Abstract
Biotin synthase (BioB) is an iron-sulfur enzyme that catalyzes the last step in biotin biosynthesis, the insertion of sulfur between the C6 and C9 carbons of dethiobiotin to complete the thiophane ring of biotin. Recent in vitro experiments suggest that the sulfur is derived from a [2Fe-2S]2+ cluster within BioB, and that the remnants of this cluster dissociate from the enzyme following each turnover. In order for BioB to catalyze multiple rounds of biotin synthesis, the [2Fe-2S]2+ cluster in BioB must be reassembled, a process that could be carried out in vivo by the ISC or SUF iron-sulfur cluster assembly systems. The bacterial ISC system includes HscA, an Hsp70-class molecular chaperone, whose yeast homolog has been shown to play an important but nonessential role in assembly of mitochondrial FeS clusters in S. cerevesiae. In the present work we show that in E. coli, HscA significantly improves the efficiency of the in vivo assembly of the [2Fe-2S]2+ cluster on BioB under conditions of low to moderate iron. In vitro, we show that HscA binds with increased affinity to BioB missing one or both FeS clusters, with a maximum of two HscA molecules per BioB dimer. BioB binds to HscA in an ATP/ADP-independent manner and a high affinity complex is also formed with a truncated form of HscA that lacks the nucleotide binding domain. Further, the BioB:HscA complex binds the FeS cluster scaffold protein IscU in a noncompetitive manner, generating a complex that contains all three proteins. We propose that HscA plays a role in facilitating the transfer of FeS clusters from IscU into the appropriate target apoproteins such as biotin synthase, perhaps by enhancing or prolonging the requisite protein:protein interaction.
Biotin synthase (BioB)1 is an AdoMet radical enzyme that catalyzes the formation of the thiophane ring of biotin (1). Active biotin synthase contains both a [4Fe-4S]2+ and a [2Fe-2S]2+ cluster (2Fe4Fe-BioB), each with a unique role in catalysis (2-4). The [4Fe-4S]2+ cluster is coordinated by the methionyl group of AdoMet (5) and facilitates reduction of the AdoMet sulfonium, generating methionine and a 5′-deoxyadenosyl radical (6). In our working model, this radical then abstracts a hydrogen atom from the substrate dethiobiotin (7), generating a substrate-centered carbon radical. We then propose that the [2Fe-2S]2+ cluster provides a sulfur atom that quenches the substrate radical through formation of a new carbon-sulfur bond (8), leading to formation of 9-mercaptodethiobiotin as a discrete intermediate (9). A similar sequence of events leads to formation of the second C-S bond, and biotin and the remnants of the [2Fe-2S]2+ cluster dissociate from the enzyme (3, 10), leaving an inactive protein containing only the [4Fe-4S]2+ cluster (4Fe-BioB). In this state, the remaining FeS cluster is extremely oxygen sensitive and may be rapidly oxidized and degraded (11), most likely resulting in the formation of a mixture of 4Fe-BioB and apoBioB following each turnover.
The minimum biotin requirements of E. coli are low, on the order of 100 – 1000 biotin molecules per cell (12), and it is possible that BioB may have evolved to undergo only a single turnover. However, measurements of biotin production and recombinant BioB levels in E. coli indicate that each BioB polypeptide chain is capable of up to 20 turnovers prior to degradation (13). In order for BioB to be returned to catalysis following each turnover, minimally the [2Fe-2S]2+ cluster, and perhaps also the [4Fe-4S]2+ cluster, must be reassembled and/or repaired. Of course, this process must also occur during the initial production of new BioB molecules, prior to the first round of catalysis. In vitro, the [4Fe-4S]2+ cluster is easily reconstituted using Fe3+, S2-, and dithiothreitol under strict anaerobic conditions (2, 14, 15). In contrast, the [2Fe-2S]2+ cluster is only partially reconstituted during prolonged aerobic dialysis with the same reagents (16, 17), and this chemically reconstituted [2Fe-2S]2+ cluster exhibits altered spectroscopic features (16) and has significantly less activity (17) than the native cluster generated within the cell. In E. coli, both FeS clusters are likely generated and/or repaired through the actions of the iron-sulfur cluster (ISC and/or SUF) assembly systems.
The various components of the ISC iron-sulfur cluster assembly system were first identified in Azotobacter vinlandii as homologs of the Nif genes required for generating the active FeS clusters in nitrogenase (18, 19). The ISC system has now been identified in most eukaryotes and eubacteria, and has been most extensively studied in S. cerevisiae and E. coli. The ISC system includes a cysteine desulfurase (IscS) that provides persulfide sulfur (S0) and possibly also sulfide (S2-) for FeS cluster assembly, two potential FeS cluster assembly scaffold proteins (IscU and IscA) that bind both [2Fe-2S]2+ and [4Fe-4S]2+ clusters, a small ferredoxin (aka. adrenodoxin) that may be involved in redox equilibration during cluster assembly, and an ATP-dependent chaperone/co-chaperone pair (HscA/HscB in E. coli, Ssq1/Jac1 in S. cerevisiae) (20). The manner in which these components interact to facilitate cluster assembly and delivery is the subject of active investigation in many laboratories.
HscA (heat shock cognate) is a homolog of the Hsp70 family of molecular chaperones, all of which have a molecular weight around 70 kDa and participate in various aspects of protein folding, including stabilization-degradation, multimer assembly-disassembly, membrane translocation, and maintaining cellular viability during stress conditions (21). In addition, most Hsp70 chaperones are heat shock proteins that are upregulated in response to cellular stress, most notably following thermal perturbation. However, HscA differs from other Hsp70 chaperones in that it is constitutively expressed in E. coli at relatively high levels under non-stress conditions and is not increased significantly during heat shock (22, 23). This implies that HscA has an important general housekeeping function within the cell. Cellular concentrations of HscA in E. coli have been estimated at ∼20 μM and comprise ∼1 % of the total cytosolic protein under normal growth conditions (23).
The specific role of HscA in E. coli remains unknown, as HscA mutants do not show a specific growth phenotype. However, mutation of the HscA homolog Ssq1 in S. cerevisiae results in defects in mitochondrial FeS cluster assembly, including a decrease in FeS clusters on Bio2 (BioB) (24), suggesting a role in FeS cluster assembly that may be partially rescued by other chaperones (25). In vitro studies demonstrate that HscA is an ATP-dependent chaperone that can form a specific complex with the co-chaperone HscB (26), as well as with the FeS cluster assembly scaffold protein IscU (27). In each case, protein binding results in stimulation of the ATPase activity of HscA (26, 28), and IscU preferentially forms a tight protein complex with the ADP-bound form of HscA (26). A specific peptide loop from IscU has been identified that binds into the canonical Hsp70 peptide binding site (29), and a structure has been determined of the HscA substrate binding domain with this IscU peptide bound (30). Based upon this data, a model for the role of HscA in FeS cluster assembly has been proposed in which HscA binds IscU and facilitates cluster assembly and/or cluster release from the scaffold protein (20).
In the present work, we present evidence that the role of HscA may be more complex. Using E. coli strains in which individual genes in the isc operon have been deleted, we demonstrate that for recombinant BioB, both the FeS cluster content and the total protein yield are significantly decreased when HscA is not present in the cell. In vitro, we use a molecular weight filtration binding assay to demonstrate that HscA forms a complex with all reconstituted states of BioB, but shows a preference towards binding apoBioB. Formation of the HscA:BioB complex is independent of MgATP and MgADP, is not ionic strength dependent, and a complex is also formed with a truncated form of HscA that contains only the substrate binding domain. The stoichiometry of this complex is maximally 2:1 HscA:BioB dimer as judged by equilibrium analytical centrifugation. Finally, we demonstrate that both IscU and BioB bind to HscA simultaneously and do not compete for the same site. This suggests a novel role for HscA in promoting an interaction of IscU with apoBioB, and perhaps with other incipient iron-sulfur proteins.
Materials and Methods
Materials
All reagents were purchased from commercial sources and used without further purification. His6-BioB was prepared as previously described (15), and the hexahistidine tag was not removed for the experiments described. The strain DH5α/pTrcHsc66 (23) was obtained from Dr. Larry E. Vickery (University of California, Irvine), and HscA was expressed and purified as previously described (23) except that the final purification step involved running HscA through a Sephacryl S-300 size exclusion column (1.6×50 cm, Pharmacia). Plasmid pTrcIscU, also obtained from Dr. Vickery, was transformed into BL21(DE3)pLysS (Novagen), and IscU was overexpressed and purified as previously described (27).
The concentration of aerobically purified BioB was estimated using ε452 = 8,400 M-1 cm-1, otherwise protein concentrations were determined using the Bradford assay with a commercial BSA standard (BioRad). Protein samples were subjected to iron analysis with bathophenanthroline and sulfide analysis with N,N-dimethyl-p-phenylenediamine using previously described modified methods (2) with ratios reported representing the average and standard deviation for 5 samples.
ApoBioB and 2Fe4Fe-BioB Preparation
Aerobic purification yields BioB containing one [2Fe-2S]2+ cluster per monomer (2Fe-BioB). BioB apoprotein (apoBioB) was prepared by anaerobic reduction of this cluster with sodium dithionite in the presence of EDTA, as previously described (31). Circular dichroism spectra and analytical centrifugation indicate that this apoprotein remains a dimer and retains ∼90 % of the secondary structure found in the active enzyme. BioB containing both [2Fe-2S]2+ and [4Fe-4S]2+ clusters (2Fe-4Fe-BioB) was prepared by anaerobic reconstitution with Fe3+, S2-, and dithiothreitol (2), followed by addition of a 4-fold excess of AdoMet and 2-fold excess of dethiobiotin, which help stabilize the [4Fe-4S]2+ cluster against spurious oxidation (11). For both apoBioB and 2Fe4Fe-BioB, excess reagents were removed by anaerobic desalting using Sephadex G-25 (1 × 30 cm, Amersham). The protein solution was then transferred to a septum-covered cuvette and a UV/visible spectrum taken to assess the extent of cluster removal or reconstitution.
