Abstract
The Aspergillus Genome Database (AspGD) is an online genomics resource for researchers studying the genetics and molecular biology of the Aspergilli. AspGD combines high-quality manual curation of the experimental scientific literature examining the genetics and molecular biology of Aspergilli, cutting-edge comparative genomics approaches to iteratively refine and improve structural gene annotations across multiple Aspergillus species, and web-based research tools for accessing and exploring the data. All of these data are freely available at http://www.aspgd.org. We welcome feedback from users and the research community at aspergillus-curator@genome.stanford.edu.
INTRODUCTION
The Aspergilli are a diverse group of fungal micro-organisms, comprising, among many other species, Aspergillus nidulans (teleomorph Emericella nidulans), a well-studied eukaryotic model organism; A. fumigatus, a deadly pathogen of immunocompromised patients; A. flavus, an agriculturally important toxin producer; and A. niger and A. oryzae, two species used in industrial processes. Diverse Aspergillus species are not only important research subjects in their own right, but they also collectively offer an opportunity to utilize comparative genomics approaches to gain insights into the genetics of the traits that allow them to inhabit diverse ecological niches and to have significant economic and human impact across industrial, agricultural and medical realms. The availability of genome sequences for several species provides new avenues for the investigation of these important fungi (1–3).
The primary mission of the Aspergillus Genome Database (AspGD) is to serve the needs of the scientific community in order to facilitate and accelerate Aspergillus research in the laboratory. To accomplish this, we provide an extensively curated data set of Aspergillus gene, protein and sequence information and easy-to-use web-based tools for accessing, analyzing and exploring these data. AspGD is based on the framework of the Saccharomyces Genome Database (SGD) and the Candida Genome Database (CGD), so the interface is already familiar to many users within the fungal research community. Initially, we are focusing on the curation of genomic information for A. nidulans, the best-characterized species of the group. In the future, we will add information for other Aspergillus species (A. fumigatus, A. flavus, A. oryzae, A. niger, A. clavatus, A. terreus and Neosartorya fischeri/A. fischerianus). We are also working to refine and optimize a genome annotation pipeline, which will be used to leverage comparative data across all incorporated genomes in order to iteratively improve gene boundary annotations.
At present, several other online resources also exist for multiple Aspergillus genomes: the Fungal Research Trust’s Aspergillus website (4), which includes the Central Aspergillus Data Repository (CADRE) database and clinical and patient-oriented information; the Aspergillus genomes site at the Broad Institute (3); and websites that focus on sequencing projects of one or several Aspergillus species. AspGD links to these resources and seeks to complement them by offering in-depth manual curation of the primary scientific literature and by continuously reviewing and improving the sequence annotation via iterative comparative analyses.
All of the data in AspGD, described in more detail subsequently, are freely available. We also have an extensive suite of online user documentation, and provide advice and user support by e-mail at aspergillus-curator@genome.stanford.edu.
Aspergillus nidulans GENE INFORMATION IN AspGD
Initially, we are concentrating our curation efforts on the experimental literature about A. nidulans, because it serves as a genetic model for the other Aspergilli and is the best represented member of the genus in scientific publications. Since the inception of the project in early 2009, we have entered 10 545 predicted protein-coding genes into AspGD and have predicted over 9900 Gene Ontology (GO) (5) annotations using orthology mappings between this gene set and experimentally characterized genes of Saccharomyces cerevisiae. We have also begun manual curation of gene descriptions, gene product functions and localization, mutant phenotypes and comprehensive reference lists from the A. nidulans literature. The curation of the entire body of literature is a large and ongoing endeavor, and we welcome suggestions from users as to papers that should be prioritized or other data that should be included.
