Abstract
Chromosomal replication results in the duplication not only of DNA sequence but also of the patterns of histone modification, DNA methylation, and nucleoprotein structure that constitute epigenetic information. Pericentromeric heterochromatin in human cells is characterized by unique patterns of histone and DNA modification. Here, we describe association of the Mi-2/NuRD complex with specific segments of pericentromeric heterochromatin consisting of Satellite II DNA located on human chromosomes 1, 9 and 16 in some, but not all cell types. This association is linked in part to DNA replication and chromatin assembly, and may suggest a role in these processes. Mi-2/NuRD accumulation is independent of Polycomb association and is characterized by a unique pattern of histone modification. We propose that Mi-2/NuRD constitutes an enzymatic component of a pathway for assembly and maturation of chromatin utilized by rapidly proliferating lymphoid cells for replication of constitutive heterochromatin.
INTRODUCTION
During development, differentiation status is reflected in patterns of gene expression. Genetic programs are thought to be established and maintained through the action of transcriptional activators and repressors. Current models postulate that these proteins bind DNA in a sequence specific manner, where they locally recruit enzymes that alter epigenetic information. For example, during B cell development, the transcriptional repressor BCL6 establishes and maintains the transcriptional profile of the germinal center B cell through recruitment of corepressors including the Mi-2/NuRD complex (Fujita et al. 2004). An alternative mechanism of transcriptional repression involves recruitment of DNA to heterochromatin, such as to the large blocks of pericentromeric heterochromatin found on human and mouse chromosomes. This mechanism is critical to the biology of B cells where loci such as Rag and TdT loci undergo nuclear repositioning to pericentromeric heterochromatin following stimulation, where they are thought to be heritably silenced by Ikaros (Brown 1999; Brown et al. 1997). Thus, the proper establishment of cell-type specific transcriptional profiles relies on both classical repression mechanisms mediated by sequence specific repressors and their associated corepressors as well as on the integrity and function of pericentromeric heterochromatin.
Consistent with this notion, disruption of pericentromeric heterochromatin, observed in ICF (Immunodeficiency, Centromeric instability and Facial Anomalies) syndrome, can have a significant impact on B cell development. Patients suffering from ICF syndrome, caused by mutations in the de novo DNA methyltransferase DNMT3B (Hansen et al. 1999; Xu et al. 1999), are characterized by a lymphoid-specific chromosome instability and by defective B cell negative selection and terminal differentiation (Blanco-Betancourt et al. 2004). It remains unclear why chromosome instability is limited to lymphoid lineages when pericentromeric heterochromatin is hypomethylated in all ICF tissues.
Here, we describe a novel, BCL6-independent accumulation of the Mi-2/NuRD complex in rapidly proliferating lymphoid cell lines. Mi-2/NuRD localizes to pericentromeric heterochromatin in a cell cycle-dependent manner and is closely associated with heterochromatin containing HP1 proteins and histone H3 trimethylated at lysine 9 (H3K9me3). These NuRD bodies are present in a variety of B lymphocyte-derived cell lines as well as primary human cells. In contrast, cells that assemble Polycomb proteins at pericentromeric heterochromatin lack NuRD bodies and differ in the composition of pericentromeric heterochromatin. The recruitment of Mi-2/NuRD to heterochromatic foci is tightly linked to DNA replication, suggesting a function for this enzyme in chromatin assembly. We propose that NuRD bodies are cytologic markers of a novel chromatin assembly pathway utilized by lymphoid cells at pericentromeric heterochromatin.
RESULTS
The Mi-2/NuRD complex localizes to nuclear structures in a cell-type specific manner
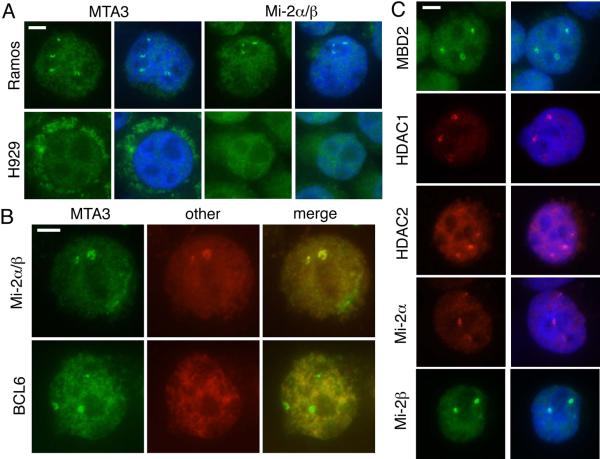
We have shown previously that MTA3 and the Mi-2/NuRD complex interact with the master regulator of B cell differentiation, BCL6, to establish the transcriptional profile of the germinal center B cell (Fujita et al. 2004). To further investigate this process, we used indirect immunofluorescence to examine the distribution of these proteins within germinal center (GC) B cell-like and plasmacytoid cell lines. We immunostained a germinal center model cell line, Ramos (Klein et al. 1975), and a plasmacytoid cell line, H929 (Gazdar et al. 1986), with antibodies directed against subunits of Mi-2/NuRD (Figure 1).
Figure 1. Mi-2/NuRD subunits are enriched in nuclear foci.

A) Ramos and H929 cells were cytocentrifuged and immunostained with antibodies against MTA3 (left) or Mi-2 (right). Primary antibodies were detected with AlexaFluor 488 conjugated secondary antibodies (green). Cells are counterstained with DAPI (blue). Scale bar (top left corner) indicates 5 μm.
B) Two color immunofluorescence in cytocentrifuged Ramos cells. In the top panel, Mi-2 was detected with AlexaFluor 568 secondary antibody; the MTA3 antibody was directly labeled with AlexaFluor 488. In the lower panel, MTA3 and BCL6 were detected with AlexaFluor 488 (green) and AlexaFluor568 (red) secondary antibodies. Scale bar indicates 5 μm.
C) Cytocentrifuged Ramos cells were immunostained with antibodies against other members of the Mi-2/NuRD complex (MBD2, HDAC1, HDAC2, Mi-2a and Mi-2b), and AlexaFluor 488 or 568 secondary antibodies. Cells were counterstained with DAPI. Scale bar indicates 5 μm.
