Abstract
The transcription factor Forkhead box M1 (FOXM1) is a key regulator of cell proliferation and is over-expressed in many forms of primary cancers, leading to uncontrolled cell division and genomic instability. To address the role of FOXM1 in chemoresistance we generated a cisplatin resistant breast cancer cell line (MCF-7-CISR), which had an elevated level of FOXM1 protein and mRNA expression relative to the parental MCF-7 cells. A close correlation was observed between FOXM1 and the expression of its proposed downstream targets that are involved in DNA repair; breast cancer associated gene 2 (BRCA2) and X-ray-cross-complementing group 1 (XRCC1) were expressed at higher levels in the resistant cell lines compared with the sensitive MCF-7 cells. Moreover, cisplatin treatment induced DNA damage repair in MCF-7-CISR and not in MCF-7 cells. Furthermore, the expression of a constitutively active FOXM1 (ΔN-FOXM1) in MCF-7 cells alone was sufficient to confer cisplatin resistance. Crucially, the impairment of DNA damage repair pathways through the siRNA knockdown inhibition of either FOXM1, or BRCA2/XRCC1 showed that only silencing of FOXM1 could significantly reduce the rate of proliferation in response to cisplatin treatment in the resistant cells. This suggests that the targeting of FOXM1 is a viable strategy in circumventing acquired cisplatin resistance. Consistently, the FOXM1 inhibitor thiostrepton also showed efficacy in causing cell death and proliferative arrest in the cisplatin resistant cells through the down-regulation of FOXM1 expression. Taken together, we have identified a novel mechanism of acquired cisplatin resistance in breast cancer cells through the induction of FOXM1.
Introduction
Platinum based chemotherapeutics, such as cisplatin, (cis-diammine-dichloro-platinum) have long been established in the routine treatment of ovarian, testicular, and non-small cell lung cancer patients under clinical settings (1). Cisplatin treatment results in the formation of intrastrand and interstrand DNA-adducts (2), triggering the nucleotide excision repair (NER) and homologous recombination pathways (3-5). Failure to activate or execute appropriate DNA repair leads to the accumulation of DNA strand breaks, and ultimately to cell death (6). Recent clinical data suggests an emerging role for platinum based chemotherapy for advanced breast cancer patients. For example, three independent phase II clinical trials involving HER2 positive or advanced metastatic breast cancer patients treated with a combination of Herceptin with cisplatin and docetaxol showed clinically significant improvement of survival rates (7-9). Furthermore, recent clinical trial data also suggests that triple negative breast cancer patients who are ER-negative, PR-negative, and with low HER2 expression levels, show better survival rates in response to cisplatin chemotherapeutic treatment (10). However, acquired cisplatin resistance is a major clinical obstacle for patients that relapse after initial favourable responses. Cisplatin resistance is a complex and multifaceted problem which involves multiple pathways including increased cisplatin efflux, inactivation of intracellular cisplatin, evasion of apoptotic pathways, replication checkpoint bypass, increased cell proliferation, and increased DNA damage repair (1, 11). A number of targets have been implicated to have a role in breast cancer cisplatin resistance including amphiregulin (12), BCL2 (13), BCL2L12 (14), cyclin D1(13), Siva-1 (15) and the miRNA regulator Dicer (16). However, a better understanding of the molecular mechanism underlying chemotherapeutic resistance is needed for the development of effective platinum-based therapeutic strategies for treatment of breast cancer. FOXM1 is a transcription factor which belongs to the wider forkhead transcription factor family (17) and is required for normal cell cycle execution during G1 to S (18, 19), G2 and M phase transitions (20), apoptosis (21, 22), angiogenesis (23), metastasis (24) and DNA damage repair (25, 26). The overexpression of FOXM1 in primary breast cancer tissues has also been associated with breast cancer tumorigenesis (27) and poor prognosis (26, 28). Intriguingly, several lines of studies have indicated that in breast cancer patients, the loss of functional BRCA2 and XRCC1 expression contributes to breast cancer development (29, 30), although this is also hypothesized to render the cells more susceptible to DNA damage and cytotoxic chemotherapies. Increasing evidence has emerged to demonstrate that re-gaining the expression of DNA repair genes such as BRCA 1, −2 and XRCC1 could lead enhance cisplatin resistance in these cells (29, 31, 32). In the present study, we investigated the role of FOXM1 and its proposed downstream DNA damage repair targets BRCA2 and XRCC1 might play in conferring cisplatin resistance.
