Abstract
Over the last 2 years the Gene3D resource has been significantly improved, and is now more accurate and with a much richer interactive display via the Gene3D website (http://gene3d.biochem.ucl.ac.uk/). Gene3D provides accurate structural domain family assignments for over 1100 genomes and nearly 10 000 000 proteins. A hidden Markov model library, constructed from the manually curated CATH structural domain hierarchy, is used to search UniProt, RefSeq and Ensembl protein sequences. The resulting matches are refined into simple multi-domain architectures using a recently developed in-house algorithm, DomainFinder 3 (available at: ftp://ftp.biochem.ucl.ac.uk/pub/gene3d_data/DomainFinder3/). The domain assignments are integrated with multiple external protein function descriptions (e.g. Gene Ontology and KEGG), structural annotations (e.g. coiled coils, disordered regions and sequence polymorphisms) and family resources (e.g. Pfam and eggNog) and displayed on the Gene3D website. The website allows users to view descriptions for both single proteins and genes and large protein sets, such as superfamilies or genomes. Subsets can then be selected for detailed investigation or associated functions and interactions can be used to expand explorations to new proteins. Gene3D also provides a set of services, including an interactive genome coverage graph visualizer, DAS annotation resources, sequence search facilities and SOAP services.
INTRODUCTION
Gene3D uses the manually curated protein domain assignments from CATH (1) to generate accurate protein domain architecture predictions for over 1000 genomes, as well as the UniProt (2) and RefSeq (3) protein sequence databases. These assignments can be queried, and the results visualized via the Gene3D website. Also displayed are other structural features, such as active sites and disordered regions from Disopred2 (4), and functional terms from UniProt, InterPro (5), the Gene Ontology (6), KEGG (7) and others (Table 1). The website enables complex sub-querying with these terms, giving the user the ability to retrieve information both for global studies and detailed analyses of function by molecular biologists. Gene3D also provides multiple web services, including DAS feature annotation sources and a set of SOAP XML services.
Table 1.
List of imported resources, type and reference
| Name | New? | Type | Description |
|---|---|---|---|
| UniProt (2) | N | SD | Protein database |
| RefSeq (3) | N | SD | Protein database |
| Ensembl metazoa (11) | Y | SD/G | Genome assemblies |
| Integr8 (14) | N | G | UniProt-based genome sets |
| CATH (1) | N | SF | Domain family classification |
| Pfama (13) | N | SF | Domain family classification |
| Superfamilya (17) | N | SF | Domain family classification |
| SMARTa (18) | N | SF | Domain family classification |
| Ensembl Variation (11) | Y | SF | Sequence polymorphisms |
| UniProt features (2) | Y (partly) | SF | Functional elements |
| TMHMM v2.0a (19) | Y | SF | Transmembrane helices |
| Sega (20) | N | SF | Low complexity regions |
| Coilsa (21) | N | SF | Coiled-coil regions |
| Panthera (22) | N | SF | Protein family classification |
| DisoPred2 (4) | Y | SF | Disordered regions |
| GO pathways (6) | N | P | Pathway descriptions |
| MINT (23) | N | P | Protein–protein interactions |
| IntAct (24) | N | P | Protein–protein interactions |
| GO cellular loc. (6) | N | P | Cellular locations. |
| KEGG pathways (7) | N | P | Pathway assignments |
| KEGG orthologue (7) | N | MF | Molecular function |
| GO molecular (6) | N | MF | Molecular function |
| UniProt descriptions (2) | Y | MF | Molecular function |
| eggNOG (16) | Y | MF | Molecular function |
| NCBI taxonomy (25) | N | T | Taxonomic hierarchy |
aObtained via SIMAP (26).
If the resource has been added to Gene3D since 2008 it is marked as ‘Y’ for ‘New’.
Types: ‘SD’: imported protein sequence databases; ‘G’: genome assemblies; ‘SF’: sequence feature annotation; ‘P’: metabolic, regulatory and biological pathways; ‘MF’: molecular function; ‘T’: taxonomy tree.