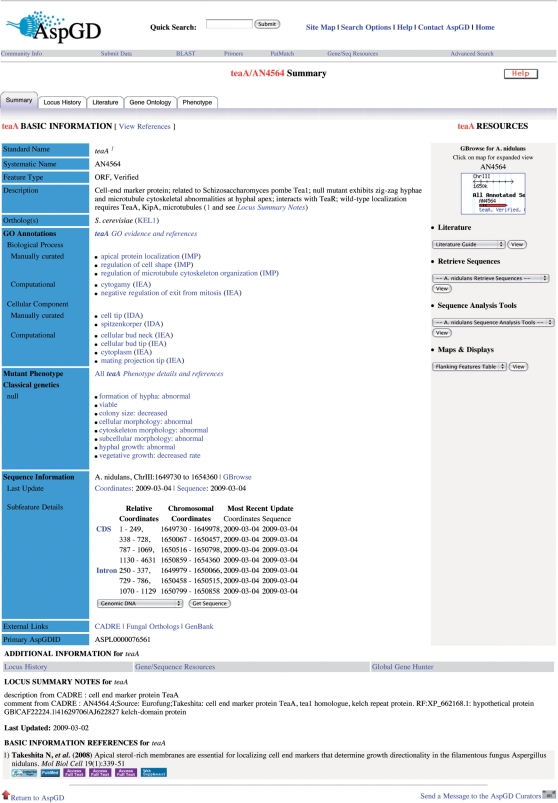
Each gene has a Locus Summary page (Figure 1), the basic organizing principle of AspGD, which contains basic information that describes the gene and provides access to tools for retrieval, analysis and visualization of data. Further links lead to pages containing additional information about the gene.
Figure 1.
Locus Summary page. The Locus Summary page is the hub around which all of the AspGD gene information is organized. This example shows the classes of data that are summarized on the page: names and aliases, the gene description, orthologs and best hits, GO annotations, phenotypes, sequence information, community annotation displayed in the locus summary notes section and the references from which the names and description have been curated. Tabs and hyperlinks access additional information and details, including the Locus History page, which explains any sequence and annotation changes affecting the gene; the Literature Guide page, which has a comprehensive listing of references that pertain to the gene; the full set of GO and phenotype annotations; and the GBrowse genome browser.
The Locus Summary page contains all of the names for each gene, including its standard genetic name (such as veA), the systematic name assigned during the genome sequence assembly and genome annotation (such as AN1052), and any other synonyms or aliases. All names and aliases are searchable, and collection of all of the aliases for each gene ensures that users can find a gene of interest even when confusion or non-standard name usage exists in the published literature.
The locus description is a free-text summary of the most important information about a gene. These summaries are written by AspGD curators, based on information collected from published literature. In the future, orthology with S. cerevisiae genes will be used to create informative descriptions for those A. nidulans genes that lack a literature-based description. References from which the descriptive information is curated are enumerated after the text of the description, and are linked to the full citations displayed at the bottom of the page. The locus summary notes in the additional information section, which is located on the lower part of the Locus Summary page, provide functional, descriptive or other information about the gene. This information was supplied to AspGD by the CADRE database, and is derived from the first update to the annotation in 2005 by The Institute for Genome Research, TIGR (now the J. Craig Venter Institute, JCVI) and the Eurofungbase community annotation effort in 2007–2008 (6).
ORTHOLOGS AND BEST HITS
Using InParanoid (7), we have generated an orthology mapping between A. nidulans and S. cerevisiae, the best characterized fungal model organism, and we provide hyperlinks on the AspGD Locus Summary pages that connect A. nidulans genes to their S. cerevisiae orthologs at SGD. The mappings and links are updated on a quarterly basis to reflect the latest gene models at AspGD and the current set of annotated genes at SGD. Aspergillus nidulans genes that do not have an S. cerevisiae ortholog are compared to the S. cerevisiae-predicted proteome using the Basic Local Alignment Search Tool (BLAST), and top matches with an E-value of 1e−5 or better are included in AspGD, labeled as ‘Best Hits’, and are linked to the corresponding pages at SGD.
Orthologs between the sequenced Aspergillus genomes were generated via a modified mutual best hit (MBH) approach, in which close paralogs are collapsed into single nodes prior to MBH clustering (8). In the near future, links will be available from the Locus Summary page to an interactive comparative visualization tool, Sybil, which allows the user to navigate ortholog clusters in their genomic context.