Molecular Weight Filtration Assay
The formation of a complex between BioB and HscA was examined using a molecular weight filtration assay. In each case, a filter was selected that would allow the smallest protein to pass through the filter, but that would not allow the protein:protein complex to pass. Examination of the filtrate by SDS polyacrylamide gel electrophoresis allows a determination of the free concentration of the smaller protein in the presence of an increasing concentration of the larger protein. For example, varying amounts of BioB (0 – 30 μM) were added to a constant amount of HscA (5 μM) in 50 mM Tris HCl, 100 mM NaCl, 1 mM TCEP, pH 7.5 (210 μl total volume). Samples were then loaded into a 0.5 ml Millipore Biomax Ultrafree 100 KDa filter and spun at 6,000 ×g for 15 – 30 sec until 20 – 25 μl had passed through the filter. Protein samples were separated on a 15% SDS polyacrylamide gel and stained with Coomassie brilliant blue R250. The gels were then destained to a very low background, optically scanned at high resolution, and the band intensity quantified using NIH Image (http://rsb.info.nih.gov/nih-image). The raw data presents as a decreasing hyperbola that is dependent on [HscAfree]. Using an HscA standard run in the absence of BioB (HscAtotal), the data were normalized, the data from 3 – 5 experiments were averaged, and the resulting data set fit to a quadratic binding isotherm:
| (equation 1) |
where [S] is the concentration of BioB and [E] is the concentration of HscA.
Binding of HscA and IscU was examined in a similar manner except that HscA (0 – 50 μM) was added to a constant amount of IscU (5 μM), and free IscU was separated by brief centrifugation using a 0.5 ml Millipore Biomax Ultrafree 50 KDa filter. In addition, to generate sharper bands for IscU, protein samples were separated by SDS polyacrylamide gel electrophoresis on 4 – 20 % gradient gels (BioRad) prior to band quantitation as described above.
Equilibrium Analytical Ultracentrifugation
All analytical ultracentrifugation experiments were performed on a Beckman XL-A Analytical Ultracentrifuge using an An60 Ti rotor. The relative concentrations of the bound and unbound forms of BioB were determined for 2Fe-BioB alone and for 2Fe-BioB mixed with HscA using equilibrium analytical ultracentrifugation. Six-channel, epon, charcoal-filled centerpieces containing 100 μl of either 7.5 μM 2Fe-BioB alone or 7.5 μM 2Fe-BioB mixed with 15 μM HscA in 50 mM Tris HCl, 100 mM NaCl, 1 mM TCEP, pH 7.5, and were subjected to centrifugation at speeds of 10,000, 12,000, and 15,000 rpm at 20 °C. Samples were equilibrated at the lowest speed for 12 hours before a scan was taken, and two hours later a duplicate scan was collected to ensure equilibration. For each successive higher speed, the samples were equilibrated for 8 hours before the first scan, followed by a second scan after 2 hours. Each individual scan is the average of 5 replicate radial absorbance scans at 452 nm. The absorbance data at 3 speeds were globally fit using the program Sedphat (available at http://www.analyticalultracentrifugation.com/sedphat) using the “Species analysis with mass conservation constraints” model (32). Approximate buoyant molecular weight values used as initial parameters in the fit were determined by velocity centrifugation experiments (data not shown).
Expression of His6-BioB in ISC deletion strains
The in vivo effect of each ISC protein on the assembly of the [2Fe-2S]2+ cluster in BioB was determined by expression of recombinant His6-BioB in strains from the E. coli Keio knockout collection. A plasmid was constructed in which His6-BioB is expressed under control of the arabinose-inducible promoter araB. The plasmid pJJ15-4A (15) was digested with XbaI and the ends blunted with Klenow polymerase. The resulting linear plasmid was digested with HindIII and the smaller fragment purified by agarose gel electrophoresis to provide the His6-BioB insert. The plasmid pGro7 (Takara Bio, Inc.) was digested with XhoI and the ends blunted with Klenow polymerase. The resulting linear plasmid was digested with HindIII and the larger fragment purified by agarose gel electrophoresis. The extracted pGro7-derived plasmid was ligated to the His6-BioB insert with T4 DNA ligase and transformed into DH5α. The resulting plasmid (pDE93) was reisolated and sequenced to verify that the correct insertion and orientation had been obtained.
Strains in which specific genes in the isc operon are replaced with a kanamycin resistance cassette (33) were obtained from the E. coli Keio knockout strain collection (34) at the Yale Coli Genetic Stock Center. Strains used in this study include the K12-derived WT strain BW25113, and knockout strains JW2510 (ΔhscA), JW2511 (ΔhscB), and as a control, JW0013 (ΔdnaK). The plasmid pDE93 was transformed into each strain and expression of His6-BioB in LB media following induction with 0.4% arabinose was confirmed by SDS polyacrylamide gel electrophoresis and Western blot analysis. Analysis of iron-sulfur cluster assembly was conducted in a defined glucose-casamino acid-M9 media that contained: M9 salts adjusted to pH 7.4, glucose (0.25 %), casamino acids (0.4 %), MgSO4 (1 mM), CaCl2 (0.1 mM), an iron-depleted trace metal supplement (35), thiamine (1 μg/ml), niacin (5 μg/ml), lipoic acid (1 μg/ml), uracil (5 μg/ml), FeCl3 (5 μM), chloramphenicol (34 μg/ml), and for each knockout strain, kanamycin (25 μg/ml). Each strain was initially grown on LB agar with chloramphenicol (34 μg/ml), and for knockout strains, kanamycin (25 μg/ml). A single colony was inoculated into a 10 ml glucose-casamino acid-M9 culture and grown overnight to saturation at 37 °C with vigorous agitation. Fresh media (1 L) was inoculated with 5 ml of the overnight culture and grown at 37 °C with agitation to an OD600 = 0.6 (about 6 h). Protein expression was induced by the addition of arabinose (0.5 %) and continued for 3 h at 37 °C with continued agitation. The cultures were pelleted by centrifugation, the cell pellets resuspended and washed with ice cold 50 mM Tris HCl, pH 8, and the cell pellets (∼2.5 g each) frozen at -80 °C overnight.
The cells were thawed and resuspended in 30 ml of cold 50 mM Tris HCl, pH 8, lysozyme was added to 1 mg/ml and the suspension incubated on ice for 15 min, and then the suspension was subjected to repeated sonication at 0 °C to lyse the cells. The crude mixture was subjected to centrifugation for 30 min at 25,000 ×g, and the soluble crude protein fraction (∼27 mL) was separated from the insoluble cell debris. The protein concentration was determined for each sample (∼4-5 mg/ml) and a volume containing 100 mg of total soluble crude protein was continued through purification, while a small sample of the crude protein was reserved for Western blot analysis. Each protein sample was loaded onto a Ni-NTA-agarose column (10 ml, Qiagen) that had been equilibrated in 20 mM imidazole, 0.5 M NaCl, 50 mM Tris HCl, pH 8, and the bound protein washed with ∼150 ml of the same buffer until a stable baseline had been achieved. The pure protein was eluted with 200 mM imidazole, 0.5 M NaCl, 50 mM Tris HCl, pH 8 in a total volume of 15 ml, concentrated in a 30 kD centrifugal concentrator, diluted in 15 mL of 50 mM Tris HCl, pH 8, and reconcentrated to a volume of 1.00 ml. For each sample, a UV/visible spectrum was collected and then an accurate determination of the polypeptide, Fe2+/3+, and S2- concentration was performed.
Results
The BioB-catalyzed formation of biotin is accompanied by the dissociation of the remnant [2Fe-1S]2+ cluster (3, 8) and additional turnovers of the enzyme likely require the repair or reassembly of the essential [2Fe-2S]2+ cluster. In vivo these assembly and repair processes are most likely catalyzed by the ISC and/or SUF iron-sulfur cluster assembly systems (18, 36). We reasoned that an interaction with the ISC system would most likely occur either through direct interaction with the scaffold proteins IscU and/or IscA, or might be mediated by the chaperone proteins HscA and/or HscB. A preliminary screen of these proteins provided no evidence for a moderate-affinity (Kd < 10 μM) complex with IscU, IscA, or HscB, but suggested a complex is formed with HscA. Initial attempts to characterize this interaction by isothermal titration calorimetry suggested the interaction had a very low enthalpy of binding and would not be amenable to this technique, while attempts to use Biacore surface plasmon resonance provided evidence for a moderate affinity complex with rapid on- and off-rates, but failed to provide definitive quantitative results. Due to the large molecular weight difference between monomeric HscA (66 kDa) and the potential complex(es) formed (≥148 kDa), we reasoned that a molecular weight separation technique that would allow rapid separation of free and bound proteins could provide interpretable equilibrium binding data.