A significant proportion of Ramos cells immunostained with antibodies against the Mi-2/NuRD complex members MTA3 and Mi-2a/b exhibited several brightly staining nuclear foci that were not evident in H929 (Figure 1a). To determine whether foci observed with individual antibodies coincided, we performed two-color immunofluorescence (Figure 1b). Colocalization between Mi-2a/b and MTA3 foci was observed in 100% of cells. To determine whether these foci coincided with known nuclear or perinuclear structures, we compared the distribution of Mi-2/NuRD and that of PML bodies (Weis et al. 1994), Cajal bodies (Cajal 1903; Monneron and Bernhard 1969), centrosomes (Zheng et al. 1991) and SC35 domains (Fu and Maniatis 1990). No significant association between Mi-2/NuRD and any nuclear structure tested was observed, suggesting that these foci represent a novel nuclear domain (data not shown). We have shown previously that, in GC B cell-like lines, the MTA3-containing form of the Mi-2/NuRD complex interacts with BCL6 to establish GC type transcriptional profiles (Fujita et al. 2004). However, there was no enrichment of BCL6 in the nuclear foci (Figure 1b), indicating that recruitment of the Mi-2/NuRD complex to these foci occurs independently of BCL6.
The components of Mi-2/NuRD are each encoded by multiple, highly similar genes and all known protein products of these genes have been associated with the complex. We have proposed that substitution of these variable subunits may confer functional specificity to the complex, directing it to a specific set of gene targets (Bowen et al. 2004). To determine whether a specific subunit resulted in targeting of the complex to these foci, we investigated the distribution of the alternative subunits MBD2, HDAC1, HDAC2, Mi-2a (CHD3), Mi-2b (CHD4), MTA1, and MTA2 in Ramos cells (Figure 1c and Supplemental Figure 1). HDAC1, HDAC2, Mi-2a and Mi-2b all exhibit a pattern of focal enrichment similar to that observed with MTA3 and Mi-2a/b antibodies (Figure 1c). MBD2 is also enriched in nuclear foci (Figure 1c); we were unable to determine whether MBD3 exhibited similar enrichment (data not shown). Both MTA1 and MTA2 showed some enrichment in the foci, although often to a lesser extent than observed for MTA3 (Supplemental Figure 1). We concluded from these experiments that Mi-2/NuRD is enriched in a novel nuclear domain in some cell types with no apparent subunit preference. We will heretofore refer to these nuclear structures as NuRD bodies.
We next sought to address whether NuRD bodies are present in other lymphoid cell types. Therefore, we immunostained a variety of B-cell lymphoma and leukemia lines with antibodies against MTA3: Daudi and Raji (Burkitt Lymphomas), HT and Pfeiffer (diffuse large B-cell lymphomas), CCRF-SB and SUP-B15 (B-cell acute lymphoblastic leukemia). We observed NuRD bodies (in 20–50% of cells) in all lymphoid cell lines tested (data not shown).
NuRD bodies localize to pericentromeric heterochromatin
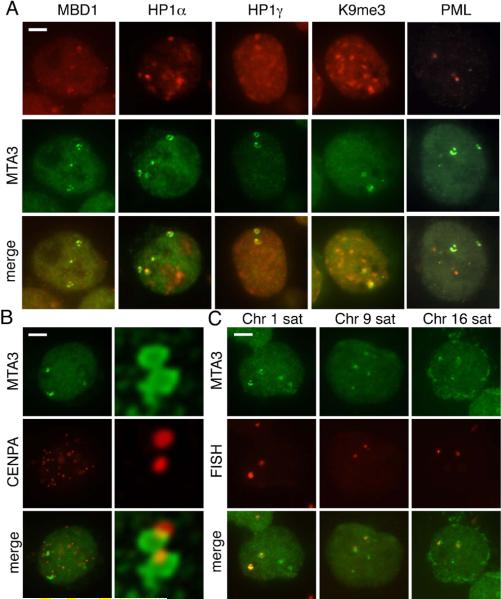
To identify the nuclear structure or DNA sequences targeted by Mi-2/NuRD, we investigated the distribution of Mi-2/NuRD subunits relative to other chromatin proteins previously reported to display a similar nuclear pattern. Although it has not been reported to associate with the Mi-2/NuRD complex, the methyl-CpG binding protein MBD1 displays a punctate localization in human interphase nuclei, similar to the pattern observed here (Fujita et al. 1999; Ng et al. 2000). This results from association of MBD1 with heterochromatin located in close proximity to centromeres, particularly on chromosome 1. Two-color immunofluorescence revealed partial colocalization of MBD1 foci with NuRD bodies (Figure 2a). We also investigated the distribution of NuRD bodies relative to the three isoforms of Heterochromatin Protein 1, HP1a, HP1b and HP1g (Figure 2a and data not shown), all of which have been reported to localize, at least in part, to pericentromeric heterochromatin in mammals (Minc et al. 2000; Nicol and Jeppesen 1994; Wreggett et al. 1994). All three HP1 isoforms showed some degree of association with NuRD bodies (88% of NuRD bodies were associated with HP1a, 45% with HP1b, and 92% with HP1g). Consistent with these data, we also observed an association between the NuRD bodies and, H3K9me3 (Figure 2a), a histone modification required for HP1 targeting (Lachner et al. 2001). We also investigated the relationship between NuRD bodies and a number of other histone modifications. We did not identify any other histone modification with any significant association with Mi-2/NuRD bodies (Supplementary Table 1). These data suggest that Mi-2/NuRD associates with centromeric or pericentromeric DNA.
Figure 2. Mi-2/NuRD localizes to pericentromeric heterochromatin.
A) Cytocentrifuged Ramos cells were immunostained with antibodies against MTA3 (green) and MBD1, HP1a, HP1g or H3K9me3 (red). NuRD bodies are closely associated with each of these proteins. NuRD bodies did not colocalize with markers of other known nuclear domains, such as PML bodies. Scale bar indicates 5 μm.
B) Cytocentrifuged Ramos cells were stained with antibodies against MTA3 (green) and CENP-A (red). Images in left side panels were acquired using conventional microscopy. Right panels show an expanded view of a large NuRD body from a second nucleus, and was acquired using confocal microscopy with deconvolution. The image presented represents a single optical section through the nucleus.
C) Immuno-FISH in Ramos cells. Cells were cytocentrifuged and stained with antibodies against MTA3 (green) with subsequent fluorescence in situ hybridization using a probe (red) directed against the chromosome 1 satellite repeat D1Z1 (left) or the chromosome 9 satellite repeat D9Z3 (right). Scale bar indicates 5 μm.