Materials and Methods
Cell lines
MCF-10A and MCF-7 cells originated from the American Type Culture Collection and were acquired from the Cell Culture Service, Cancer Research UK (London, UK), where they were tested and authenticated. These procedures include cross species checks, DNA authentication and quarantine. Cell lines used in the present study were in culture for less than 6 months. MCF10-A were cultured in DMEM HAM-F12/1:1 mix (Sigma UK, Poole), supplemented with 5% (v/v) horse serum, 10 mg/ml insulin, 5 mg/ml hydrocortisol, 100 ng/ml cholorotoxin and 20 ng/ml of Epidermal Growth Factor (EGF), 100 u/ml penicillin and 100 μg/ml streptomycin (Sigma, Poole, UK). MCF-7 cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM) (Sigma UK) supplemented with 10% (v/v) FCS, 2 mM glutamine, 100u/ml penicillin and 100 μg/ml streptomycin at 37°C. The MCF-7-ΔN-FOXM1 cell line was established by co-transecting MCF-7 cells with a CMV-driven expression vector of ΔN-FOXM1 and pBABEpuro (Addgene) that contains a puromycin selection marker. Cells were then selected at 1.5 μg/ml of puromycin (Autogen Bioclear Ltd, UK) and maintained at 0.75 μg/ml of puromycin (Invitrogen, Paisley, UK). The MCF-7-CISR cell line is a cisplatin resistant cell line derived from parental MCF-7 cells. MCF-7 were subjected to increasing concentrations of cisplatin (Onco-tain® DBL, Leamington Spa, UK) until the MCF-7 cells acquire resistance to 0.112 M of cisplatin. MCF-7-CISR cell line was then maintained in 0.1M of cisplatin.
Western blot analysis and antibodies
Cells were lysed and SDS-PAGE gel electrophoresis was performed as previously described. The antibodies against FOXM1 (c-20), β-tubulin (H-235), CDC25B (C-20) and PLK-1 (F-8) were purchased from Santa Cruz Biotechnology (Autogen Bioclear, Wiltshire, UK), whereas FOXO3a (06-951) and BRCA2 (5.23) were from Upstate (Dundee, UK). Antibodies against phospho-Akt (Ser473), total Akt, phospho-FOXO3a (Thr32) were from Cell Signaling Technologies (Hitchin, UK) and XRCC1 (AHP832) was from AbD Serotec (Kidlington, UK). Primary antibodies were detected using horseradish peroxidase linked anti-mouse, anti-goat or anti-rabbit conjugates as appropriate (DAKO, Ely, UK.), and visualized using the ECL detection system (Amersham Biosciences, Amersham, UK). Protein expression levels were quantified using the software ImageJ to detect intensity of the protein bands.
Real time quantitative PCR (RTQ-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagen, Crawley, UK), and cDNA prepared using the SuperScript™ III reverse transcriptase and random primers (Invitrogen). For RTQ-PCR, 100 ng of cDNA was added to SYBER-Green Master Mix (Applied BioSystems, Foster City, CA, USA) and run in 7900 HT Fast Real-time PCR System (Applied BioSystems). The cycling programme was 95°C for 20 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. Each sample was assayed in triplicates, and the results were normalized to the level of ribosomal protein L19 RNA. The forward and reverse primers used were: FOXM1-F, 5′-TGCAGCTAGGGATGTGAATCTTC-3′,FOXM1-R,3′-GGAGCCCAGTCCATCAGAACT-5′; L19-F, 5′- GCGGAGAGGGTACAGCCAAT-3′, L-19-R, 3′-GCAGCCGGCGCAAA-5′; BRCA2-F,5′-GCTGGCTTCAACTCCAATAATATTC;3′BRCA2-R GTCTACTATTGGGAACATTCCTTCCT; XRCC1-F,5′AAAGGGAAGAGGAAGTTGGATTTG;XRCC1-R3′-GCAATTTAGGTCTCTTGGGAACA.