HscA Forms a Complex with ApoBioB
Formation of a complex between HscA and apoBioB was examined by addition of an increasing concentration of apoBioB to a fixed concentration of HscA, followed by separation of free HscA from the bulk sample using a 100 kDa centrifugal concentrator. Unbound monomeric HscA passed through the filter and was visualized by SDS polyacrylamide gel electrophoresis with Coomassie stain. The amount of free HscA was determined by digitally scanning the gel bands, integrating the pixel intensities using NIH Image, and comparing to HscA standards of known concentration. This MW filtration method provides a convenient method for sampling the free protein concentration without significantly altering the bulk sample volume or concentrations, and therefore the data can be analyzed using standard equilibrium binding equations.
ApoBioB (0 – 30 μM final concentration) and HscA (5 μM) were mixed in 50 mM Tris HCl, 100 mM NaCl, 1 mM TCEP, pH 7.5 and a sample removed to verify the total protein concentrations (Figure 1A). The sample was then subjected to brief centrifugation in a 100 kDa MW cutoff centrifugal concentrator, until ∼10% of the initial volume had been collected. This sample contains free HscA (65.5 kDa, filter was >95% permissive) and some of the free BioB dimer (82.4 kDa, filter was only ∼70% permissive, not shown) from the initial samples (Figure 1B). Using control HscA samples run in the absence of apoBioB, the pixel intensity was converted to a concentration and normalized to reflect the fractional saturation of the HscA:BioB complex. This data was fit with a quadratic binding isotherm to give a Kd = 1.3 ± 0.4 μM (Figure 1C, filled circles). In addition, due to the relatively tight binding observed, we can use the curve fit to estimate that 4.8 μM of apoBioB dimer is required to saturate 5.0 μM HscA, or a 0.96:1 binding ratio. However, since the cutoff filter would hold back a 1:1 HscA:BioB complex (148 kDa) and also any larger complexes, we cannot rule out a larger complex based on this data.
Figure 1.
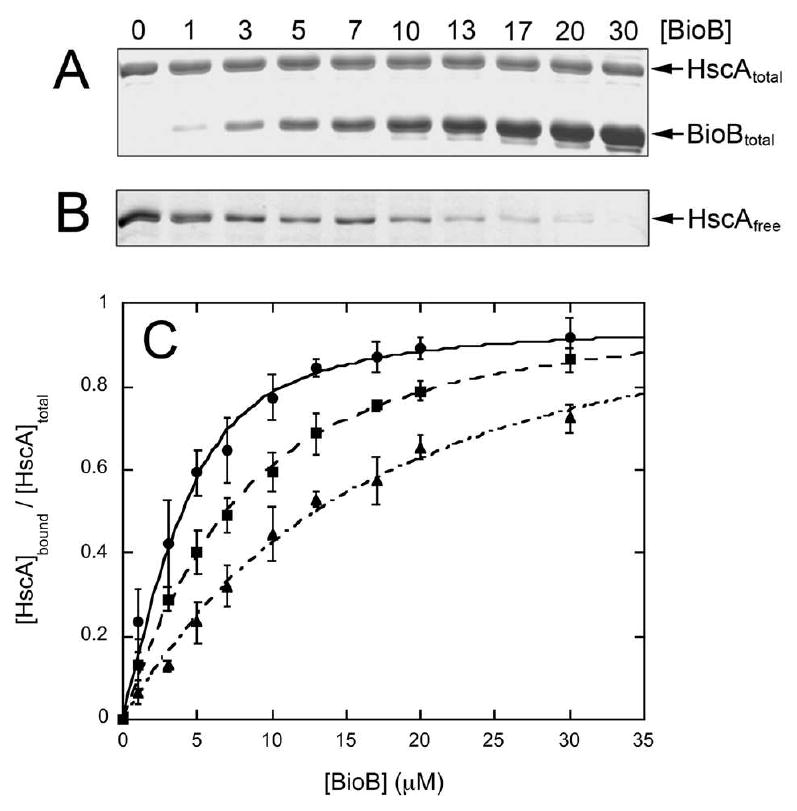
Formation of a complex between HscA and apoBioB, 2Fe-BioB, and 2Fe4Fe-BioB in 50 mM Tris.HCl (pH 7.5), 100 mM NaCl, and 1 mM TCEP. (A) Total HscA (5 μM) and apoBioB (0 – 30 μM) in the initial mixture, as detected by SDS polyacrylamide gel electrophoresis with Coomassie R250 stain. (B) Free HscA that passes through a 100 kDa centrifugal concentrator, as detected by SDS polyacrylamide gel electrophoresis. (C) Fractional saturation of the HscA:BioB complex, calculated from total and free HscA concentrations. Data shown for apoBioB (●), 2Fe-BioB (■), and 2Fe4Fe-BioB (▲) are an average of three trials each. Data are fit to a quadratic binding isotherm as described in the Materials and Methods, yielding values of KdapoBioB = 1.3 μM, Kd2Fe-BioB = 4.7 μM, and Kd2Fe4Fe-BioB = 12.9 μM.
While the MW filtration assay seemed a simple and straightforward analytical method for measuring binding to HscA, we had no independent method for verifying the binding constants obtained for BioB. However, the interaction of HscA with IscU has been studied directly using Biacore surface plasmon resonance by Vickery and coworkers: Kd = 9.3 μM (28), as well as indirectly through the IscU stimulation of the HscA ATPase activity: Km = 33.7 μM (27). As a control to verify the accuracy of our method, we examined the formation of a complex between apoIscU (10 μM dimer) and HscA (0 – 50 μM). Since the IscU dimer (27.4 kDa) is the smaller of the two proteins, we used a 50 kDa MW cutoff centrifugal concentrator and examined depletion of the concentration of free IscU as the concentration of HscA was increased (Figure 2). We observe the formation of a complex with an affinity similar to that previously reported, KdIscU = 10.3 ± 2.5 μM.
Figure 2.
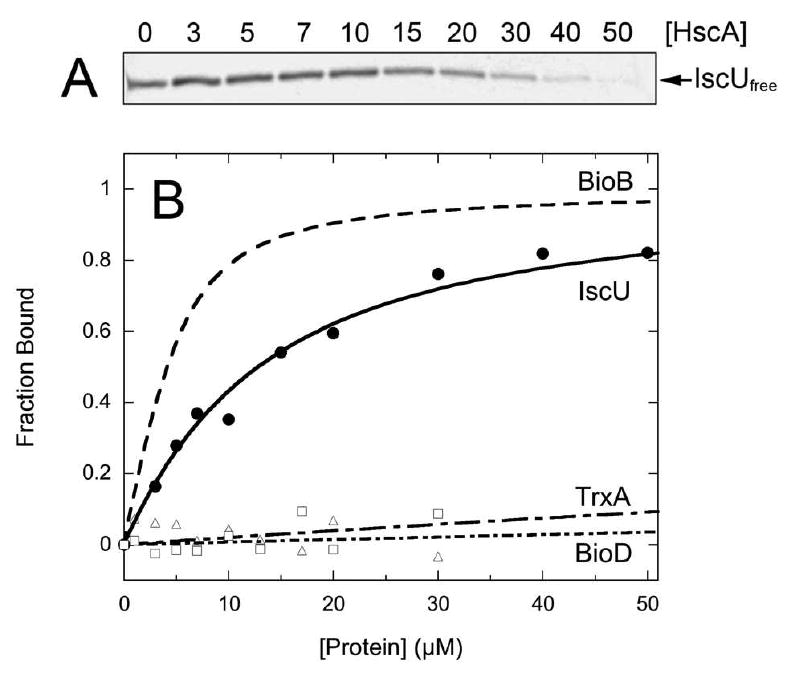
Formation of a complex between IscU (5 μM) and HscA (0 – 50 μM) in 50 mM Hepes (pH 7.5), 150 mM NaCl, 10 mM MgCl2, 1 mM ADP, and 1 mM DTT. (A) Free IscU that passes through a 50 kDa centrifugal concentrator, as detected by SDS polyacrylamide gel electrophoresis on a 4-20 % gradient gel with Coomassie R250 stain. (B) Fractional saturation of the HscA:IscU complex (●), calculated from total and free IscU concentrations. Data are fit to a quadratic binding isotherm, yielding a value of KdIscU = 10.3 μM. Also shown are controls in which thioredoxin (TrxA, 5 μM, □) or BioD (5 μM, △) were titrated with HscA (0 – 30 μM). From these data we can estimate a lower limit for indiscriminate binding (Kd ≥ 500 μM). The binding curve for apoBioB from Figure 1 is also reproduced for reference.
Although typical Hsp70 chaperones bind to numerous peptide and protein substrates (21), HscA is atypical in that it has high sequence specificity for binding a loop from IscU in the canonical Hsp70 peptide binding site (29). HscA also binds the cochaperone HscB and, as demonstrated in this work, binds BioB at a new site. Since this new protein binding site was previously unrecognized and the specificity is unknown, we examined whether HscA could bind to other proteins that were readily available in our lab, including the other enzymes from the biotin biosynthetic pathway, flavodoxin and flavodoxin:NADP+ oxidoreductase (used in the BioB assay), thioredoxin and thioredoxin reductase, and biotin protein ligase. We found no evidence that HscA forms a high affinity complex with these proteins. Two representative examples are shown in Figure 2B. E. coli thioredoxin (TrxA) is a 14 kD monomeric protein involved in disulfide reduction, and dethiobiotin synthase (BioD) is a 24 kD monomeric enzyme that utilizes ATP to close the biotin ureido ring. We can estimate a lower limit on the affinity of HscA for these proteins at KdTrxA ≥ 500 μM and KdBioD ≥ 1100 μM.