To directly address this question, we performed two color immunofluorescence in Ramos cells using antibodies against the kinetochore protein CENP-A and MTA3 as a marker for the NuRD body. MTA3 foci were identified and scored for association with one or more CENP-A signals. We observed a significant degree of association between CENP-A signals and NuRD bodies (86% of NuRD bodies were closely associated with at least one CENP-A signal, Figure 2b), further supporting the hypothesis that Mi-2/NuRD may be targeted to centromeric or pericentromeric heterochromatin in these cells.
The pericentromeric heterochromatin on chromosomes 1, 9 and 16 contain megabase blocks of Satellite II DNA, representing one of the highest concentrations of methylated DNA in the human genome (Lubit et al. 1976). Because the MBD2 subunit of Mi-2/NuRD is known to preferentially bind methylated DNA (Hendrich and Bird 1998), we reasoned that the complex may localize to methylated Satellite II repeats at these chromosomes. To investigate this possibility, we used immunofluorescence against the MTA3 subunit of the complex combined with fluorescence in situ hybridization (FISH) directed against chromosome 1, 9 and 16 satellite repeats (Figure 2c). The X chromosome alpha satellite repeat DXZ1 was used as a negative control. The results of this analysis are summarized in Table 1. As predicted, Mi-2/NuRD was targeted to pericentromeric heterochromatin of chromosomes 1, 9 and 16.
Table 1.
| FISH probe | # visible FISH signals | % FISH signals associated with NuRD |
|---|---|---|
| Chr 1 (D1Z1 - classical) | 143 | 77% |
| Chr 9 (D9Z3 - classical) | 116 | 52% |
| Chr 16 (D16Z1 - alpha) | 203 | 47% |
| Chr X (DXZ1 -alpha) | 75 | 17% |
Formation of NuRD bodies occurs primarily during S phase
The Mi-2/NuRD complex was localized to pericentromeric heterochromatin in only a subset of Ramos cells (43% of cells in an asynchronous culture), exhibiting a generalized nuclear localization in the remainder of nuclei. This suggested that targeting may occur only at specific points in the cell cycle. Therefore, cells were pulsed for 15 minutes with 5-bromodeoxyuridine (BrdU) to mark replicating cells, and subsequently stained with antibodies against CENP-F (to identify cells in G2), BrdU and MTA3 (as a marker for the NuRD body). Cells with NuRD bodies (n = 75) were then scored for cell cycle distribution, on the basis of BrdU incorporation and CENP-F staining. The majority of cells containing NuRD bodies were in S phase (59% of cells), with lesser amounts in G2 (22% of cells) and G1 (19% of cells) (Figure 3a). Overall, approximately half of cells in the asynchronous culture were in some stage of S phase (51%), irrespective of the presence of NuRD bodies. Furthermore, NuRD bodies present in S phase cells generally colocalized with (74% of NuRD bodies) or were closely associated with (20% of NuRD bodies) BrdU signals, suggesting that recruitment of the Mi-2/NuRD complex to pericentromeric heterochromatin is linked to active DNA replication. NuRD bodies were not detected in every replicating cell; rather, approximately two-thirds of cells in S phase did not contain these structures (data not shown). Given that heterochromatic sequences characteristically replicate late in S phase and that Mi-2/NuRD localization to pericentromeric heterochromatin is closely associated with their replication, we inferred that NuRD bodies are primarily observed in late S phase cells. To address this more directly, we synchronized Ramos cells at the G1/S boundary and released them into S phase. Cells were harvested at several timepoints after release, immunostained with antibodies against MTA3, and scored for NuRD bodies (Figure 3b). While NuRD bodies were observed with some frequency at all timepoints, they were most prevalent during late S phase (4.5 hours and 6 hours after release).
Figure 3. Mi-2/NuRD enrichment at pericentromeric heterochromatin is tightly linked to DNA replication and chromatin assembly.
A) Graph shows the proportion of cells with NuRD bodies in each stage of the cell cycle, on the basis of BrdU incorporation and CENP-F staining. Cells containing NuRD bodies were identified and scored as to whether they were in S, G1 or G2 phase. Overall, 51% of cells in the asynchronous culture were scored as being in S phase, irrespective of the presence of NuRD bodies.
B) Graph shows the proportion of synchronized cells that contain NuRD bodies. Cells were synchronized at the G1/S border, released into S phase, and analyzed at several time points (unsynchronized, 0 hrs, 1.5 hrs, 3 hrs, 4.5 hrs, 6 hrs and 8 hrs after release). Cells were scored as to whether Mi-2/NuRD foci were observed. Left side of chart shows DNA content analysis after flow sorting.
C) Ramos cells were pulsed for 15 minutes with BrdU prior to cytocentrifugation, fixation and detection with MTA3 (red), BrdU (green) and PCNA (AlexaFluor 647, purple) antibodies. The lower panel shows an expanded view of two brightly staining foci in the nucleus pictured in the upper panel. Scale bar indicates 5 μm.
D) Ramos cells were cytocentrifuged and stained with MTA3 (directly labeled with AlexaFluor 568) and CAF-1 p150 (detected with AlexaFluor 488 secondary antibodies). Scale bar indicates 5 μm.
E. Ramos cells were cytocentrifuged and immunostained with antibodies against MTA3 (green) and HP1g (red). Images were acquired using confocal microscopy with subsequent deconvolution. Panel shows an expanded view of one NuRD body. The image presented is a summed Z-series. Scale bar indicates 2.5 μm.
To further characterize the association of Mi-2/NuRD with active replication forks, we stained BrdU-pulsed Ramos cells with antibodies against MTA3 (as a marker for Mi-2/NuRD), BrdU, and PCNA (as a marker for replication forks). In general, the BrdU and PCNA distributions overlapped and were difficult to distinguish. However, regions could be identified that stained with the PCNA antibody, but not the BrdU antibody, differentiating the site of DNA synthesis from nascently replicated DNA. In 63% of nuclei (n = 70) scored, differential PCNA and BrdU staining was observed in the region of the NuRD body. In this subset of nuclei, we generally observed colocalization between PCNA and MTA3 (84% of this subset of cells, Figure 3c). Furthermore, immunofluoresence comparing the distribution of MTA3 and the replication-coupled chromatin assembly protein CAF-1 demonstrated that the Mi-2/NuRD foci coincide with sites of chromatin assembly (Figure 3d). Finally, Mi-2/NuRD foci are often observed in a characteristic ring (or toroid) formation surrounding heterochromatin markers, like HP1g (Figure 3e). This pattern is reminiscent of that observed in previous studies of DNA replication and chromatin assembly at pericentromeric heterchromatin in other mammalian cells (Dimitrova and Berezney 2002; Fox et al. 1991; O'Keefe et al. 1992; Quivy et al. 2004; Taddei et al. 1999). Taken together, these data indicate that during S phase, the Mi-2/NuRD complex associates with chromatin during, or immediately following DNA replication at a time period coinciding with chromatin assembly.