GO
The GO is a structured vocabulary used to describe three aspects of gene products: their molecular or catalytic activity, the broader biological role or context in which they participate, and the cellular location in which they reside (5). Because the GO is rigorously structured, it enables powerful computational approaches to analysis of genomic data sets, and is in wide use across the community of model organism databases (9–16). Each term assignment is associated with an evidence code that describes the type of data used to make the assignment, and with a reference from which the inference was made. We initially loaded a total of 2529 GO term assignments that were made during the TIGR and Eurofungbase annotation efforts for 977 unique A. nidulans genes (excluding annotations with the evidence code IEA, ‘Inferred from Electronic Annotation’). As we systematically curate the primary literature, these GO assignments are being replaced or augmented with updated annotations, and the community annotations are being archived in the Locus Summary notes section for reference.
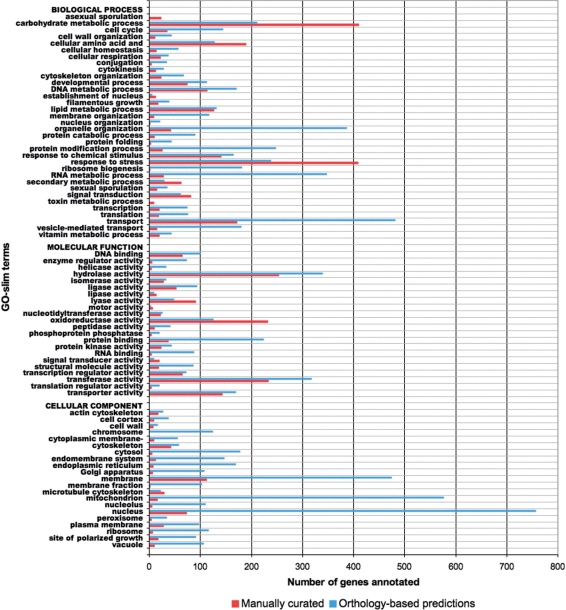
In addition to assigning annotations during manual curation, we use an automated pipeline to predict GO annotations based on experimental characterization of the S. cerevisiae orthologs of A. nidulans genes, using the procedure developed at CGD for Candida albicans genes (17). Briefly, if an A. nidulans gene has an S. cerevisiae ortholog with an experimentally based GO annotation at SGD, and that annotation is not redundant with a term already assigned to the A. nidulans gene at AspGD, the orthology-based prediction will be entered into AspGD with evidence code IEA along with a reference that describes this procedure in detail. These orthology-based predictions are updated quarterly to ensure that they remain current, reflecting the latest gene models at AspGD as well as the latest S. cerevisiae annotations at SGD. AspGD GO annotations are summarized in Figure 2.
Figure 2.
Statistics of GO term annotation for A. nidulans in AspGD. Red bars represent the number of A. nidulans genes at AspGD that are annotated to each of the selected GO slim terms based on published literature (annotated directly to each term itself, or annotated to one of its more granular child terms). Blue bars represent the number of A. nidulans genes annotated to each GO slim term based on predictions made from the annotation of orthologous, characterized S. cerevisiae genes at the SGD. As of 6 August 2009, AspGD contains a total of 14 804 GO annotations for 3297 unique genes.
All of the GO annotations for each gene, derived from in-house literature-based curation, orthology-based prediction, community curation and previous annotation efforts, are listed in brief on the Locus Summary page. The complete set of GO information for each gene, including the term, the full name of the evidence code, the entire citation for the reference and the source of the annotation (e.g. AspGD, Eurofung or TIGR), is displayed on the GO details page, which can be accessed from the Locus Summary page via the tab labeled ‘GO’ or from the hyperlink labeled ‘GO evidence and references’ in the GO summary section.
The GO annotations are also used to assign a feature type to each protein-coding gene, which is an at-a-glance indication of whether or not a particular gene has been characterized experimentally or whether all characterization is predicted by similarity. A gene with an experimentally based GO term assignment is classified as ‘Verified,’ meaning that it appears to encode an expressed, functional gene product, whereas genes without experimental characterization are classified as ‘Uncharacterized.’ The Genome Snapshot, linked from the AspGD home page, provides a genome-wide overview of feature-type assignments and the current GO annotation in AspGD.