The CATH domain database provides a compendium of domains found in the protein structures deposited in the wwPDB (8), grouping them into a hierarchy based on homology relationships and structural features. Domain boundaries and superfamily assignments are determined using manual curation and automated evidence-gathering tools. For each superfamily (the CATH ‘H(omology)-level’) one or more representatives are selected and a set of sequence profiles generated. From these hidden Markov model (Hmmer)3 (http://hmmer.janelia.org/) models are built and, in combination with a recently developed in-house method for resolving conflicting matches (DomainFinder 3), accurate domain architectures are assigned. DomainFinder 3 is a novel algorithm that uses heaviest-weighted clique finding to optimize the selection of matches into a multi-domain architecture, and has significantly improved the overall quality of the assignments (paper submitted).
Over the last 2 years, the underlying technology and software powering the Gene3D resource have been re-implemented, allowing faster generation of releases, easier updates of imported annotations and quicker, more sophisticated querying. These and other improvements are described in the following sections.
IMPROVED DOMAIN ASSIGNMENT ACCURACY
A total of 10 013 non-redundant domain representatives were selected for all 2386 superfamilies in CATH v.3.3.0, and profiles built by using each sequence to seed a SAM Target-2K (9) iterative search (strategy 10). A mask was added to the resulting sequence alignment, so that only columns that align with the original seed sequence are used to build a profile HMM with HMMER 3’s ‘hmmbuild’. The Gene3D v9 sequence database was generated by merging UniProt, RefSeq and Ensembl 55 (11), creating a set of 9.5 million distinct protein sequences; ‘hmmsearch’ was then used to find matches with E-value <0.001.
Once all significant matches have been gathered, DomainFinder is used to resolve them into a set of confident domain assignments. DomainFinder has been significantly redeveloped (version 3; paper submitted). This version significantly reduces the number of false negatives (missed domains) by using a more sophisticated representative match selection protocol based around searching graphs of overlapping matches with the exact-weighted clique finding algorithm of Cliquer (12). This translated into a 15% increase in the number of domains confidently identified by Gene3D in large-scale sequence databases.
For Gene3D v9.0.0, a match overlap of 30 or less residues was permitted, while if a sub-graph of matches contained more than 5900 nodes, then the old algorithm was used to reduce computational time (<0.005% of all sequences affected). In total, 9 050 048 domains were identified in 5 186 382 million sequences, a mean of 1.7 domains per annotated sequence, and an overall sequence coverage of 55%. By using the benchmark set generated for testing DomainFinder, we estimated the minimum and maximum bounds for the rate of false-positive predictions for Gene3D (for details see Supplementary Data) to be between 0.2% and 0.6% of all domains. However, many of these ‘false positives’ may signify complex relationships between two superfamilies or errors in the CATH classification due to incomplete information.
A NEW WEBSITE FOR LARGE AND SMALL QUERIES
The Gene3D website supports a wide range of query terms, from protein and gene identifiers to superfamily codes, species names and function codes. It returns either a detailed description of the protein of interest or a summary view of the selected set of proteins. Previously, the size of a returned set of proteins was restricted to 5000, meaning that only subsets of the biggest superfamilies could be retrieved. The new site is now capable of retrieving annotations, species assignments and architectures for the huge P-loop hydrolase superfamily (34 050 300), containing over 530 000 domains in 440 000 sequences—although a few seconds can be required to render some of the results tabs in the browser.
Both the single protein and protein collection (i.e. a family or genome) views present a similar set of integrated information grouped into five categories: (i) taxonomic and genome data; (ii) sequence features and domain architectures; (iii) molecular and biochemical function; (iv) interactions and pathways; and (v) sequence database cross-references. The querying system then allows the user to select identifiers—for instance, the superfamily a protein domain belongs to, or proteins in the human genome with a specific GO term—and retrieve that subset or superset for further investigation. A query chain can be built up and followed by means of the ‘breadcrumb trail’ on the front summary tab in the results views. Imported annotations and descriptions, such as Pfam families (13), are also linked to the source database.