The Affinity of the HscA:BioB Complex is Increased by the Absence of FeS Clusters
One possible purpose for the formation of a complex between HscA and BioB could be to identify and stabilize apoBioB and perhaps to enhance the delivery of FeS clusters from other components of the ISC system. In this scenario, HscA might be expected to preferentially bind apBioB as compared with protein forms that contain FeS clusters. We repeated the MW filtration binding assay as described above, but using 2Fe-BioB and 2Fe4Fe-BioB, where the latter is the correctly assembled holoprotein containing the full complement of FeS clusters. For 2Fe4Fe-BioB, the binding and filtration experiments were performed in a nitrogen-filled glove box to minimize oxidation of the [4Fe-4S]2+ cluster. We observed formation of a complex between HscA and both forms of BioB (Figure 1C), but with decreased affinity as compared to apoprotein: 2Fe-BioB, Kd = 4.7 ± 1.0 μM, and 2Fe4Fe-BioB, Kd = 12.9 ± 3.0 μM. In both cases, the complex is again saturated at an approximate 1:1 ratio of HscA to BioB dimer.
Equilibrium Analytical Ultracentrifugation Characterization of the BioB:HscA Complex
The MW filtration assay provided a convenient method for determining the affinity of the HscA:BioB complex, but this method could not distinguish between a 1:1 complex and higher order complexes. To distinguish these possibilities, we used equilibrium analytical ultracentrifugation to examine HscA alone, 2Fe-BioB alone, and a mixture containing 2Fe-BioB (7.5 μM monomer) and a 2-fold excess of HscA (15 μM) in 50 mM Tris HCl, 100 mM NaCl, 1 mM TCEP, pH 7.5. The radial distribution of HscA alone (15 μM) was measured by radial absorbance scans at 280 nm and the data at 3 different centrifugation speeds were globally fit to a single protein component with MWexp = 66,515 Da (data not shown, MWcalc = 65,521 Da) and this experimental value was used for HscA in fitting the HscA:BioB complex. The radial distribution of 2Fe-BioB alone (7.5 μM monomer) was measured by radial absorbance scans at 452 nm (Figure 3A), and the data were fit to a nonequilibrating mixture of BioB dimer with MWexp = 90,680 Da (90.5 % of the total absorbance, MWcalc = 82,630 Da) and BioB tetramer with MWexp = 173,860 Da (9.5 % of the total absorbance, MWcalc = 165,260 Da). The nonphysiologic tetrameric form of BioB had previously been detected by gel filtration chromatography and is likely due to nonreducible covalent crosslinks between two molecules of the native dimeric protein.
Figure 3.
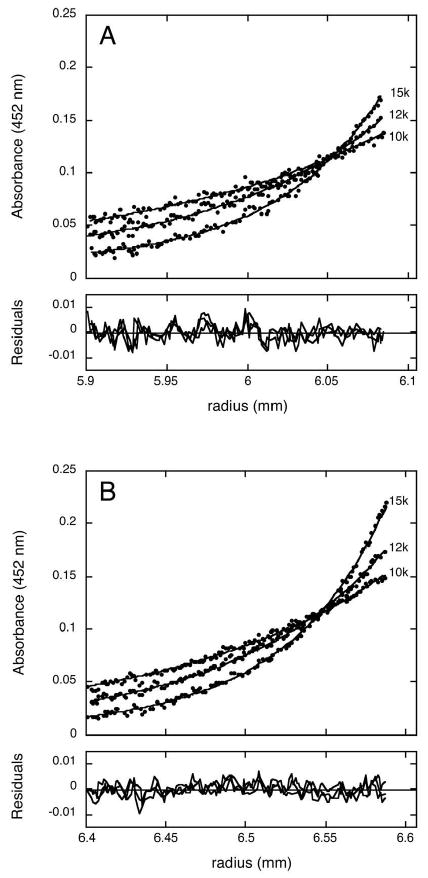
Characterization of the BioB:HscA complex using equilibrium analytical ultracentrifugation. Protein samples were equilibrated in 50 mM Tris HCl (pH 7.5), 100 mM NaCl, 1 mM TCEP, centrifuged at speeds of 10,000, 12,000, and 15,000 rpm for 8 h, and radial absorbance scans were collected at 452 nm. At this wavelength, only protein species that include BioB are detected. (A) 2Fe-BioB (7.5 μM). The data are fit to a two-component model with M1 = 90,680 (90.5 % of the mixture) and M2 = 173,860. (9.5 %), corresponding to BioB2 and BioB4, respectively. (B) 2Fe-BioB (7.5 μM) and HscA (15 μM). The data are fit to a three-component model with M1 = 81,410 (50.6 %), M2 = 139,980 (33.3 %), and M3 = 217,920 (16.1 %), corresponding to BioB2, BioB2:HscA, and BioB2:HscA2 respectively.
The equilibrium analytical ultracentrifugation of a mixture of 2Fe-BioB (7.5 μM monomer) and HscA (15 μM) was monitored by radical absorbance at 452 nm (Figure 3B); at this wavelength HscA alone is spectroscopically invisible and only protein complexes that include 2Fe-BioB with or without HscA need be included in fitting the data. The data at 3 centrifugation speeds were globally fit to a 3 component model: component 1, 81,410 Da, 50.6% of the total absorbance; component 2, MWexp = 139,980 Da, 33.3% of the total; and component 3, MWexp = 217,920 Da, 16.1% of the total (rmsd = 0.002603, runs test Z = 6.16, Figure 3B). Our interpretation of these components is that they represent dimeric BioB (MWcalc = 82,630 Da), dimeric BioB with 1 equivalent of HscA bound (MWcalc = 148,150 Da), and dimeric BioB with 2 equivalents of HscA bound (MWcalc = 213,670 Da). In addition, this mixture likely also contains minor amounts of tetrameric BioB with and without HscA bound, although we have neglected these components in the present fit. We also attempted to fit the data in Figure 3B using a two-component model: component 1, MWexp = 81,410 Da, 44.6 % of the total; and component 2, MWexp = 148,125 Da, 55.4 % of the total (rmsd = 0.002604, runs test Z = 6.42). However, the higher runs test Z-value suggests that for this model the nonrandom variation in residuals is likely due to a missing component, in this case a high MW component that likely corresponds to BioB dimer with 2 equivalents of HscA bound. Finally, as a control we also attempted to model the data to a mixture of BioB dimer (90 %) and BioB tetramer (10 %) with no HscA interaction, but found the fit to be significantly worse (rmsd = 0.003203, runs test Z = 8.84), demonstrating that major components were not accounted for and that addition of HscA to 2Fe-BioB was clearly generating one or more high molecular weight BioB:HscA complexes.
Assuming that the 3-component fit best models the mixture, we can use an independent-site binding model to estimate the binding constants necessary to generate the ratio of species observed in Figure 3B. Assuming that HscA binds with equal affinity to each monomer of the BioB dimer, we can approximately model the ratio of components in the centrifugation data with a Kd = 12 μM (51 %, 31 %, and 19% for each component). Alternatively, if we assume that the first and second equivalents of HscA bind with different affinities, we can generate a slightly improved model for the ratio of components with Kd1 = 10 μM and Kd2 = 15 μM (51 %, 33 %, and 16 % for each component). These affinities are slightly higher than those determined by MW filtration analysis (Kd = 4.4 μM), but certainly within the overall error of the multiple data analysis steps required for this comparison.
Formation of the BioB:HscA Complex Requires Only the HscA Substrate Binding Domain
Based upon homology to other Hsp70 chaperones, HscA likely consists of two discrete domains: a nucleotide-binding domain (residues 1-378) is connected via a short helical linker to a substrate binding domain (residues 391-616) (37). Vickery and coworkers have demonstrated that the HscA substrate binding domain forms a complex with IscU, and have solved the structure of this domain in complex with an IscU-derived peptide (30). In order to determine which HscA domain BioB interacts with, we obtained a sample of recombinant HscA substrate binding domain (SBD-HscA, residues 389-616) from Dr. Larry Vickery (UC Irvine). SBD-HscA (5 μM) and 2Fe-BioB (0 – 30 μM) were equilibrated in 50 mM Tris HCl, 100 mM NaCl, 1 mM TCEP, pH 7.5 and free SBD-HscA separated by passing through a 100 kDa MW cutoff centrifugal concentrator and analyzing using SDS-polyacrylamide gel electrophoresis (Figure 4). We observe depletion of free SBD-HscA consistent with the formation of a 1:1 complex between SBD-HscA and 2Fe-BioB (Figure 3), with Kd = 0.55 ± 0.30 μM, which is roughly 10-fold higher affinity than was observed for full length HscA binding to 2Fe-BioB (Figure 1). The observed increase in the binding affinity might suggest that the BioB binding site on the HscA substrate binding domain is partially shielded by the N-terminal ATPase domain.
Figure 4.