Relationship of Mi-2/NuRD to other components of pericentromeric heterochromatin: BMI1 and Ikaros
The Polycomb core complex, PRC1, localizes, in a cell-cycle dependent manner, to pericentromeric heterochromatin in a variety of cell lines in structures called Polycomb bodies (Saurin et al. 1998; Voncken et al. 1999). BMI1 (a component of the PRC1 complex) foci are visible during G1 and early/mid S phase, disappearing in late S and M phase. To investigate the relationship between the NuRD body and Polycomb, we immunostained Ramos (GC) and H929 (plasmacytoid) cell lines with antibodies against MTA3 and BMI1 (Figure 4a). In H929, we observed obvious Polycomb bodies (but no NuRD bodies) in a significant proportion of cells (nearly 100% of cells). In contrast, Ramos cells exhibited NuRD bodies but no obvious enrichment of BMI1. The PRC1 complex is recruited to heterochromatin through an interaction with histone H3 trimethylated at lysine 27 (H3K27me3) (Cao et al. 2002). In order to determine whether this histone modification exhibited a differential association with pericentromeric heterochromatin in the two cell lines, we performed FISH coupled with immunofluorescence using antibodies against H3K27me3. As expected, the chromosome 1 Satellite II repeat in H929 was enriched for H3K27me3. However, Ramos cells, which lack obvious Polycomb bodies, showed no association between H3K27me3 and chromosome 1 pericentromeric heterochromatin, in fact, this modification was generally depleted in the region of the FISH signals (Figure 4b). These data indicate the presence of at least two alternate heterochromatin types at the pericentromeric satellite repeat: one NuRD-associated/K27me3-depleted (as in Ramos) and one BMI1-associated/K27me3-enriched (as in H929). In order to determine whether this either/or pattern was observed in other cell types, we analyzed several additional cell lines, many of which form Polycomb bodies at pericentromeric heterochromatin (Saurin et al. 1998; Voncken et al. 1999). We observed obvious Polycomb bodies in some proportion of A375, WI-38, MCF7 and U2-OS cells (Figure 4c). However, consistent with our hypothesis, none of these cell lines exhibited obvious NuRD accumulation at pericentromeric heterochromatin. Interestingly, some cell lines did not fit either pattern. For example, the lymphoblast cell line GM09146 did not have obvious NuRD bodies or Polycomb bodies (Figure 4c). Consistent with the absence of Polycomb bodies, immuno-FISH experiments in this cell line demonstrated that chromosome 1 pericentromeric heterochromatin was not enriched for H3K27me3 (data not shown). This finding indicates that the composition of constitutive heterochromatin varies substantially between cell types.
Figure 4. NuRD bodies and Polycomb bodies appear to be mutually exclusive.
A) Ramos (left) and H929 (right) cells were cytocentrifuged and immunostained with antibodies against MTA3 (top) and BMI1 (middle). Scale bar indicates 5 μm. B) Ramos (left) and H929 (right) cells were subjected to immuno-FISH with an antibody against H3K27me3 (top) and a FISH probe directed against the chromosome 1 satellite II repeat (middle). Arrowheads indicate regions of K27me3 depletion in pericentromeric heterochromatin. Scale bar indicates 5 μm. C) A variety of cell lines (A375, WI-38, MCF7, U2OS and GM09146) were stained with antibodies against Mi-2 (MCF7) or MTA3 (others, green) and BMI1 (red). Polycomb bodies were clearly visible in some proportion of cells in most cell lines, except GM09146. NuRD bodies were largely absent from all five cell lines. Scale bar indicates 5 μm.
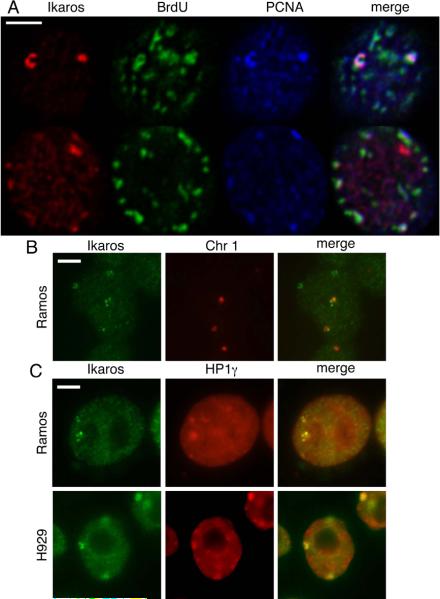
The zinc finger protein Ikaros is recruited to pericentromeric heterochromatin in cycling B and T lymphocytes. Ikaros is known to interact with Mi-2 in mouse T cells, and can be observed in toroidal structures at the pericentromeric heterochromatin of activated T cells in G1 and S phase (Kim 1999). Thus, we investigated the relationship between Ikaros and the NuRD body in Ramos and H929. We immunostained BrdU-pulsed Ramos cells with antibodies against Ikaros, BrdU and PCNA. Ikaros was closely associated with BrdU and PCNA in about half of cells with obvious Ikaros foci (Figure 5a). In many cases, Ikaros foci were not clearly associated with BrdU (Figure 5a). Like Mi-2/NuRD, Ikaros also associated with HP1g and chromosome 1 pericentromeric heterochromatin (Figure 5B and 5C). These data are consistent with the notion that Mi-2/NuRD and Ikaros localize to pericentromeric heterochromatin in a similar manner. Interestingly, Ikaros also associates with pericentromeric heterochromatin in H929, indicating that the presence of Ikaros is not sufficient to recruit Mi-2/NuRD to these sequences.
Figure 5. Ikaros localizes to pericentromeric heterochromatin with similar kinetics.
A) Ramos cells were seeded on poly-L-lysine coated slides, pulsed with BrdU for 30 minutes and immunostained with antibodies against Ikaros, BrdU and PCNA. Images presented represent a single plane of a z-stack acquired on the confocal microscope. An example is presented of a cell where Ikaros appears to be closely associated with replication (top) and one where it does not (bottom). Scale bar indicates 5 μm.
B) Immuno-FISH of cytocentrifuged Ramos cells shows colocalization between Ikaros foci and chromosome 1 pericentromeric heterochromatin. Scale bar indicates 5 μm.