Sulforhodamine B (SRB) assay
Approximately 3000 cells were seeded in each well of the 96 well plates. After culture, 100 μl of trichloroacetic acid was added to each well and incubated for 1 h at 4°C. The plates were then washed with deionised water for three times, before incubation at RT for 1 h with 0.4% SRB in 1% acetic acid. The plates were then washed with deionised water and air-dried. 10mM Tris base was then added to the wells to solubilise the bound SRB dye, and the plates were then read at 492 nm using the Anthos 2001 plate read (Jencons Scientific Ltd, Leighton Buzzard, UK).
Cell cycle analysis
Cell cycle analysis was performed by PI staining, as previously described (33). The cell cycle profile was analyzed using Cell Diva software (Becton Dickinson UK Ltd, Oxford, UK).
siRNA transfection
MCF-7 and MCF-7-CISR cells were transfected with 100 nM FOXM1 siRNA (Dhamarcon) or 100 nM XRCC1 or 100 nM BRCA2 siRNA using oligofectamine (Invitrogen). 24 h post transfection, transfected cells were treated with 0.1 μM of cisplatin and viable cell counts were quantified by SRB assay.
Phospho-γH2AX immunofluorescent staining and quantification
MCF-7, MCF-7-CISR and MCF-7-ΔN-FOXM1 cells were transfected with 100 nM FOXM1 siRNA (Dhamarcon) or 100 nM XRCC1 or 100 nM BRCA2 siRNA by oligofectamine (Invitrogen). After 24 h, cells were then treated with 0.1 μM of cisplatin for 6 h (34, 35). Subsequently, cells were then subjected to anti-γH2AX (Ser139) staining. Briefly, cells were fixed with 4% paraformaldehyde (Sigma), permeabilized with 0.1% Triton X-100 in 10% FCS for 10 min. Samples were then blocked with 5% goat serum in 10% FCS for 30 min and then incubated overnight with the primary rabbit anti-γH2AX (Ser139) (1:120; Cell Signalling). Following washes with PBS, secondary goat anti-rabbit IgG-fluorescein isothiocyanate (1:500, Invitrogen) was added to the samples for an hour. Cells were the counterstained with 4′, 6′-diamidino-2-phenylindole (DAPI) prior to mounting. Images were captured and quantified using the Zeiss Axiovert 100 confocal laser scanning microscope and software Zeiss LSM 500 (Zeiss Ltd, Welwyn Garden City, UK). DNA damage foci were selected based on the following parameters - DNA foci: min width: 0.05μm, max width: 8μm, intensity above background: 200 gray levels. Nuclei: min width 10μm, max width: 20μm, intensity above background: 400 gray levels.