PHENOTYPE
AspGD utilizes the phenotype curation and display system that was recently introduced at SGD (18). The mutant phenotypes curated for each gene are summarized briefly on the Locus Summary page, and the full set of phenotype information for the gene is displayed on the phenotype details page, accessible by the tab labeled ‘Phenotype’ or the ‘Phenotype details and references’ hyperlink on the Locus Summary page. A single phenotype annotation comprises an experiment-type assignment (e.g. classical genetics or large-scale survey), a description of the mutant allele, the phenotype itself and a description of the abnormality (e.g. ‘conidiation: decreased’ or ‘sporulation: absent’), associated relevant experimental details or conditions, and the reference from which the phenotype was curated. While phenotype and GO curation can overlap somewhat in the information they provide, the phenotype controlled vocabulary is designed to describe and capture the actual observations that are made, whereas the GO annotations describe conclusions or inferences about biological attributes of gene products made from the observation. For example, the curated phenotype entry may report sensitivity to cell wall-perturbing agents, whereas the GO Biological Process term that is inferred from this mutant phenotype may be ‘cell wall organization.’ The phenotype terms themselves are organized into a hierarchical structure that can be viewed using the button labeled ‘Browse phenotype terms,’ which is located near the top of any phenotype details page. The terms are hyperlinked to informational pages that display a list of all of the phenotype assignments made to that term, so it is possible to view at a glance all of the genes that share a particular mutant phenotype.
SEQUENCE INFORMATION
The chromosomal sequence coordinates and exon–intron structure of a gene are displayed on its Locus Summary page. The genomic sequence spanning the gene, coding sequence, intron sequences and translated protein sequence are available for direct retrieval from the Locus Summary page, by using the pull-down menu in the sequence information section. As updates are made to the sequence of the gene or the structural annotation of the gene model, these changes are described in detail on the gene’s Locus History page, which is accessible via the ‘Locus History’ tab near the top of the Locus Summary page or from a link near the bottom of the Locus Summary page.
COMMUNITY GENE ANNOTATION
Notes from the community gene annotation and from the annotation revision performed at TIGR, which were supplied to AspGD by the CADRE database, are displayed on the lower part of the Locus Summary page. The external links section provides access to information about the gene in other databases, and includes links to CADRE (4), to GenBank records and to the fungal ortholog clusters site at the Broad Institute (19).
USING COMPARATIVE GENOMICS TO IMPROVE MULTISPECIES ANNOTATION
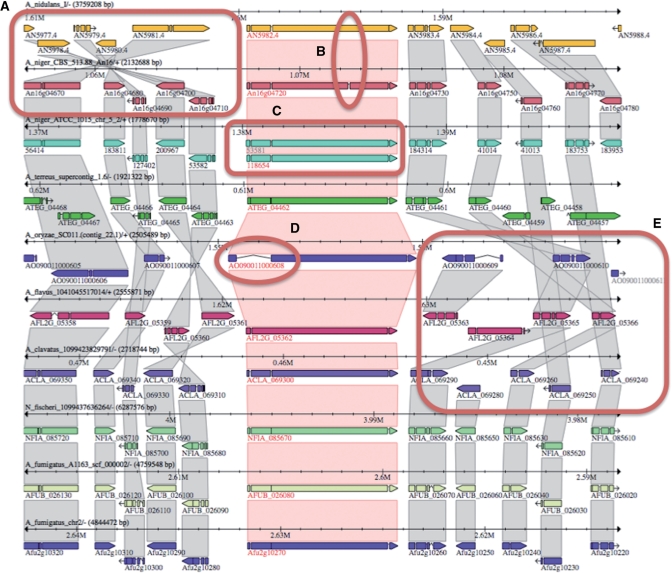
An integral and distinctive component of AspGD is the use of comparative genomics to refine the structural annotation of genes across the Aspergilli (Figure 3). Aspergillus genome sequences have been generated over an extended period of time at multiple institutes using different sequencing platforms, and have been structurally annotated using various methods. Hence, the generation of a consistent set of structural annotations using the same methodologies across species will likely improve upon prior annotation attempts and will facilitate meaningful comparisons among the genomes. For example, to find genes that may be responsible for a unique hallmark of a particular species—such as pathogenicity in an animal host, toxin generation or exceptional production of citric acid under industrial conditions—consistency of annotation is essential to ensure that differences in the gene complement reflect actual biological differences, rather than variations among different gene-calling algorithms. The annotation improvements will benefit annotations across multiple Aspergillus genomes, and will be frequently updated to leverage new data from diverse sources, including new genomic sequence, new cDNA data sets and gene model improvements that will be made as the published literature is subject to manual curation.