C) Ramos and H929 cells immunostained with antibodies against Ikaros (green) and HP1g (red). Scale bar indicates 5 μm.
Mi-2/NuRD associates with pericentromeric heterochromatin in human primary cells
We have demonstrated that in GC B cell-like lines, Mi-2/NuRD associates with pericentromeric heterochromatin in a cell cycle-dependent manner. To address this phenomenon in primary cells, we isolated germinal center B cells from human tonsil biopsies, and immunostained them with an antibody against Mi-2α/β, in order to capture all NuRD complexes, regardless of subunit composition. We observed clearly visible NuRD foci in primary germinal center B cells. These data support the hypothesis that the association of Mi-2/NuRD with pericentromeric heterochromatin occurs in human tissues, and is not an artifact limited to immortalized cell lines.
DISCUSSION
A defining characteristic of an epigenetic state is faithful transmission to daughter cells after cell division. When the replication fork passes through chromatin, existing, modified nucleosomes are distributed among the two daughter chromosomes. Chromatin assembly proteins, such as CAF-1, fill gaps in the daughter chromosomes with newly synthesized histones, which carry characteristic deposition marks, such as histone H4 acetylated at lysines 5 and 12 (Sobel et al. 1995). Established chromatin states are then reconstructed on daughter chromosomes through the process of chromatin maturation: histone deposition marks are removed, proper nucleosomal spacing is restored, and parental patterns of histone modification are re-established on newly acquired nucleosomes (reviewed in Polo and Almouzni 2006).
In this study, we have shown that the Mi-2/NuRD complex localizes to pericentromeric heterochromatin primarily during late S phase. This may indicate a role in heterochromatin replication. Thus, we propose that Mi-2/NuRD functions in the maturation of heterochromatin in rapidly proliferating lymphoid cells. Mi-2/NuRD is unique amongst well-characterized chromatin remodeling complexes in that it possesses the ability both to mobilize nucleosomes through the Mi-2 ATPase subunit (Guschin et al. 2000) and to deacetylate histones through the HDAC subunit (Wade et al. 1998; Xue et al. 1998; Zhang et al. 1998), making it an excellent candidate for this process. This model predicts that Mi-2/NuRD removes histone deposition marks such as H4K5Ac and H4K12Ac. The removal of these marks permits histone methyltransferase enzymes, such as SUV39H or SETDB1, to re-establish trimethylation of K9 on histone H3, and for the subsequent recruitment of downstream structural components of heterochromatin, such as HP1. Although the data we have presented here suggest a role for Mi-2/NuRD in the replication of constitutive heterochromatin, the biology revealed predicts a more general function for Mi-2/NuRD in, both euchromatin and developmentally regulated regions of facultative heterochromatin.
While all pericentromeric heterochromatin can be characterized by H3K9 trimethylation, we have uncovered evidence for the presence of at least three cell type-specific subtypes: NuRD-enriched/H3K27me3-depleted, PRC1-enriched/H3K27me3-enriched, and a third that is associated with neither complex and is depleted for H3K27me3. That an essential nuclear structure, pericentrometric heterochromatin, would exhibit this degree of diversity in composition is somewhat surprising. Rapidly proliferating lymphoid cells, exemplified by the germinal center B lymphocyte, are characterized by a significantly shortened interphase and accelerated S phase (Hanna 1964; Liu et al. 1991; Zhang et al. 1988). The Mi-2/NuRD pathway may be utilized in such cell types to rapidly re-establish functional heterochromatin in a limited time window. In contrast, other cell types may use more complex mechanisms, such as the recruitment of PRC1 and the assembly of the nucleoprotein structures associated with this complex (Sewalt et al. 2002), in order to achieve a heterochromatin configuration with sufficient stability for a more generous interphase. It is important to note that although this study primarily focuses on the association between Mi-2/NuRD and pericentromeric heterochromatin during its replication, this may also serve a purpose outside of S phase, as NuRD bodies are observed, albeit at lesser frequencies, in both G1 and G2.
ICF syndrome is a rare genetic disorder, many cases of which are caused by mutations in the de novo DNA methyltransferase DNMT3B. A constant feature of ICF is hypomethylation of Satellite II repeats, the very same DNA underlying the Mi-2/NuRD foci described in this work. Given that defects in chromatin maturation after DNA replication would be predicted to destabilize pericentromeric heterochromatin and that Mi-2/NuRD is known to associate with methylated DNA (Feng and Zhang 2001), the failure to recruit Mi-2/NuRD to these densely methylated sequence blocks likely contributes to the chromosomal instability observed in ICF syndrome. Furthermore, the observation that some lymphoid-derived cells assemble heterochromatin at these sequence elements in a manner distinct from most other cell types analyzed may explain one perplexing feature of ICF syndrome; although all cell types exhibit hypomethylation of pericentromeric heterochromatin, chromosome decondensation and the formation of characteristic radial chromosome figures is restricted to lymphoid-derived cell types (Maraschio et al. 1989).
The pathway for heterochromatin assembly and maturation outlined in this work may have a fundamental impact on B lymphocyte pathology. The failure of these cells to incorporate more complex nucleoprotein architecture at pericentromeric heterochromatin may predispose them to failure of these critical structures. Chromosomal translocations are a recurrent etiologic agent in both leukemias and lymphomas; a subset of characterized translocation events involve Satellite II DNA (Berger and Busson-Le Coniat 1999; Busson-Le Coniat et al. 1999; Sawyer et al. 1995; Sawyer et al. 1998). The presence of a heterochromatin assembly mechanism that fundamentally differs from other cell types may provide a means by which lymphocytes are predisposed to chromosomal accidents resulting in neoplasia.
METHODS
Cell lines
The following cell lines were obtained from the American Type Culture Collection (ATCC): Raji, Ramos, and Daudi (Burkitt's lymphoma, BL); H929 (multiple myeloma, MM); HT and Pfeiffer (diffuse large B-cell lymphoma, DLBCL); SUP-B15 and CCRF-SB (acute lymphoblastic leukemia, ALL), MCF7 (adenocarcinoma), A-375 (malignant melanoma), WI-38 (fetal lung fibroblast) U2OS (osteosarcoma). GM09146 (lymphoblast cell line, EBV transformed) was obtained from the Coriell Cell Repository. Lymphoid cell lines were cultured in RPMI-1640 media (Gibco) supplemented with 10%–15% fetal bovine serum (FBS).