New genome assemblies and displays
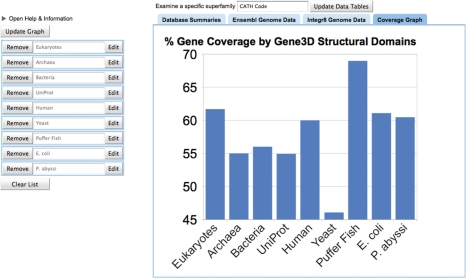
The Ensembl Metazoa genome assemblies have been added to the Gene3D sequence database (currently Ensembl 55). All transcripts are included and individually scanned. Users may query using Ensembl gene, transcript and protein identifiers; mappings to UniProt and RefSeq identifiers are also shown. Gene3D annotates two sets of sequenced genomes: eukaryotic transcripts from Ensembl metazoan (49 species) and UniProt-based assemblies for archaea, bacteria and eukaryotes (1085 species) from Integr8 (14). In addition, a new dynamic web application for querying and visualizing genome coverage for specific superfamilies and genomes is available at: http://gene3d.biochem.ucl.ac.uk:8090/GenomeCoverageGraphs (Figure 1). As can be seen, coverage for different genomes can vary significantly, although this could reflect biases in gene annotation in addition to variation in domain content. Phylogenetic profiles for Ensembl 55 metazoa and Integr8 prokaryotes for the CATH-Gene3D superfamilies are available for download from the Gene3D FTP site (ftp://ftp.biochem.ucl.ac.uk/pub/gene3d_data/CURRENT_RELEASE/).
Figure 1.
The Gene3D genome coverage visualizer. Genome coverage—the number of genes with a match to a CATH superfamily—can be viewed using the new visualization web tool (http://gene3d.biochem.ucl.ac.uk:8090/GenomeCoverageGraphs/). Coverage for a specific superfamily can be retrieved using the top query bar. In the left-hand column, species can be removed from the graph or their species names manually edited. New species can be added via tables found by selecting the ‘Database Summaries’, ‘Ensembl Genome Data’ and ‘Integr8 Genome Data’ tab headers.
A new aspect to the genome annotation is the inclusion of non-synonymous single-nucleotide polymorphisms (SNPs) and other sequence variants from Ensembl Variation (11). These are included in the protein feature display, allowing the correlation of altered activity or phenotype with other annotations, such as the location of binding sites or domains.
Sequence features and domain architectures
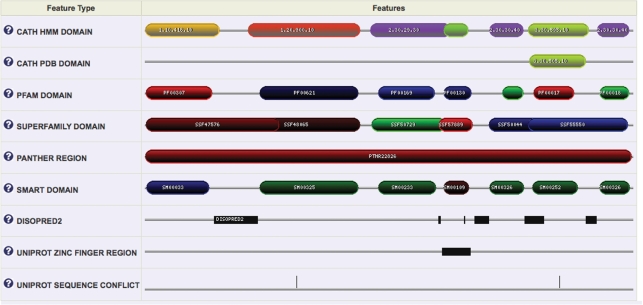
The Gene3D website uses a graphics package kindly provided by Pfam to draw sequence feature annotations (Figure 2), including domains, active sites, SNP locations, transmembrane regions, signal peptides and others. This has now been localized to the Gene3D server, greatly improving rendering speeds and site stability. For the single protein view (see above) a detailed description of the protein is provided (the ‘Features’ tab), with one ‘track’ per annotation type. A second visualization view (‘Sequence Image’) amalgamates all the annotations into a single ‘track’. The ‘Sequence Image’ tab is also available in the collection view (e.g. for a superfamily), along with a tab displaying the set of distinct multi-domain architectures found (‘Multi-Domain Architectures’). A useful facility of Gene3D is the inclusion of CATH domains based on the Protein Data Bank (PDB). We now use the SIFTS PDB-to-UniProt mapping, which has increased the number of domains we are able to map to Gene3D (15).
Figure 2.
Sequence feature annotation for human proto-oncogene vav. Displayed is the feature annotation returned for UniProt identifier VAV_HUMAN. Laying out each type and source of annotation against each other allows the user to easily cross-reference functional information. Clicking on domains brings up available functional descriptions and a link to the original resource. Detailed tables of assignments can be found in separate tabs. To aid users each family is given a unique colour that is consistent across the website; for CATH-Gene3D domains the colour-picking algorithm makes common superfamilies brighter, and ensures superfamilies within the same fold have similar hues.
Molecular function, interactions, pathways and orthologues
The parsing and import of UniProt records has been improved, leading to more descriptive terms being imported along with more associated information. We have also added EggNOG orthologue assignments (16). EggNog orthologue families are generated by large-scale clustering of protein sequences using an extension of the COGS protocol and applying a rule-based annotation system.
NEW DAS AND OTHER WEB SERVICES
The current set of DAS sources provided by Gene3D and CATH can be viewed at: http://cathdb.info:9000. Recently added is the ‘gene3d_ensembl’ track, which can be queried with Ensembl protein identifiers to retrieve domain annotations and the results viewed in a DAS client.
