Nat. Genet. 2005;37:1175–1180. doi: 10.1038/ng1666. [DOI] [PubMed] [Google Scholar]
- 5.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, Carter NP, Church DM, Felsenfeld A, Guyer M, Lee C, et al. Completing the map of human genetic variation. Nature. 2007;447:161–165. doi: 10.1038/447161a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 7.Yang H, Bell TA, Churchill GA, Pardo-Manuel de Villena F. On the subspecific origin of the laboratory mouse. Nat. Genet. 2007;39:1100–1107. doi: 10.1038/ng2087. [DOI] [PubMed] [Google Scholar]
- 8.International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bhangale TR, Rieder MJ, Livingston RJ, Nickerson DA. Comprehensive identification and characterization of diallelic insertion-deletion polymorphisms in 330 human candidate genes. Hum. Mol. Genet. 2005;14:59–69. doi: 10.1093/hmg/ddi006. [DOI] [PubMed] [Google Scholar]
- 10.Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion-deletion polymorphisms. Nat. Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- 11.Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C, Pittard WS, Devine SE. An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res. 2006;16:1182–1190. doi: 10.1101/gr.4565806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kvikstad EM, Chiaromonte F, Makova KD. Ride the wavelet: a multiscale analysis of genomic contexts flanking small insertions and deletions. Genome Res. 2009;19:1153–1164. doi: 10.1101/gr.088922.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clark TG, Andrew T, Cooper GM, Margulies EH, Mullikin JC, Balding DJ. Functional constraint and small insertions and deletions in the ENCODE regions of the human genome. Genome Biol. 2007;8:R180. doi: 10.1186/gb-2007-8-9-r180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Druker R, Whitelaw E. Retrotransposon-derived elements in the mammalian genome: a potential source of disease. J. Inherit. Metab. Dis. 2004;27:319–330. doi: 10.1023/B:BOLI.0000031096.81518.66. [DOI] [PubMed] [Google Scholar]
- 17.Van de Lagemaat LN, Landery JR, Mager DL, Medstrand P. Transposable elements in mammals promote regulatory variation and diversification of genes with specialized functions. Trends Genet. 2003;19:530–536. doi: 10.1016/j.tig.2003.08.004. [DOI] [PubMed] [Google Scholar]
- 18.Belancio VP, Hedges DJ, Deininger P. LINE-1 RNA splicing and influences on mammalian gene expression. Nucleic Acids Res. 2006;34:1512–1521. doi: 10.1093/nar/gkl027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Akagi K, Li J, Stephens RM, Volfovsky N, Symer DE. Extensive variation between inbred mouse strains due to endogenous L1 retrotransposition. Genome Res. 2008;18:869–880. doi: 10.1101/gr.075770.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egan CM, Sridhar S, Wigler M, Hall IM. Recurrent DNA copy number variation in the laboratory mouse. Nat. Genet. 2007;39:1384–1389. doi: 10.1038/ng.2007.19. [DOI] [PubMed] [Google Scholar]
- 21.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.She X, Cheng Z, Zollner S, Church DM, Eichler EE. Mouse segmental duplication and copy number variation. Nat. Genet. 2008;40:909–914. doi: 10.1038/ng.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thomas DJ, Trumbower H, Kern AD, Rhead BL, Kuhn RM, Haussler D, Kent WJ. Variation resources at UC Santa Cruz. Nucleic Acids Res. 2007;35:D716–D720. doi: 10.1093/nar/gkl953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum. Mutat. 2006;27:323–329. doi: 10.1002/humu.20307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manaster C, Zheng W, Teuber M, Wachter S, Doring F, Schreiber S, Hampe J. InSNP: a tool for automated detection and visualization of SNPs and InDels. Hum. Mutat. 2005;26:11–19. doi: 10.1002/humu.20188. [DOI] [PubMed] [Google Scholar]
- 27.Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- 28.Bennett EA, Coleman LE, Tsui C, Pittard WS, Devine SE. Natural genetic variation caused by transposable elements in humans. Genetics. 2004;168:933–951. doi: 10.1534/genetics.104.031757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maksakova IA, Romanish MT, Gagnier L, Dunn CA, van de Lagemaat LN, Mager DL. Retroviral elements and their hosts: insertional mutagenesis in the mouse germ line. PLoS Genet. 2006;2:e2. doi: 10.1371/journal.pgen.0020002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hubbard TJ, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE Mouse Genome Database Group. The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35:D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gelfand Y, Rodriguez A, Benson G. TRDB – the Tandem Repeats Database. Nucleic Acids Res. 2007;35:D80–D87. doi: 10.1093/nar/gkl1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Agrafioti I, Stumpf MP. SNPSTR: a database of compound microsatellite-SNP markers. Nucleic Acids Res. 2007;35:D71–D75. doi: 10.1093/nar/gkl806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. Detection of large-scale variation in the human genome. Nat. Genet. 2004;36:949–951. doi: 10.1038/ng1416. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Feuk L, Duggan GE, Khaja R, Scherer SW. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 2006;115:205–214. doi: 10.1159/000095916. [DOI] [PubMed] [Google Scholar]
- 37.Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE, Rosenbloom KR, Rhead B, Raney BJ, Pohl A, Pheasant M, et al. The UCSC Genome Browser Database: update 2009. Nucleic Acids Res. 2009;37:D755–D761. doi: 10.1093/nar/gkn875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smit AFA, Hubley R, Green P. 2009. RepeatMasker Open-3.0. ( http://www.repeatmasker.org/) [Google Scholar]
- 39.Wu TD, Watanabe CK. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 40.Kent WJ. BLAT – the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Y, Maksakova IA, Gagnier L, van de Lagemaat LN, Mager DL. Genome-wide assessments reveal extremely high levels of polymorphism of two active families of mouse endogenous retroviral elements. PLoS Genet. 2008;4:e1000007. doi: 10.1371/journal.pgen.1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hinrichs AS, Karolchik D, Baertsch R, Barber GP, Bejerano G, Clawson H, Diekhans M, Furey TS, Harte RA, Hsu F, et al. The UCSC Genome Browser Database: update 2006. Nucleic Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Akagi K, Suzuki T, Stephens RM, Jenkins NA, Copeland NG. RTCGD: retroviral tagged cancer gene database. Nucleic Acids Res. 2004;32:D523–D527. doi: 10.1093/nar/gkh013. [DOI] [PMC free article] [PubMed] [Google Scholar]