Antibodies
The following primary antibodies were used in this study: Mouse monoclonal: PCNA (F-2, Santa Cruz). BCL6 (PG-B6p, Dako). CHD3/Mi-2a (611846, BD Transduction Laboratories). HP1a (MAB3584, Chemicon) HP1b (MAB3448, Chemicon) HP1g (MAB3450, Chemicon). BrdU-fluorescein (Roche). BMI1 (clone F6, Upstate), CENP-A (3–19, Stressgen). SC35 (obtained from Dr. Maureen Powers, Emory University)
Rabbit polyclonal
MTA3, as described (Fujita et al. 2003). Mi-2, as described (Wade et al. 1999). MTA2, as described (Fujita et al. 2003). CENP-F (NB 500-101, Novus Biologicals). HDAC1 (H-51, Santa Cruz), HDAC2 (H-54, Santa Cruz). MTA1 (Bethyl Labs). CAF-1 p150 (H-300, Santa Cruz). H3K9me3 (07-523, Upstate), H3K27me3 (07-449, Upstate), Ikaros (H-100, Santa Cruz). H2A.X (ab4176, Abcam), H3K4me2 (ab7766, Abcam), H3K4me3 (ab8580, Abcam), H3K9Ac (07-352, Upstate), K36me2 (07-369, Upstate), H3K79me2 (05-835, Upstate). Rabbit anti-Coilin, rabbit anti-PML were a generous gift from Dr. Maureen Powers. Goat polyclonal: MBD1 (G-18, Santa Cruz), MBD2 (D-15, Santa Cruz).
Immunofluorescence and Fluorescence In Situ Hybridization
Lymphoid cell lines were washed with PBS, resuspended at a concentration of 3×106 cells/ml in PBS, and attached to slides by cytospinning at 200 rpm for 10 minutes in a Shandon Cytospin 4 cytocentrifuge. U2OS, MCF7, WI-38 and A-375 were grown directly on slides. Slides were fixed and permeabilized for 10 minutes in 4% formaldehyde/0.1% Triton-X100 in PBS before a 1 hour incubation with primary antibodies diluted in PBS-Tween/1% BSA. Detection was achieved using 1:50–1:200 dilutions of the appropriate secondary antibodies (donkey anti-rabbit IgG conjugated with AlexaFluor 488 or AlexaFluor 568, donkey anti-mouse IgG conjugated with AlexaFluor 488, AlexaFluor 568 or AlexaFluor 647, or donkey anti-goat IgG conjugated with rhodamine. AlexaFluor conjugated secondaries obtained from Invitrogen/Molecular Probes, rhodamine conjugated secondary obtained from Jackson ImmunoResearch). For experiments comparing the distributions of two rabbit polyclonal primary antibodies, one of the antibodies was conjugated to AlexaFluor 488 or AlexaFluor 568 using the Zenon Rabbit IgG labeling kit (Invitrogen/Molecular Probes). Slides were fixed again after secondary detection before incubation with Zenon conjugated antibodies. Following secondary detection, mounting media was added (Vectashield + DAPI, Vector Laboratories). Immunostaining of BrdU-pulsed cells was achieved by first fixing and staining as described above, before proceeding as described by Spector (1998).
Fluorescence In Situ Hybridization (FISH) was performed after secondary detection and fixation. Slides were denatured in 70% formamide/2× SSC for 10 min at 83°C, dehydrated with a 70–80–100% ethanol series, and then air dried. Direct labeled (rhodamine or Spectrum Orange) probes against chromosomes 1, 9, 16 and X (D1Z1, D9Z3, D16Z1 and DXZ1) were obtained from MP Biomedicals or Vysis. The probes were denatured for 5 min at 72°C (Chr 1 probe) or 83°C (Chr 9, 16 and X probes) before placing on ice, and hybridization was carried out overnight in a humid chamber at 37°C. Slides were washed twice in 50% formamide/2× SSC for 10 min at 42°C and once in 2x SSC at 42°C before addition of mounting media.
Imaging
Images were collected on a Zeiss Axiovert 200 imaging system equipped with an AxioCam MR digital camera controlled by AxioVision software. Confocal images were collected on a Zeiss LSM 510 NLO META laser-scanning confocal microscope controlled by Zeiss LSM software. Deconvolution of confocal images was performed using the Huygens 3D image deconvolution software.
Cell synchronization
Ramos cells were grown in culture to a density of 5×105 cells/ml. Aphidicolin was added to culture media at a final concentration of 1 μg/ml. Cells were cultured for twelve hours, washed with media and resuspended in media at a concentration of 3.7×105 cells/ml. Cells were then cultured in the absence of aphidicolin. After ten hours of culture, aphidicolin was again added to a final concentration of 1 μg/ml. Cells were cultured in the presence of aphidicolin for twelve additional hours. Finally, cells were washed with fresh media (release into S phase) and cultured for eight hours. An aliquot of cells was removed at each time point (0, 1.5, 3, 4.5, 6 and 8 hours after release) and cytocentrifuged onto slides.
Isolation of human germinal center B cells
Tonsil samples were processed into single cell suspension and resuspended in PBS with 2% FBS and 0.02% NaN3 (staining buffer). T cells were depleted by incubating sample with purified mouse anti-human CD4 (clone OKT4, eBioscience) and anti-CD8 (clone OKT8, eBioscience) antibodies, follow by sheep anti-mouse IgG Dynal beads (Invitrogen). T cell depleted tonsil samples were stained with PE-IgD (BD Pharmingen), PECy7-CD38 (clone HIT2, ebioscience), and Pacific Blue-CD20 (clone 2H7, eBioscience) for FACS purification of germinal center B cells. Germinal center B cells were defined as CD20+CD38+IgD− cells.
Supplementary Materials
Figure 6. NuRD bodies are observed in primary human germinal center B cells.
Germinal center B cells were isolated from human tonsil samples using FACS. Cells were then stained with antibodies against Mi-2a/b and counterstained with DAPI. Three examples are shown. Scale bar indicates 5 μm.
ACKNOWLEDGEMENTS
We thank Drs. Huntington Willard (Duke University), Judd Rice (University of Southern California), Stephen Smale (UCLA) and Maureen Powers (Emory University) for the generous gift of antibodies used in this study. We are grateful to Dr. Beth Sullivan for excellent suggestions and to Jeff Reece and the NIEHS Confocal Microscopy Center for assistance with confocal imaging. This manuscript was substantially improved by critical comments from Drs. Karen Adelman and Harriet Kinyamu. We thank the various members of the Wade, Adelman and Archer laboratories for useful discussions during the course of this work. This work was supported in part by the Intramural Research Program of the National Institute of Environmental Health Sciences (Project number Z01ES101965), NIH (PW), and by grants from the National Institutes of Health (GM073120 to BPC, DK60647 to DLJ).