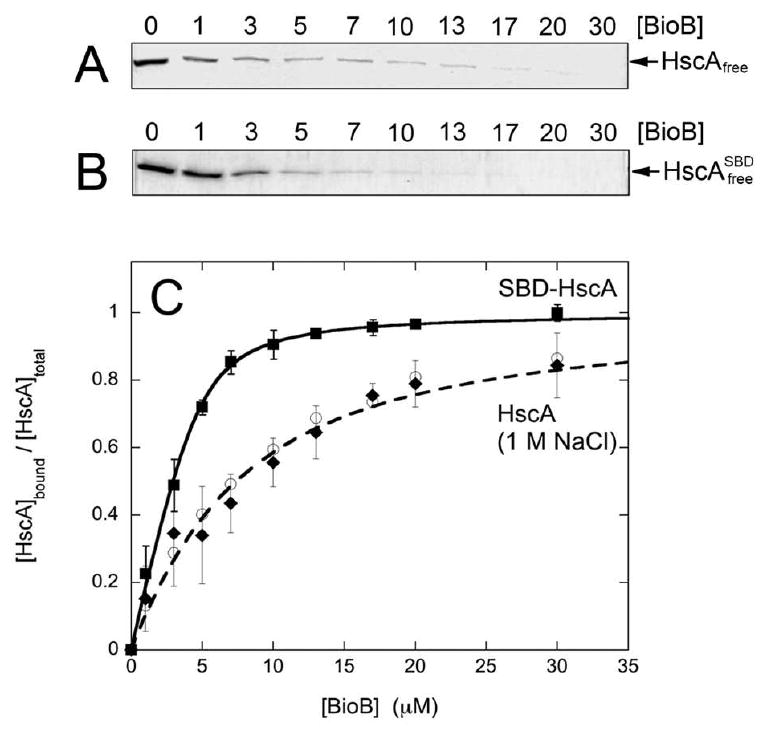
Binding studies that probe the nature of the complex between HscA and 2Fe-BioB in 50 mM Tris HCl (pH 7.5) and 1 mM TCEP (A) High ionic strength conditions: free HscA that passes through a 100 kDa centrifugal concentrator after equilibration with 2Fe-BioB in 1 M NaCl, as detected by SDS polyacrylamide gel electrophoresis. (B) No nucleotide-binding domain: free SBD-HscA (substrate binding domain) that passes through a 100 kDa centrifugal concentrator after equilibration with 2Fe-BioB in 0.1 M NaCl, as detected by SDS polyacrylamide gel electrophoresis. (C) Fractional saturation of the HscA:BioB complex, calculated from total and free HscA concentrations. Data shown for SBD-HscA:2Fe-BioB in 0.1 M NaCl (■), HscA:2Fe-BioB in 1 M NaCl (◆), and HscA:2Fe-BioB (○, from Figure 1) are an average of three trials each. Data are fit to a quadratic binding isotherm as described in the Materials and Methods, yielding values of KdSBD-HscA = 0.56 μM and KdHscA(1M NaCl) = 5.6 μM.
To further probe the nature of the BioB:HscA interaction, we examined the ionic strength and nucleotide dependence of complex formation. If the interaction between HscA and BioB involved a significant number of salt bridges between ionic residues, or is perhaps even due to nonspecific ionic attractions between surface residues on each protein, then increasing the ionic strength of the buffer should significantly reduce the affinity of the interaction. When the formation of a complex between HscA and 2Fe-BioB was examined in 1.0 M NaCl (Figure 3), the affinity was found to be unaffected by ionic strength (Kd = 5.5 ± 4 μM). We also examined whether the presence of MgATP vs MgADP in the nucleotide binding domain would affect the affinity of the complex. Similar to other Hsp70 proteins, the affinity of peptides and proteins for the canonical peptide binding site is modulated by the nucleotide binding site, with the presence of MgATP leading to decreased binding affinity (28). Nucleotide-dependent modulation of the protein:protein interaction has been observed for the formation of a complex between IscU and HscA (28), where interaction with the peptide binding site of HscA has been demonstrated through mutagenesis (38) and x-ray crystallography (30). In contrast, we did not observe any effect of nucleotide on the interaction between 2Fe-BioB and HscA (data not shown), with a Kd of ∼5 μM observed for HscA alone (Figure 1) and also in the presence of MgADP or MgATP. Taken together, these results suggest that BioB interacts with the substrate binding domain of HscA, but not at the canonical peptide binding site but rather at an alternate previously unidentified binding site. Further, the protein:protein interface involves no ionic interactions and is most likely dominated by nonionic hydrogen bonds and hydrophobic interactions.
BioB Does Not Stimulate the ATPase Activity of HscA
In a manner similar to other Hsp70 chaperones, the nucleotide binding domain of HscA has an intrinsic ATPase activity that is stimulated by the presence of a peptide or protein substrate in the canonical binding site of the substrate binding domain (26). Using a colorimetric coupled-enzyme assay to monitor Pi release, the effect on the HscA ATPase activity of apoBioB and IscU were compared. The basal turnover rate of HscA alone is slow (0.059 min-1), but this activity is stimulated ∼3-fold by IscU alone (0.16 min-1) and is further stimulated ∼340-fold by IscU plus the co-chaperone HscB (20 min-1). Stimulation of the ATPase activity by IscU and HscB has previously been reported (26), and it has been suggested that nucleotide hydrolysis and exchange may play an important role in effecting FeS cluster release from IscU. In contrast to the effect of IscU, the ATPase activity of HscA is not significantly stimulated by apoBioB alone (0.066 min-1, 1.1×). The stimulation by apoBioB in combination with HscB (0.38 min-1, 6.5×) is not significantly more than is observed for HscA and HscB in the absence of a protein substrate (0.30 min-1, 5.1×). Finally, apoBioB does not compete with IscU for the canonical peptide binding site. When apoBioB is added to a mixture of HscA, HscB, and IscU, the ATPase activity (18 min-1, 305×) is similar to that observed in the absence of apoBioB. Since BioB does not affect the ATPase activity of HscA, and the presence of nucleotide does not affect the affinity of HscA for BioB, then we can conclude that BioB is likely not binding at the canonical peptide binding site on HscA.
HscA, IscU, and BioB Form a Three Component Complex
The observation that BioB does not compete with IscU in stimulation of the HscA ATPase activity strongly suggested that IscU can remain bound to HscA even in the presence of BioB, indicating that a three component protein complex can be formed. To further probe whether a three component HscA:IscU:BioB complex is formed, or whether IscU and BioB are competitive for the same site on HscA, we turned back to the MW filtration assay using a 100 kDa cutoff centrifugal concentrator. Each protein alone is capable of passing through this filter (Figure 5, lanes 1-3) with >80% permissibility for BioB dimer, and >95% for HscA monomer and IscU dimer. When BioB (5 μM dimer) and HscA (10 μM) are mixed, ∼90% of the protein is retained by the filter, consistent with the formation of a complex with Kd ∼ 5 μM (lane 4). When HscA (10 μM) is mixed with IscU (10 μM dimer), ∼70% of the protein is retained by the filter, consistent with the formation of a complex with Kd ∼ 10 μM (lane 5). When HscA (10 μM) is mixed with BioB (5 μM dimer) and IscU (10 μM dimer), even more HscA and IscU is retained by the filter (lane 6). If BioB were in competition with IscU for the same or overlapping binding sites on HscA, then the amount of free IscU should have significantly increased in this last sample. We can conclude that HscA can simultaneously bind IscU to the canonical peptide binding site and bind BioB to another site located elsewhere on the substrate binding domain.
Figure 5.
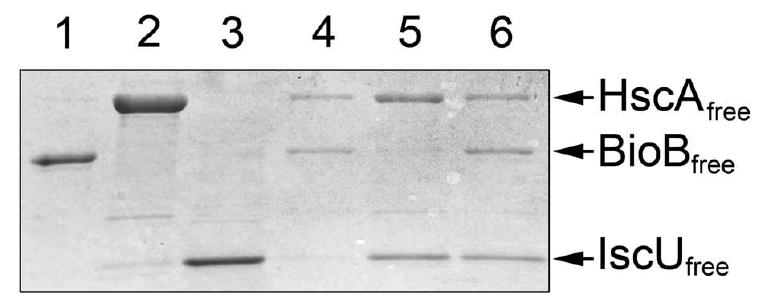
Evidence for formation of a three-component complex involving HscA, BioB, and IscU. Each individual protein or mixtures of proteins was equilibrated in 50 mM Tris HCl (pH 7.5), 100 mM NaCl, and 1 mM TCEP and filtered through a 100 kDa centrifugal concentrator; the amount of free unbound proteins is then detected by SDS polyacrylamide gel electrophoresis. Each protein is at 10 μM monomer in all six samples. Lane 1, BioB; Lane 2, HscA; Lane 3, IscU; Lane 4, BioB + HscA; Lane 5, IscU + HscA; Lane 6, BioB + IscU + HscA.
HscA is Important for the Efficient In Vivo Assembly of the [2Fe-2S]2+ Cluster on BioB
The finding that BioB forms a three-component complex with HscA and IscU in vitro suggests that this complex may play a functional role in FeS cluster assembly on or cluster transfer into apoBioB. However, many Hsp70-class chaperones are known to bind numerous proteins with low to moderate affinity. For example, DnaK is a heat shock protein in E. coli that plays a role in protein folding and/or stabilization under stress and that has significant homology with HscA, and prior studies have suggested interactions of DnaK with numerous polypeptide sequences in vitro (39, 40) and have suggested a role for DnaK in the folding of at least 94 different proteins in vivo (41). Prior studies in S. cerevisiae have demonstrated that mutation of the HscA homolog Ssq1 results in an 80-90 % decrease in the Fe content of biotin synthase (Bio2) within mitochondria, suggesting an important functional (although nonessential) role for Ssq1 in the transfer of FeS clusters from Isu1 to Bio2 in yeast.