Gene3D also underpins the FuncNet pipeline for analysis of protein function (paper submitted). This resource integrates various orthogonal methods that predict functional associations between proteins. Gene3D provides the Gene Expression Comparison, homology inherited Protein–Protein Interactions and Co-occurrence of Domains Analysis services, which can be queried directly via SOAP services or together through the FuncNet front-end service. Please refer to the FuncNet website for more information (http://funcnet.eu/).
RELEASE SCHEDULE AND VERSIONING
As of Gene3D_v9.0.0 release numbering system has been stabilized to use a three-part identifier—‘Gene3D_v#.#.#’. The first part signifies a major release, and is incremented if there is a potential change to the domain assignments for a given protein. This would be the case if there is a new CATH release or if there is a change to the domain-identification protocol. The second number is incremented if new sequences are added to the database, such as a new set of Ensembl genomes; while the third is incremented if any imported annotation is updated or added. As such, there is no predefined schedule for releases, but instead is done ‘as needed’. However, major releases are expected two to three times a year, while sequences will be updated more often.
DISCUSSION
The primary purpose behind the changes made to Gene3D is to help support the investigations of the biological and medical research communities. To this extent, Gene3D actively collaborates within several teams and networks, providing data and tools, including the FuncNet function prediction web services as well as a member of the InterPro consortium of protein family databases and the IMPACT network, and provides targets for structural determination through the NIH PSI2 project. We also provide individualized domain predictions and linked data sets on request. Possible examples are lists of domains that contain annotated active sites, or are adjacent to transmembrane regions. Through these collaborations we hope to extend the web platform to provide data in formats that can be used by the various research communities.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The European Commission’s BioSapiens and EMBRACE Networks of Excellence funded under the Framework Program 6 (grant numbers LSHG-CT-2003-503265 to C.Y., LSHG-CT-2004-512092 to A.C.); National Institutes of Health’s PSI2 initiative (grant number DE-AC02-065CH11357); ENFIN (grant number LSHG-CT-2005-518254 to J.L.). Funding for open access charge: European Commission's EMBRACE NoE; LSHG-CT-2004-512092.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the CATH team for their help; SIMAP for providing significant computational services and annotations; Prof. David Jones for providing Disopred2 predictions; InterPro for reviews of the data quality; Alison Cuff for advice on distant homologies between superfamilies; and the advice and support of collaborators in the BioSapiens, EMBRACE, ENFIN and IMPACT networks
REFERENCES
- 1.Cuff AL, Sillitoe I, Lewis T, Redfern OC, Garratt R, Thornton J, Orengo CA. The CATH classification revisited – architectures reviewed and new ways to characterize structural divergence in superfamilies. Nucleic Acids Res. 2009;37:D310–D314. doi: 10.1093/nar/gkn877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.UniProt Consortium. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Karplus K, Karchin R, Draper J, Casper J, Mandel-Gutfreund Y, Diekhans M, Hughey R. Combining local-structure, fold-recognition, and new-fold methods for protein structure prediction. Proteins: Struct. Funct. Gen. 2003;B:491–496. doi: 10.1002/prot.10540. [DOI] [PubMed] [Google Scholar]
- 10.Sillitoe I, Dibley M, Bray J, Addou S, Orengo C. Assessing strategies for improved superfamily recognition. Protein Sci. 2005;7:1800–1810. doi: 10.1110/ps.041056105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ostergard PRJ. A fast algorithm for the maximum clique problem. Discr. Appl. Math. 2002;120:197–207. [Google Scholar]
- 13.Finn RD, Tate J, Mistry J, Coggill PC, Hotz JS, Ceric HR, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen LJ, Julien P, Kuhn M, von Mering C, Muller J, Doerks T, Bork P. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36:D250–D254. doi: 10.1093/nar/gkm796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M, Chothia C, Gough J. SUPERFAMILY– sophisticated comparative genomics, data mining, visualization and phylogeny. Nucleic Acids Res. 2009;37:D380–D386. doi: 10.1093/nar/gkn762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 20.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 21.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 22.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R, et al. IntAct – open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rattei T, Tischler P, Arnold R, Hamberger F, Krebs J, Krumsiek J, Wachinger B, Stümpflen V, Mewes W. SIMAP – structuring the network of protein similarities. Nucleic Acids Res. 2008;36:D289–D292. doi: 10.1093/nar/gkm963. [DOI] [PMC free article] [PubMed] [Google Scholar]