REFERENCES
- Berger R, Busson-Le Coniat M. Centric and pericentric chromosome rearrangements in hematopoietic malignancies. Leukemia. 1999;13:671–678. doi: 10.1038/sj.leu.2401365. [DOI] [PubMed] [Google Scholar]
- Blanco-Betancourt CE, Moncla A, Milili M, Jiang YL, Viegas-Pequignot EM, Roquelaure B, Thuret I, Schiff C. Defective B-cell-negative selection and terminal differentiation in the ICF syndrome. Blood. 2004;103:2683–2690. doi: 10.1182/blood-2003-08-2632. [DOI] [PubMed] [Google Scholar]
- Bowen NJ, Fujita N, Kajita M, Wade PA. Mi-2/NuRD: multiple complexes for many purposes. Biochim Biophys Acta. 2004;1677:52–57. doi: 10.1016/j.bbaexp.2003.10.010. [DOI] [PubMed] [Google Scholar]
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG. Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell. 1999;3:207–217. doi: 10.1016/s1097-2765(00)80311-1. [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG. Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell. 1997;91:845–854. doi: 10.1016/s0092-8674(00)80472-9. [DOI] [PubMed] [Google Scholar]
- Busson-Le Coniat M, Salomon-Nguyen F, Dastugue N, Maarek O, Lafage-Pochitaloff M, Mozziconacci MJ, Baranger L, Brizard F, Radford I, Jeanpierre M, Bernard OA, Berger R. Fluorescence in situ hybridization analysis of chromosome 1 abnormalities in hematopoietic disorders: rearrangements of DNA satellite II and new recurrent translocations. Leukemia. 1999;13:1975–1981. doi: 10.1038/sj.leu.2401587. [DOI] [PubMed] [Google Scholar]
- Cajal SRy. Un sencillo metodo de coloracion seletiva del reticulo protoplasmatico y sus efectos en los diversos organos nerviosos de vertebrados e invertebrados. Trab Lab Invest Biol. 1903;2:129–221. [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science (New York, NY. 2002;298:1039–1043. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- Dimitrova DS, Berezney R. The spatio-temporal organization of DNA replication sites is identical in primary, immortalized and transformed mammalian cells. J Cell Sci. 2002;115:4037–4051. doi: 10.1242/jcs.00087. [DOI] [PubMed] [Google Scholar]
- Feng Q, Zhang Y. The MeCP1 complex represses transcription through preferential binding, remodeling, and deacetylating methylated nucleosomes. Genes Dev. 2001;15:827–832. doi: 10.1101/gad.876201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox MH, Arndt-Jovin DJ, Jovin TM, Baumann PH, Robert-Nicoud M. Spatial and temporal distribution of DNA replication sites localized by immunofluorescence and confocal microscopy in mouse fibroblasts. J Cell Sci. 1991;99(Pt 2):247–253. doi: 10.1242/jcs.99.2.247. [DOI] [PubMed] [Google Scholar]
- Fu XD, Maniatis T. Factor required for mammalian spliceosome assembly is localized to discrete regions in the nucleus. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Geigerman C, Akyildiz A, Mooney MR, Boss JM, Wade PA. MTA3 and the Mi-2/NuRD complex regulate cell fate during B lymphocyte differentiation. Cell. 2004;119:75–86. doi: 10.1016/j.cell.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Fujita N, Jaye DL, Kajita M, Geigerman C, Moreno CS, Wade PA. MTA3, a Mi-2/NuRD complex subunit, regulates an invasive growth pathway in breast cancer. Cell. 2003;113:207–219. doi: 10.1016/s0092-8674(03)00234-4. [DOI] [PubMed] [Google Scholar]
- Fujita N, Takebayashi S, Okumura K, Kudo S, Chiba T, Saya H, Nakao M. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–6426. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Oie HK, Kirsch IR, Hollis GF. Establishment and characterization of a human plasma cell myeloma culture having a rearranged cellular myc proto-oncogene. Blood. 1986;67:1542–1549. [PubMed] [Google Scholar]
- Guschin D, Wade PA, Kikyo N, Wolffe AP. ATP-Dependent histone octamer mobilization and histone deacetylation mediated by the Mi-2 chromatin remodeling complex. Biochemistry. 2000;39:5238–5245. doi: 10.1021/bi000421t. [DOI] [PubMed] [Google Scholar]
- Hanna MG., Jr. An Autoradiographic Study of the Germinal Center in Spleen White Pulp During Early Intervals of the Immune Response. Lab Invest. 1964;13:95–104. [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci U S A. 1999;96:14412–14417. doi: 10.1073/pnas.96.25.14412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houston SI, McManus KJ, Adams MM, Sims JK, Carpenter PB, Hendzel MJ, Rice JC. Catalytic function of the PR-Set7 histone H4 lysine 20 monomethyltransferase is essential for mitotic entry and genomic stability. J Biol Chem. 2008;283:19478–19488. doi: 10.1074/jbc.M710579200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Sif S, Jones B, Jackson A, Koipally J, Heller E, Winandy S, Viel A, Sawyer A, Ikeda T, Kingston R, Georgopoulos K. Ikaros DNA-binding proteins direct formation of chromatin remodeling complexes in lymphocytes. Immunity. 1999;10:345–355. doi: 10.1016/s1074-7613(00)80034-5. [DOI] [PubMed] [Google Scholar]
- Klein G, Giovanella B, Westman A, Stehlin JS, Mumford D. An EBVgenome-negative cell line established from an American Burkitt lymphoma; receptor characteristics. EBV infectibility and permanent conversion into EBV-positive sublines by in vitro infection. Intervirology. 1975;5:319–334. doi: 10.1159/000149930. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Liu YJ, Zhang J, Lane PJ, Chan EY, MacLennan IC. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur J Immunol. 1991;21:2951–2962. doi: 10.1002/eji.1830211209. [DOI] [PubMed] [Google Scholar]
- Lubit BW, Pham TD, Miller OJ, Erlanger BF. Localization of 5-methylcytosine in human metaphase chromosomes by immunoelectron microscopy. Cell. 1976;9:503–509. doi: 10.1016/0092-8674(76)90032-5. [DOI] [PubMed] [Google Scholar]