To probe whether HscA plays a similar role in the assembly of FeS clusters on BioB in E. coli, we characterized BioB expressed and purified from E. coli knockout strains (34) in which individual genes have been replaced by a kanamycin resistance cassette. We cloned His6-BioB into a plasmid under control of the araB promotor, which could be tuned to provide a moderate, but not excessive, level of protein expression. We conducted these studies in a defined M9 minimal media containing glucose, casamino acids, and 5.0 μM FeCl3. The relative amounts of glucose (0.25 %) and arabinose (0.5 %) were carefully adjusted since glucose metabolites are known to repress the arabinose uptake system in E. coli, and casamino acids were required during arabinose-induced protein expression to provide a reasonable yield of the recombinant protein. The iron concentration in the media (5 μM) was adjusted to the minimum amount that would provide >0.9 equiv. of the [2Fe-2S]2+ cluster in His6-BioB grown in the WT strain. Too little iron (≤500 nM) resulted in essentially no FeS clusters in the recombinant protein in any strain, while excessive iron (≥50 μM) resulted in no significant differences between strains. Along these same lines, our analyses indicate that LB media contains 50 – 100 μM total iron (manufacturer and batch dependent) and no significant effects of any of the isc knockouts is observed in LB or related rich culture media.
His6-BioB was expressed following arabinose induction at 37 °C for 3 h in the E. coli K12-derived WT strain BW25113 in 1 liter of glucose-casamino acid-M9 media, yielding 105 mg of total soluble crude protein. The crude protein (100 mg) was passed through a small Ni-NTA-agarose column, eluted with imidazole, and concentrated to 1.0 mL, yielding 3.24 ± 0.11 mg of pure His6-BioB. The UV/visible spectrum (Figure 6A) is consistent with the presence of the air-stable [2Fe-2S]2+ cluster, while the air-sensitive [4Fe-4S]2+ cluster is apparently lost during the aerobic purification. Based upon our previously determined extinction coefficient, the spectrum is consistent with the presence of 1.03 equiv. of [2Fe-2S]2+ cluster per monomer, while iron analysis provides a value of 0.94 ± 0.07 equiv. of [2Fe-2S]2+ cluster per monomer.
Figure 6.
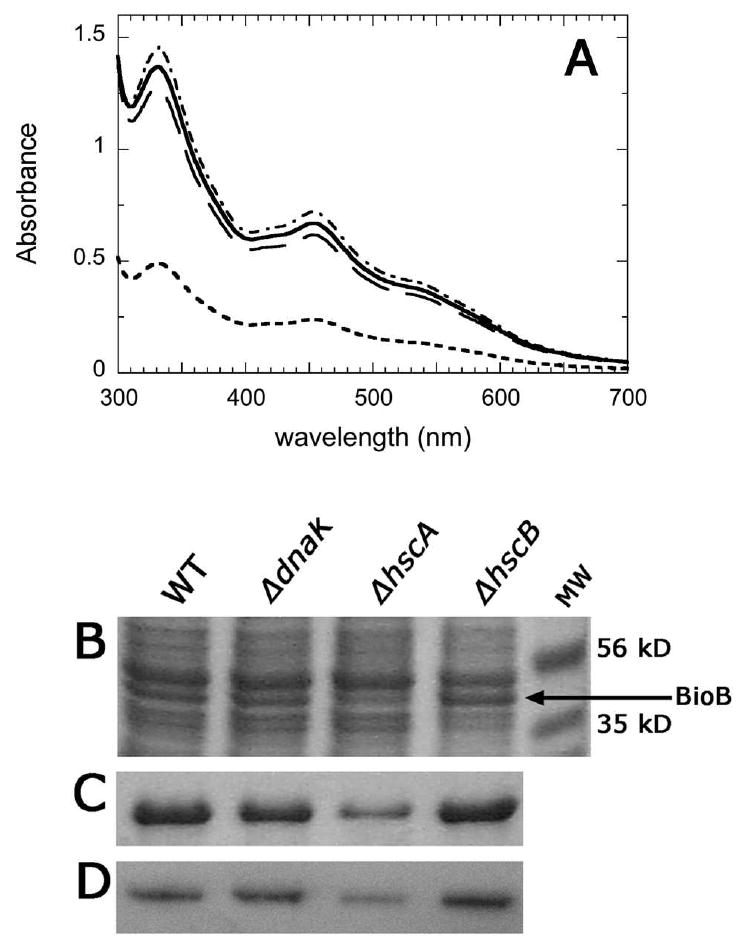
BioB expression in E. coli Keio knockout strains lacking HscA, HscB, or DnaK. (A) His6-BioB was purified from an E. coli K12 WT strain BW25113 (––––––), or from knockout strains ΔdnaK (–– –– ––), ΔhscA (- - - - - -), or ΔhscB (– - – - – -). Each strain carried plasmid pDE93, with the bioB gene under control of the araB promoter. Strains were grown at 37 °C in 1 L of a defined glucose-casamino acid-M9 medium containing 5 μM FeCl3. Protein expression was induced with 0.5% arabinose at OD600 = 0.6 and continued for 3 h at 37 °C. The crude soluble protein was isolated from the lysate, and the His6-BioB in 100 mg of total protein from each strain was purified on a Ni-NTA-agarose column and concentrated to 1.0 mL for these spectra. The absorbance at 452 nm reflects a combination of the total polypeptide yield and the [2Fe-2S]2+ cluster content of each BioB dimer. (B) SDS polyacrylamide gel electrophoresis of the crude soluble protein (20 μg per lane) isolated from each strain demonstrates that the amount of soluble full-length BioB polypeptide is decreased in the ΔhscA strain. (C) SDS polyacrylamide gel electrophoresis of the pure His6-BioB isolated from each strain confirms that ∼50% less His6-BioB polypeptide is present in the sample purified from the ΔhscA strain. Each lane contains 0.5 μl from the samples in panel A. (D) A Western blot of the crude soluble protein (250 ng per lane) isolated from each strain further confirms that there is a marked decrease in soluble His6-BioB and no evidence for stable immunoreactive proteolysis fragments. Protein samples were separated on SDS polyacrylamide gel electrophoresis, transferred to a PVDF membrane, and developed with anti-BioB rabbit IgG, peroxidase-fused donkey anti-rabbit IgG, and tetramethylbenzidene stain as previously described (31).
The mutant strains were analyzed in parallel with the WT strain using exactly the same protocol. The ΔdnaK and ΔhscB strains provided similar amounts of total pure His6-BioB (ΔdnaK: 3.33 ± 0.14 mg, ΔhscB: 3.15 ± 0.19 mg), had nearly indistinguishable UV/visible spectra (Figure 6A), and based upon Fe analysis contained similar levels of [2Fe-2S]2+ cluster (ΔdnaK: 0.82 ± 0.04 equiv., ΔhscB: 0.95 ± 0.06 equiv). In contrast, protein expressed in the ΔhscA strain was deficient in both protein yield and FeS cluster content. The yield of pure His6-BioB from an equivalent 100 mg of total soluble crude proteins was decreased 52% to 1.56 ± 0.12 mg. Both SDS polyacrylamide gel electrophoresis (Figure 6B) and Western blot analysis (Figure 6D) of the crude soluble protein are also consistent with the presence of significantly less His6-BioB in the cytosol following expression in the ΔhscA strain. The decrease in total cytosolic BioB is likely due to degradation of apoBioB that has failed to obtain an FeS cluster, as our prior studies have demonstrated that apoBioB is rapidly proteolyzed both in vitro and in vivo (31). Furthermore, even the small amount of protein that was purified contains significantly less than stoichiometric levels of [2Fe-2S]2+ cluster. Based upon the UV/visible spectrum (Figure 6A), we estimate His6-BioB purified from the ΔhscA strains contained 0.50 equiv. of [2Fe-2S]2+ cluster, while Fe analysis indicates the presence of 0.52 ± 0.13 equiv. of [2Fe-2S]2+ cluster, perhaps suggesting that each surviving dimer contains one [2Fe-2S]2+ cluster in one monomer, while the other monomer is vacant. Overall these results suggest that in E. coli, HscA plays a significant, although nonessential, role in enhancing the assembly or transfer of the [2Fe-2S]2+ cluster in BioB.
Discussion
Iron-sulfur clusters are found in numerous proteins and enzymes, where they function as stable cofactors in electron transport, as catalysts in hydration/dehydration reactions, and as sensors of iron levels, redox status, and oxidative stress. In contrast, biotin synthase is among only three known enzymes that appear to utilize a [2Fe-2S]2+ cluster as a substrate that provides sulfur for formation of new C-S bonds. At the end of each turnover of biotin formation, dissociation of the remaining Fe2+ and S2- yields inactive BioB that has an empty FeS cluster binding site located deep within the core of the (αβ)8 barrel. In order to regenerate active BioB in vivo, the [2Fe-2S]2+ cluster must be reassembled and deposited into this binding site, most likely by components of the ISC iron-sulfur cluster assembly system.
In vivo genetic studies in S. cerevisiae implicate a chaperone/co-chaperone pair as being important in transferring FeS clusters from IscU to mitochondrial proteins. Using biotin synthase (Bio2) as a target protein, Lill and coworkers demonstrate that an Isu1 mutant (yeast IscU homolog) deposits <10% of the normal FeS cluster content in immunoprecipitated Bio2 (42). They have also demonstrated that in an Ssq1 mutant (yeast HscA homolog), Bio2 is expressed with <20% of the normal FeS cluster content (24). The residual cluster content may be attributable to rescue by another Hsp70 homolog. In the Ssq1 mutant, the homologous Hsp70 chaperone Ssc1 is upregulated and appears to suppress the cold-sensitive growth phenotype associated with FeS cluster deficiency, suggesting Ssc1 is partially replacing the functional role of Ssq1 (25). Thus in yeast, biotin synthase obtains FeS clusters specifically from the scaffold protein Isu1, and cluster transfer from Isu1 into biotin synthase is significantly enhanced by the specific chaperone/co-chaperone pair Ssq1 and Jac1.