Results
FOXM1 and targets BRCA2 and XRCC1 are up-regulated in cisplatin resistant MCF-7 cells
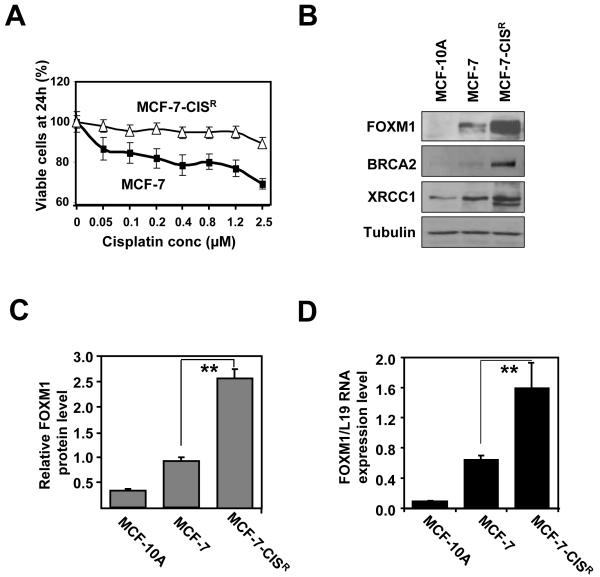
Previously, it has been demonstrated that the over-expression of FOXM1 is indicative of poor prognosis in breast cancer patients (26, 28). FOXM1 has also been reported to regulate the expression of the DNA damage repair genes, BRCA2 and XRCC1 (25). Hitherto, the role of FOXM1 in cisplatin resistance through the repair of cisplatin-DNA adducts resistance has not been established. In the first instance, we established a new cisplatin resistance cell line, MCF-7-CISR, through repeated exposures of MCF-7 cells to successive rounds of cisplatin until resistance up to 1.2 μM was reached as indicated by SRB proliferation assay (Figure 1A). Subsequent western blot analysis reveals that MCF-7 cells expressed a higher level of FOXM1 relative to the untransformed MCF-10A breast epithelial cells. Interestingly, MCF-7-CISR showed an even higher FOXM1 level compared with the parental MCF-7 cells (Figure 1B). Furthermore, MCF-7-CISR also had higher levels of DNA repair proteins BRCA2 and XRCC1. Relative FOXM1 protein expression level was on average 2.5 fold higher in MCF-7-CISR cells compared with MCF-7 cells (Figure 1C). The results were mirrored at mRNA level, where MCF-7-CISR had a 2-fold increase (Figure 1D).
Figure 1. Cisplatin resistant cell line shows elevated FOXM1 protein and mRNA expression levels.
A) MCF-7 and MCF-7-CISR cells were treated with increasing concentrations of cisplatin and their rates of proliferation measured by SRB assay. B) Western blot analysis determining the relative protein expression levels of FOXM1, BRCA2 and XRCC1 in MCF-10A, MCF-7 and MCF-7-CISR cells. C) FOXM1 protein expression level was quantified using ImageJ normalized against tubulin levels. D) FOXM1 mRNA transcript levels were being determined by RTQ-PCR analysis. Results shown were derived from at least three independent experiments. The error bars show the standard deviation (mean ± SD). Statistical analysis was performed using Student’s t tests. **, P ≤ 0.01, significant.
FOXM1 and DNA repair are up-regulated in the resistant MCF-7-CISR cells but not in MCF-7 cells
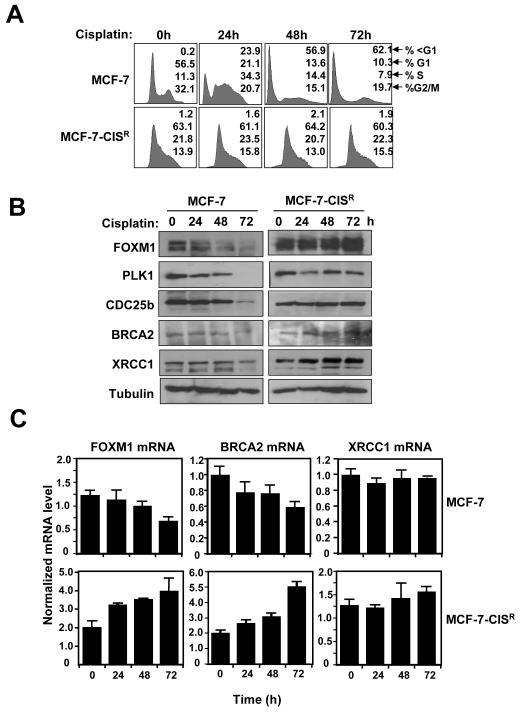
Next, we sought to determine molecular mechanism which confers acquired cisplatin resistance in breast cancer cell lines. Cell cycle analysis showed that following cisplatin treatment (100 nM; 0-72 h) high numbers of MCF-7 cells contained sub-G1 DNA content, indicative of DNA fragmentation and cell death, whilst no significant changes in sub-G1 population were observed for MCF-7-CISR cells (Figure 2A). A series of short time courses revealed that no significant changes in FOXM1, BRCA2 and XRCC1 levels occurred prior to 24 h