Figure 3.
Comparative analysis and refinement of gene models. Comparative alignments of orthologous proteins from 10 Aspergillus genomes, shown in the Sybil viewer. (A) Structural rearrangement among Aspergillus genomes (here A. nidulans and A. niger CBS 513.88 are emphasized). (B) Presence or absence of introns and different numbers of exons. (C) Gene duplications (paralogs) in one species relative to other species. (D) Intron structure, such as inordinately long introns relative to orthologs in other species. (E) Gene deletion (or creation) in syntenic blocks among species. Differences in structural annotation among species that can be illuminated with the Sybil view, thus leading to refinement of gene models, include missed gene calls, gene truncations, failure to predict small exons and incorrect intron predictions leading to spurious exons or gene merges.
TOOLS
GO tools
A ‘GO slim’ comprises a reduced set of high-level GO terms, selected from each of the three aspects of the ontology (Molecular Function, Biological Process and Cellular Component), which broadly categorize the biological attributes of a particular organism. We have generated a GO slim set for Aspergillus. The GO Slim Mapper tool categorizes a list of genes by comparison to a GO slim; by displaying shared high-level GO term categories general commonalities can become apparent. A second analysis tool that uses the GO to identify commonalities among genes on a list is GO::TermFinder (20), which provides a list of GO terms that are statistically overrepresented among their annotations for a given list of genes. Both tools have particular utility in the interpretation of large-scale experimental results. By highlighting characteristics shared by a set of genes that may be co-expressed or otherwise associated with each other in an experiment, GO Slim Mapper and GO::TermFinder can aid in interpretation of results and in formulation of hypotheses for follow-up studies.
Bulk data search and download
AspGD provides bulk search and download functionality through a Batch Download tool, and a browsable download site. The Batch Download tool allows retrieval of sequence and other information for a list of chromosomal features (e.g. protein-coding genes or tRNA genes). The AspGD download site includes files of basic gene information, sequence information, GO annotations, phenotype curation, interspecies homology mappings and other types of data. Available files are listed on the download contents page, and can also be viewed directly by browsing the download directories. Downloadable files are regenerated regularly, so data are up-to-date. We also provide custom AspGD data files upon request.
Sequence search, viewing and analysis tools
The main sequence search and retrieval tool is called Gene/Sequence Resources. The tool allows retrieval of the genomic or coding sequence of any gene with a user-defined amount of upstream and downstream flanking sequence, or protein translation of any ORF. The tool also supports individual or batch searches to obtain sequence of a region within any set of chromosomal coordinates. The query output can be obtained in several sequence formats or sent for analysis as the input to other tools at AspGD, including GBrowse, Batch Download (for information about annotated features within the region), BLAST, the computational restriction mapping tool or the primer design tool.
The BLAST tool in AspGD supports protein or nucleotide BLAST searches against A. nidulans chromosomal, genomic, coding sequence or translated ORF data sets. The BLAST links from Locus Summary pages go to a BLAST query form with the sequence already displayed in the query box, ready to submit. Alternatively, the BLAST input form may be accessed directly for submission of any user-defined query sequence. The BLAST results page displays a graphical summary as well as individual alignments, with links to the Locus Summary page, genome browser, sequence retrieval tool and the AspGD literature citation list for each ‘hit’.
The GBrowse genome browser allows a user to scan regions along the chromosome and visualize the genomic context of annotated sequence features, including nearby genes, GC content and six-frame translation (21). Each Locus Summary page links to a GBrowse view of the chromosomal region, showing the gene and the other features nearby. In the future, AspGD GBrowse will also provide tracks showing expressed sequence tags (ESTs) aligned to the genome to provide an at-a-glance summary of the experimental evidence that exists for each gene model in A. nidulans.