In E. coli, there is no obvious growth phenotype associated with deletion of HscA or HscB. However, our studies suggest that HscA does play a significant role in the assembly of the [2Fe-2S]2+ cluster on BioB, with deletion of HscA resulting in a 52% decrease in total protein and a 50% decrease in the cluster content of the purified protein. If we assume that the decreased protein yield is due to degradation of apoprotein, then the total decrease in FeS cluster assembly on BioB was ∼75%. However, this cluster assembly deficiency would be predicted to have no effect on growth rates, since the residual active BioB would be more than sufficient to meet the minimal biotin demands of the cell. We have also observed that the effect of the HscA deletion on FeS cluster assembly is highly dependent on the culture medium. In glucose minimal media, the effect is increased at lower iron concentrations (<500 nM), but is more difficult to detect this effect using our current protocols due to the much lower overall FeS cluster content on BioB. At high iron concentrations (>50 μM), and in commercially-available rich media with high iron content, the effect is completely suppressed, with no detectable differences between BioB expressed in mutant and WT strains. Under these conditions, iron uptake may overwhelm the iron storage and transport systems, and spontaneous FeS cluster assembly could proceed without the need for specific scaffold proteins or chaperones.
Surprisingly, we find no effect of deletion of the co-chaperone HscB on FeS cluster assembly on BioB, suggesting that this protein may be dispensable or replaceable in the cluster assembly process. Vickery and coworkers have suggested that the role of HscB is to steer IscU into an interaction with HscA and to stimulate the ATPase activity of HscA, resulting in formation of a long-lived IscU:HscA:MgADP complex (20). Perhaps these functions of HscB increase the efficiency of HscA but are not essential for the overall FeS cluster assembly process; or conversely, perhaps another cochaperone is upregulated to fulfill the function of HscB. Moreover, the differential effect of HscA vs. HscB deletions raises the possibility that some FeS proteins, such as BioB, may only require a subset of the ISC system for efficient cluster assembly. To address this question, we are broadening our in vivo studies reported in this paper to include other E. coli genes that have been implicated in FeS cluster assembly. While these experiments are ongoing, it is clear at this point that, whereas IscU is essential for FeS cluster assembly on BioB, the other proposed scaffold protein IscA is not required (Fugate, Hu, and Jarrett, unpublished data).
Hsp70 chaperones typically have a nucleotide binding domain that binds MgATP and catalyzes slow hydrolysis to MgADP, inducing an altered conformation of the protein that is propagated through to the substrate binding domain (37). Vickery and coworkers have demonstrated that the MgADP-bound state of HscA has a high affinity and specificity for binding IscU (26) to the canonical peptide binding site found in the substrate binding domain. The interaction with IscU involves a specific peptide loop from IscU and, in contrast with other Hsp70s, HscA has high specificity for this peptide loop and does not bind unrelated peptides or proteins in the substrate binding site (29, 38). Vickery and coworkers have proposed a kinetic cycle for HscA/HscB in which ATP hydrolysis is coupled to a conformational change that repeatedly binds and releases IscU (20, 43). However, the possible in vivo function of this binding and release cycle is not yet clear.
We have explored the interaction of BioB with components of the ISC system, and we find a detectable complex is formed only with HscA. Although one might initially suspect that BioB is simply interacting with the canonical peptide binding site on HscA (the IscU binding site), several experimental details suggest that this is not the case. For example, the interaction of BioB with HscA does not stimulate the ATPase activity of the nucleotide binding domain. Conversely, the presence of ATP or ADP in the nucleotide binding domain of HscA does not alter the affinity for BioB. Finally, BioB does not compete with IscU for the peptide binding site. Each of these experiments indicates that BioB is not binding to the Hsp70 canonical peptide binding site, but is instead interacting with another site on the HscA substrate-binding domain. In addition, we have identified specific peptide loops from BioB that interact with HscA; these peptides compete with BioB binding and do not compete with IscU, establishing that the BioB peptides are not binding at the canonical peptide binding site (M. Reyda and J. Jarrett, unpublished data). Together, these data strongly suggest the formation of multicomponent complex that involves minimally HscA, IscU, and BioB, and, although we have not specifically addressed the possibility in this work, might also include the co-chaperone HscB.
We propose that the role of this protein complex with HscA is to enhance the efficiency of the handoff of an FeS cluster from IscU to apoBioB, possibly by simply promoting or stabilizing the interaction of IscU with BioB. Our in vitro studies suggest that IscU does not have a significant affinity for BioB, and the lack of a specific interaction would force the in vivo cluster assembly process to proceed through a random second-order collision of IscU and BioB. However, since both proteins bind to HscA, this complex could enhance the interaction between IscU and BioB. Given the relatively high concentration of HscA in the bacterial cytosol (∼20 μM (23)), apoBioB is likely saturated with bound HscA under most in vivo conditions. Thus the interaction of the apoBioB/HscA complex with [2Fe-2S]2+-IscU, as stimulated by HscB and ATP, may be most relevant in understanding the FeS cluster transfer mechanism for biotin synthase.
The finding of a specific interaction between HscA and BioB raises the question of whether there is an as-yet unidentified binding epitope through which HscA can identify and bind to other incipient FeS proteins. E. coli contains at least 110 known FeS proteins and possibly another 100 uncharacterized predicted FeS proteins (44). Could all of these proteins have a shared epitope that facilitates cluster delivery? On the other hand, BioB is among only a handful of proteins that require a constant supply of FeS clusters to maintain catalytic activity. In the case of BioB, inefficient cluster assembly leads to rapid targeted protein degradation (31). Perhaps the interaction of BioB with HscA is a unique adaptation of this enzyme to maintain the active enzyme state and facilitate optimal cell growth under nutrient-deprived conditions.
Acknowledgments
We thank Dr. Larry Vickery (University of California – Irvine) for providing the HscA and IscU expression plasmids and SBD-HscA protein, and for many helpful discussion regarding interpretation of our data. We also thank Dr. Jason T. Wan for assistance in adapting the MW filtration binding assay, and Dr. James Lear for advice concerning the analytical ultracentrifugation experiments. Plasmid pDE93 was constructed by Dr. Devrim Eren.
Footnotes
This research has been supported by the NIH (R01 GM59175 to J.T.J.) and the David and Lucille Packard Foundation (J.T.J.).
Abbreviations: AdoMet, S-adenosyl-L-methionine; BioB, biotin synthase; apoBioB, BioB lacking both iron-sulfur clusters; 2Fe-BioB, containing only a [2Fe-2S]2+ cluster; 2Fe4Fe-BioB, containing both FeS clusters; 5′-dAH, 5′-deoxyadenosine; 5′-dA•, 5′-deoxyadenosyl radical; DTB, dethiobiotin; DTT, dithiothreitol; EDTA, ethylenediaminetetraacetic acid; HscA, Hsp70 chaperone found in the Isc operon; HscA-SBD, peptide/protein-binding domain of HscA; ISC, E. coli iron-sulfur cluster assembly proteins; 9-MDTB, 9-mercaptodethiobiotin; TCEP, tris(2-carboxyethyl)-phosphine; Tris, tris(hydroxymethyl)aminomethane.