of cisplatin treatment (Supplemental Figure S1). However, MCF-7 cells treated with cisplatin (0-72 h) showed a decrease in FOXM1 expression, and that of its downstream targets CDC25B and PLK, in addition to the DNA repair proteins XRCC1 and BRCA2 (Figure 2B). In contrast, FOXM1 and BRCA2 expression levels were further increased following cisplatin treatment in MCF-7-CISR cells, whilst CDC25B, PLK and XRCC1 levels remained relatively constant. Consistently, RTQ-PCR analysis revealed that in MCF-7 cells FOXM1 mRNA level decreased by 50% at 72 h, whilst FOXM1 transcript level increased by 2-fold in MCF-7-CISR cells (Figure 2C), suggesting that the ability to maintain elevated FOXM1 expression in acquired cisplatin resistant breast cancer cell lines is mediated at least partially at transcriptional level. Interestingly, although BRCA2 mRNA levels closely mirrored FOXM1 mRNA levels, XRCC1 mRNA levels did not change significantly in both MCF-7 and MCF-7-CISR cells, this suggests that an increase in FOXM1 expression level could stabilize XRCC1 expression indirectly through its other downstream targets. We next performed the immunostaining of phosphorylated histone H2AX loci to assay for DNA damage in response to cisplatin in the drug sensitive and resistant MCF-7 cells. γH2AX staining was examined at the earlier 6 h time point to avoid cell loss due to cell cycle arrest and cell death as a result of DNA damage induced by cisplatin (34, 35). Quantification of γH2AX staining (Figure 3A) demonstrated that MCF-7 cells had significantly higher levels of DNA damage after cisplatin treatment compared with MCF-7-CISR cells, indicating that MCF-7-CISR are more efficient than MCF-7 cells in the repair of damaged DNA induced by cisplatin, which correlates with a much lower amount of apoptosis.
Figure 2. Elevated levels of FOXM1 correlate with enhanced DNA damage repair in MCF-7-CISR cells.
A) MCF-7 and MCF-7-CISR cells were treated with 0.1 μM of cisplatin for 0 to 72 h and FACS analysis was performed after propidium iodide staining. Percentage of cells in each phase of the cell cycle (sub-G1, G1, S, and G2/M) is indicated. Representative data from three independent experiments are shown. B) MCF-7 and MCF-7-CISR cells were treated with 0.1 μM of cisplatin and western blot analysis was performed to determine the protein expression levels of FOXM1, CDC25B, PLK1, BRCA2, XRCC1 and β-tubulin. C) FOXM1, BRCA2 and XRCC1 mRNA transcript levels were determined by RTQ-PCR and normalized to L19 RNA expression. Results of three independent experiments in triplicate and means ± SD are shown.
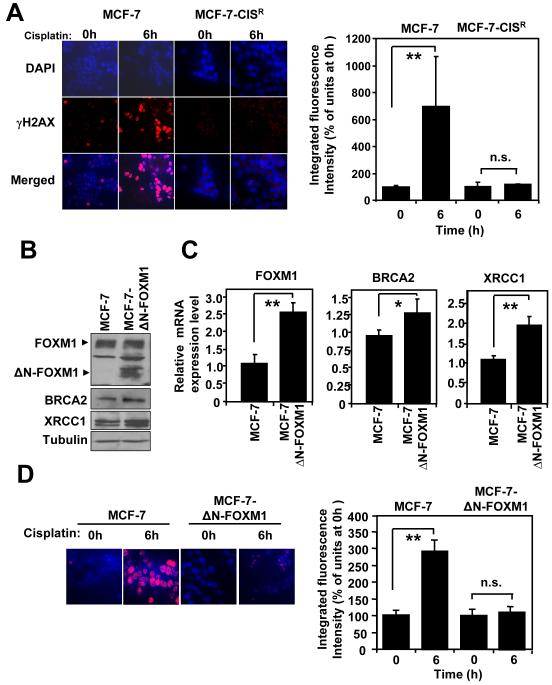
Figure 3. Overexpression of ΔN-FOXM1 is sufficient to confer cisplatin resistance by enhancing DNA repair pathways in reducing DNA damage.