COMMUNITY FUNCTIONS
As a community database, AspGD’s mission encompasses a broader community service role than mere development of the gene model pipeline and curation of the experimental literature. We aim to be a unifying and positive force in the community, helping to facilitate collaboration among research groups. AspGD provides a colleague registry and Aspergillus labs page, whereby community members may share their contact information and research interests. Participation is entirely voluntary, and there are currently 195 colleagues and 37 Aspergillus labs listed in the registry. We provide web pages that list meetings, conferences, workshops and job opportunities in the field. In future, AspGD could help to promote nomenclature standards for newly published gene names, if the community wishes and provides a strong mandate for us to do so. We are working closely with the CADRE database to share annotation updates, exchange data and maintain reciprocal hyperlinks, thereby ensuring that both resources serve complementary roles in supporting the research community while reflecting the most up-to-date annotation information.
Members of the AspGD staff welcome questions and help requests by e-mail or via submissions using the ‘Contact AspGD’ form on our web site. AspGD looks forward to a productive collaboration with the Aspergillus research community as we work together to develop and provide this curated, community resource for Aspergillus genomics and molecular biology research.
FUNDING
National Institute of Allergy and Infectious Diseases at the US National Institutes of Health (R01 AI077599 to G.S. and J.R.W.). Funding for open access charge: National Institute of Allergy and Infectious Diseases at the US National Institutes of Health (R01 AI077599 to G.S. and J.R.W.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank CADRE for data exchange and support, SGD for providing their code base and the Aspergillus research community for giving us their support and the opportunity to serve them.
REFERENCES
- 1.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 2.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 3.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 4.Mabey Gilsenan JE, Atherton G, Bartholomew J, Giles PF, Attwood TK, Denning DW, Bowyer P. Aspergillus genomes and the Aspergillus cloud. Nucleic Acids Res. 2009;37:D509–D514. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, et al. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 2009;46(Suppl. 1):S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in paralogs from pairwise species comparisons. J. Mol. Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 8.Crabtree J, Angiuoli SV, Wortman JR, White OR. Sybil: methods and software for multiple genome comparison and visualization. Methods Mol. Biol. 2007;408:93–108. doi: 10.1007/978-1-59745-547-3_6. [DOI] [PubMed] [Google Scholar]
- 9.Consortium TGO. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aslett M, Wood V. Gene ontology annotation status of the fission yeast genome: preliminary coverage approaches 100% Yeast. 2006;23:913–919. doi: 10.1002/yea.1420. [DOI] [PubMed] [Google Scholar]
- 11.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong EL, Balakrishnan R, Dong Q, Christie KR, Park J, Binkley G, Costanzo MC, Dwight SS, Engel SR, Fisk DG, et al. Gene ontology annotations at SGD: new data sources and annotation methods. Nucleic Acids Res. 2008;36:D577–D581. doi: 10.1093/nar/gkm909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhee SY, Beavis W, Berardini TZ, Chen G, Dixon D, Doyle A, Garcia-Hernandez M, Huala E, Lander G, Montoya M, et al. The Arabidopsis Information Resource (TAIR): a model organism database providing a centralized, curated gateway to Arabidopsis biology, research materials and community. Nucleic Acids Res. 2003;31:224–228. doi: 10.1093/nar/gkg076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007–easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Arnaud MB, Costanzo MC, Shah P, Skrzypek MS, Sherlock G. Gene ontology and the annotation of pathogen genomes: the case of Candida albicans. Trends Microbiol. 2009;17:295–303. doi: 10.1016/j.tim.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Costanzo MC, Skrzypek MS, Nash RS, Wong ED, Binkley G, Engel SR, Hitz BC, Hong EL, Cherry JM. New mutant phenotype data curation system in the Saccharomyces Genome Database. Database. 2009;2009 doi: 10.1093/database/bap001. Article ID bap001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 20.Boyle EI, Weng S, Gollub J, Jin H, Botstein D, Cherry JM, Sherlock G. GO::TermFinder–open source software for accessing gene ontology information and finding significantly enriched gene ontology terms associated with a list of genes. Bioinformatics. 2004;20:3710–3715. doi: 10.1093/bioinformatics/bth456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]