- Maraschio P, Tupler R, Dainotti E, Piantanida M, Cazzola G, Tiepolo L. Differential expression of the ICF (immunodeficiency, centromeric heterochromatin, facial anomalies) mutation in lymphocytes and fibroblasts. J Med Genet. 1989;26:452–456. doi: 10.1136/jmg.26.7.452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minc E, Courvalin JC, Buendia B. HP1gamma associates with euchromatin and heterochromatin in mammalian nuclei and chromosomes. Cytogenet Cell Genet. 2000;90:279–284. doi: 10.1159/000056789. [DOI] [PubMed] [Google Scholar]
- Monneron A, Bernhard W. Fine structural organization of the interphase nucleus in some mammalian cells. J Ultrastruct Res. 1969;27:266–288. doi: 10.1016/s0022-5320(69)80017-1. [DOI] [PubMed] [Google Scholar]
- Ng HH, Jeppesen P, Bird A. Active repression of methylated genes by the chromosomal protein MBD1. Mol Cell Biol. 2000;20:1394–1406. doi: 10.1128/mcb.20.4.1394-1406.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicol L, Jeppesen P. Human autoimmune sera recognize a conserved 26 kD protein associated with mammalian heterochromatin that is homologous to heterochromatin protein 1 of Drosophila. Chromosome Res. 1994;2:245–253. doi: 10.1007/BF01553325. [DOI] [PubMed] [Google Scholar]
- O'Keefe RT, Henderson SC, Spector DL. Dynamic organization of DNA replication in mammalian cell nuclei: spatially and temporally defined replication of chromosome-specific alpha-satellite DNA sequences. J Cell Biol. 1992;116:1095–1110. doi: 10.1083/jcb.116.5.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo SE, Almouzni G. Chromatin assembly: a basic recipe with various flavours. Curr Opin Genet Dev. 2006;16:104–111. doi: 10.1016/j.gde.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Quivy JP, Roche D, Kirschner D, Tagami H, Nakatani Y, Almouzni G. A CAF-1 dependent pool of HP1 during heterochromatin duplication. Embo J. 2004;23:3516–3526. doi: 10.1038/sj.emboj.7600362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saurin AJ, Shiels C, Williamson J, Satijn DP, Otte AP, Sheer D, Freemont PS. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer JR, Swanson CM, Koller MA, North PE, Ross SW. Centromeric instability of chromosome 1 resulting in multibranched chromosomes, telomeric fusions, and “jumping translocations” of 1q in a human immunodeficiency virus-related non-Hodgkin's lymphoma. Cancer. 1995;76:1238–1244. doi: 10.1002/1097-0142(19951001)76:7<1238::aid-cncr2820760722>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Sawyer JR, Tricot G, Mattox S, Jagannath S, Barlogie B. Jumping translocations of chromosome 1q in multiple myeloma: evidence for a mechanism involving decondensation of pericentromeric heterochromatin. Blood. 1998;91:1732–1741. [PubMed] [Google Scholar]
- Sewalt RG, Lachner M, Vargas M, Hamer KM, den Blaauwen JL, Hendrix T, Melcher M, Schweizer D, Jenuwein T, Otte AP. Selective interactions between vertebrate polycomb homologs and the SUV39H1 histone lysine methyltransferase suggest that histone H3-K9 methylation contributes to chromosomal targeting of Polycomb group proteins. Mol Cell Biol. 2002;22:5539–5553. doi: 10.1128/MCB.22.15.5539-5553.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JK, Houston SI, Magazinnik T, Rice JC. A trans-tail histone code defined by monomethylated H4 Lys-20 and H3 Lys-9 demarcates distinct regions of silent chromatin. J Biol Chem. 2006;281:12760–12766. doi: 10.1074/jbc.M513462200. [DOI] [PubMed] [Google Scholar]
- Sobel RE, Cook RG, Perry CA, Annunziato AT, Allis CD. Conservation of deposition-related acetylation sites in newly synthesized histones H3 and H4. Proc Natl Acad Sci U S A. 1995;92:1237–1241. doi: 10.1073/pnas.92.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spector DL, Goldman RD, Leinwand LA. Cells: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1998. [Google Scholar]
- Taddei A, Roche D, Sibarita JB, Turner BM, Almouzni G. Duplication and maintenance of heterochromatin domains. J Cell Biol. 1999;147:1153–1166. doi: 10.1083/jcb.147.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voncken JW, Schweizer D, Aagaard L, Sattler L, Jantsch MF, van Lohuizen M. Chromatin-association of the Polycomb group protein BMI1 is cell cycle-regulated and correlates with its phosphorylation status. J Cell Sci. 1999;112(Pt 24):4627–4639. doi: 10.1242/jcs.112.24.4627. [DOI] [PubMed] [Google Scholar]
- Wade PA, Gegonne A, Jones PL, Ballestar E, Aubry F, Wolffe AP. Mi-2 complex couples DNA methylation to chromatin remodelling and histone deacetylation. Nat Genet. 1999;23:62–66. doi: 10.1038/12664. [DOI] [PubMed] [Google Scholar]
- Wade PA, Jones PL, Vermaak D, Wolffe AP. A multiple subunit Mi-2 histone deacetylase from Xenopus laevis cofractionates with an associated Snf2 superfamily ATPase. Curr Biol. 1998;8:843–846. doi: 10.1016/s0960-9822(98)70328-8. [DOI] [PubMed] [Google Scholar]
- Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- Wreggett KA, Hill F, James PS, Hutchings A, Butcher GW, Singh PB. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]
- Xu GL, Bestor TH, Bourc'his D, Hsieh CL, Tommerup N, Bugge M, Hulten M, Qu X, Russo JJ, Viegas-Pequignot E. Chromosome instability and immunodeficiency syndrome caused by mutations in a DNA methyltransferase gene. Nature. 1999;402:187–191. doi: 10.1038/46052. [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Cote J, Wang W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell. 1998;2:851–861. doi: 10.1016/s1097-2765(00)80299-3. [DOI] [PubMed] [Google Scholar]
- Zhang J, MacLennan IC, Liu YJ, Lane PJ. Is rapid proliferation in B centroblasts linked to somatic mutation in memory B cell clones? Immunol Lett. 1988;18:297–299. doi: 10.1016/0165-2478(88)90178-2. [DOI] [PubMed] [Google Scholar]
- Zhang Y, LeRoy G, Seelig HP, Lane WS, Reinberg D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell. 1998;95:279–289. doi: 10.1016/s0092-8674(00)81758-4. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.