References
- 1.Marquet A, Tse Sum Bui B, Florentin D. Biosynthesis of biotin and lipoic acid. Vitam Horm. 2001;61:51–101. doi: 10.1016/s0083-6729(01)61002-1. [DOI] [PubMed] [Google Scholar]
- 2.Ugulava NB, Gibney BR, Jarrett JT. Biotin synthase contains two distinct iron-sulfur cluster binding sites: Chemical and spectroelectrochemical analysis of iron-sulfur cluster interconversions. Biochemistry. 2001;40:8343–8351. doi: 10.1021/bi0104625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ugulava NB, Sacanell CJ, Jarrett JT. Spectroscopic changes during a single turnover of biotin synthase: Destruction of a [2Fe-2S] cluster accompanies sulfur insertion. Biochemistry. 2001;40:8352–8358. doi: 10.1021/bi010463x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ugulava NB, Surerus KK, Jarrett JT. Evidence from Mössbauer Spectroscopy for Distinct [2Fe-2S]2+ and [4Fe-4S]2+ Cluster Binding Sites in Biotin Synthase from Escherichia coli. J Am Chem Soc. 2002;124:9050–9051. doi: 10.1021/ja027004j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cosper MM, Jameson GN, Davydov R, Eidsness MK, Hoffman BM, Huynh BH, Johnson MK. The [4Fe-4S]2+ cluster in reconstituted biotin synthase binds S-adenosyl-L-methionine. J Am Chem Soc. 2002;124:14006–14007. doi: 10.1021/ja0283044. [DOI] [PubMed] [Google Scholar]
- 6.Guianvarc'h D, Florentin D, Tse Sum Bui B, Nunzi F, Marquet A. Biotin synthase, a new member of the family of enzymes which uses S-adenosylmethionine as a source of deoxyadenosyl radical. Biochem Biophys Res Commun. 1997;236:402–406. doi: 10.1006/bbrc.1997.6952. [DOI] [PubMed] [Google Scholar]
- 7.Escalletes F, Florentin D, Tse Sum Bui B, Lesage D, Marquet A. Biotin Synthase Mechanism: Evidence for Hydrogen Transfer from the Substrate into Deoxyadenosine. J Am Chem Soc. 1999;121:3571–3578. [Google Scholar]
- 8.Jarrett JT. The novel structure and chemistry of iron-sulfur clusters in the adenosylmethionine-dependent radical enzyme biotin synthase. Arch Biochem Biophys. 2005;433:312–321. doi: 10.1016/j.abb.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Taylor AM, Farrar CE, Jarrett JT. 9-Mercaptodethiobiotin is formed as a competent catalytic intermediate by Escherichia coli biotin synthase. Biochemistry. 2008;47:9309–9317. doi: 10.1021/bi801035b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jameson GNL, Cosper MM, Hernandez HL, Johnson MK, Huynh BH. Role of the [2Fe-2S] Cluster in Recombinant Escherichia coli Biotin Synthase. Biochemistry. 2004;43:2022–2031. doi: 10.1021/bi035666v. [DOI] [PubMed] [Google Scholar]
- 11.Ugulava NB, Frederick KK, Jarrett JT. Control of Adenosylmethionine-Dependent Radical Generation in Biotin Synthase: A Kinetic and Thermodynamic Analysis of Substrate Binding to Active and Inactive Forms of BioB. Biochemistry. 2003;42:2708–2719. doi: 10.1021/bi0261084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronan JE., Jr The biotinyl domain of Escherichia coli acetyl-CoA carboxylase. Evidence that the “thumb” structure id essential and that the domain functions as a dimer. J Biol Chem. 2001;276:37355–37364. doi: 10.1074/jbc.M106353200. [DOI] [PubMed] [Google Scholar]
- 13.Choi-Rhee E, Cronan JE. Biotin Synthase Is Catalytic In Vivo, but Catalysis Engenders Destruction of the Protein. Chem Biol. 2005;12:461–468. doi: 10.1016/j.chembiol.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 14.Duin EC, Lafferty ME, Crouse BR, Allen RM, Sanyal I, Flint DH, Johnson MK. [2Fe-2S] to [4Fe-4S] Cluster Conversion in Escherichia coli Biotin Synthase. Biochemistry. 1997;36:11811–11820. doi: 10.1021/bi9706430. [DOI] [PubMed] [Google Scholar]
- 15.Ugulava NB, Gibney BR, Jarrett JT. Iron-Sulfur Cluster Interconversions in Biotin Synthase: Dissociation and Reassociation of Iron during Conversion of [2Fe-2S] to [4Fe-4S] Clusters. Biochemistry. 2000;39:5206–5214. doi: 10.1021/bi9926227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cosper MM, Jameson GN, Hernandez HL, Krebs C, Huynh BH, Johnson MK. Characterization of the cofactor composition of Escherichia coli biotin synthase. Biochemistry. 2004;43:2007–2021. doi: 10.1021/bi0356653. [DOI] [PubMed] [Google Scholar]
- 17.Tse Sum Bui B, Benda R, Schunemann V, Florentin D, Trautwein AX, Marquet A. Fate of the (2Fe-2S)2+ cluster of Escherichia coli biotin synthase during reaction: a Mössbauer characterization. Biochemistry. 2003;42:8791–8798. doi: 10.1021/bi034426c. [DOI] [PubMed] [Google Scholar]
- 18.Frazzon J, Dean DR. Formation of iron-sulfur clusters in bacteria: an emerging field in bioinorganic chemistry. Curr Opin Chem Biol. 2003;7:166–173. doi: 10.1016/s1367-5931(03)00021-8. [DOI] [PubMed] [Google Scholar]
- 19.Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu Rev Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- 20.Vickery LE, Cupp-Vickery JR. Molecular chaperones HscA/Ssq1 and HscB/Jac1 and their roles in iron-sulfur protein maturation. Crit Rev Biochem Mol Biol. 2007;42:95–111. doi: 10.1080/10409230701322298. [DOI] [PubMed] [Google Scholar]
- 21.Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv Prot Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4. [DOI] [PubMed] [Google Scholar]
- 22.Silberg JJ, Hoff KG, Vickery LE. The Hsc66-Hsc20 chaperone system in Escherichia coli: chaperone activity and interactions with the DnaK-DnaJ-GrpE system. J Bacteriol. 1998;180:6617–6624. doi: 10.1128/jb.180.24.6617-6624.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vickery LE, Silberg JJ, Ta DT. Hsc66 and Hsc20, a new heat shock cognate molecular chaperone system from Escherichia coli. Prot Sci. 1997;6:1047–1056. doi: 10.1002/pro.5560060511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muhlenhoff U, Gerber J, Richhardt N, Lill R. Components involved in assembly and dislocation of iron-sulfur clusters on the scaffold protein Isu1p. EMBO J. 2003;22:4815–4825. doi: 10.1093/emboj/cdg446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Voisine C, Schilke B, Ohlson M, Beinert H, Marszalek J, Craig EA. Role of the mitochondrial Hsp70s, Ssc1 and Ssq1, in the maturation of Yfh1. Mol Cell Biol. 2000;20:3677–3684. doi: 10.1128/mcb.20.10.3677-3684.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silberg JJ, Tapley TL, Hoff KG, Vickery LE. Regulation of the HscA ATPase Reaction Cycle by the Co-chaperone HscB and the Iron-Sulfur Cluster Assembly Protein IscU. J Biol Chem. 2004;279:53924–53931. doi: 10.1074/jbc.M410117200. [DOI] [PubMed] [Google Scholar]
- 27.Hoff KG, Silberg JJ, Vickery LE. Interaction of the iron-sulfur cluster assembly protein IscU with the Hsc66/Hsc20 molecular chaperone system of Escherichia coli. Proc Nat Acad Sci U S A. 2000;97:7790–7795. doi: 10.1073/pnas.130201997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Silberg JJ, Hoff KG, Tapley TL, Vickery LE. The Fe/S assembly protein IscU behaves as a substrate for the molecular chaperone Hsc66 from Escherichia coli. J Biol Chem. 2001;276:1696–1700. doi: 10.1074/jbc.M009542200. [DOI] [PubMed] [Google Scholar]
- 29.Hoff KG, Ta DT, Tapley TL, Silberg JJ, Vickery LE. Hsc66 substrate specificity is directed toward a discrete region of the iron-sulfur cluster template protein IscU. J Biol Chem. 2002;277:27353–27359. doi: 10.1074/jbc.M202814200. [DOI] [PubMed] [Google Scholar]
- 30.Cupp-Vickery JR, Peterson JC, Ta DT, Vickery LE. Crystal Structure of the Molecular Chaperone HscA Substrate Binding Domain Complexed with the IscU Recognition Peptide ELPPVKIHC. J Mol Biol. 2004;342:1265–1278. doi: 10.1016/j.jmb.2004.07.025. [DOI] [PubMed] [Google Scholar]
- 31.Reyda MR, Dippold R, Dotson ME, Jarrett JT. Loss of iron-sulfur clusters from biotin synthase as a result of catalysis promotes unfolding and degradation. Arch Biochem Biophys. 2008;471:32–41. doi: 10.1016/j.abb.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vistica J, Dam J, Balbo A, Yikilmaz E, Mariuzza RA, Rouault TA, Schuck P. Sedimentation equilibrium analysis of protein interactions with global implicit mass conservation constraints and systematic noise decomposition. Anal Biochem. 2004;326:234. doi: 10.1016/j.ab.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 33.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mansy SS, Cowan JA. Iron-sulfur cluster biosynthesis: Toward an understanding of cellular machinery and molecular mechanism. Acc Chem Res. 2004;37:719–725. doi: 10.1021/ar0301781. [DOI] [PubMed] [Google Scholar]
- 37.Jiang J, Prasad K, Lafer EM, Sousa R. Structural Basis of Interdomain Communication in the Hsc70 Chaperone. Mol Cell. 2005;20:513. doi: 10.1016/j.molcel.2005.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hoff KG, Cupp-Vickery JR, Vickery LE. Contributions of the LPPVK Motif of the Iron-Sulfur Template Protein IscU to Interactions with the Hsc66-Hsc20 Chaperone System. J Biol Chem. 2003;278:37582–37589. doi: 10.1074/jbc.M305292200. [DOI] [PubMed] [Google Scholar]
- 39.Gragerov A, Zeng L, Zhao X, Burkholder W, Gottesman ME. Specificity of DnaK-peptide binding. J Mol Biol. 1994;235:848–854. doi: 10.1006/jmbi.1994.1043. [DOI] [PubMed] [Google Scholar]
- 40.Rudiger S, Germeroth L, Schneider-Mergener J, Bukau B. Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 1997;16:1501–1507. doi: 10.1093/emboj/16.7.1501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B. Trigger Factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol. 2003;47:1317–1328. doi: 10.1046/j.1365-2958.2003.03370.x. [DOI] [PubMed] [Google Scholar]
- 42.Muhlenhoff U, Gerl MJ, Flauger B, Pirner HM, Balser S, Richhardt N, Lill R, Stolz J. The Iron-Sulfur Cluster Proteins Isa1 and Isa2 Are Required for the Function but Not for the De Novo Synthesis of the Fe/S Clusters of Biotin Synthase in Saccharomyces cerevisiae. Eukaryotic Cell. 2007;6:495–504. doi: 10.1128/EC.00191-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bonomi F, Iametti S, Morleo A, Ta D, Vickery LE. Studies on the mechanism of catalysis of iron-sulfur cluster transfer from IscU[2Fe2S] by HscA/HscB chaperones. Biochemistry. 2008;47:12795–12801. doi: 10.1021/bi801565j. [DOI] [PubMed] [Google Scholar]
- 44.Fontecave M. Iron-sulfur clusters: ever-expanding roles. Nat Chem Biol. 2006;2:171–174. doi: 10.1038/nchembio0406-171. [DOI] [PubMed] [Google Scholar]