A) MCF-7 and MCF-7-CISR cells were treated with 0.1 μM of cisplatin for 0h or 6h and stained with γH2AX antibodies and DAPI. Images were visualized by confocal microscopy and the average integrated fluorescence intensity quantified by Zeiss Axiovert 100 confocal laser scanning microscope using Zeiss LSM 500 software. Images: original magnification X 40. The relative expression levels of FOXM1, BRCA2 and XRCC1 in MCF-7 and MCF-7-ΔN-FOXM1 were determined by B) Western blotting and C) RTQ-PCR analysis, respectively. D) MCF-7 and MCF-7-ΔN-FOXM1 cells were treated with 0.1 μM of cisplatin for 0h or 6h and stained with γH2AX antibodies and DAPI. Images visualized by confocal microscopy; the average integrated fluorescence intensities are shown. Original magnification X 40. The average results of three independent experiments in triplicates are shown. Mean ± SD. Statistical analyses were performed using Student’s t tests. *, P ≤ 0.05; **, P ≤ 0.01 significant.
Overexpression of ΔN-FOXM1 is sufficient confer cisplatin resistance by enhancing DNA repair pathways in reducing DNA damage
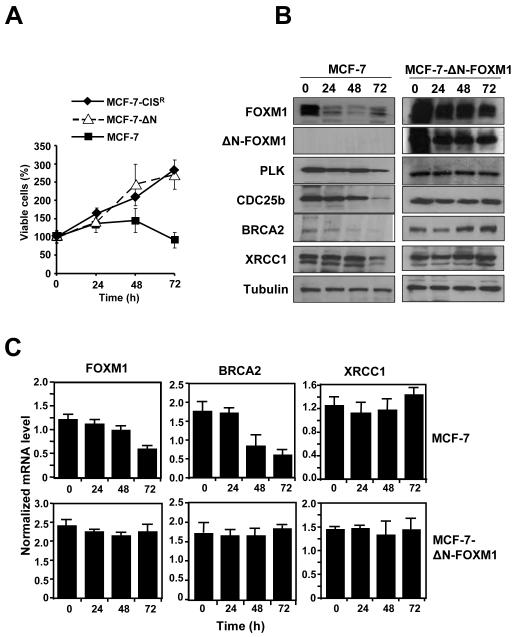
To prove definitively that FOXM1 is important in acquired cisplatin resistance, we employed the use of the MCF-7-ΔNFOXM1 cell line previously described, which over-express a constitutively active form FOXM1 (33). Consistently, expression of ΔN-FOXM1 was accompanied by slightly higher levels of BRCA2 and XRCC1 at both the protein and mRNA level (Figures 3B and 3C). We next examined the level of cisplatin-induced (0.1 μM) DNA damage in MCF-7 and MCF-7-ΔN-FOXM1 cells. A 3.5 fold increase in DNA damage was observed in MCF-7 cells, whilst no significant increase in DNA damage was observed following cisplatin treatment in MCF-7-ΔN-FOXM1 cells (Figure 3D). Therefore, MCF-7-ΔNFOXM1 demonstrated an enhanced ability for DNA repair. Significantly, SRB assay also revealed that the over-expression of ΔN-FOXM1 alone was sufficient to confer resistance to MCF-7 (Figure 4A, Figure 2 and Supplemental Figure S2). Furthermore, the expression levels of FOXM1, BRCA2 and XRCC1 were maintained at both the protein and mRNA levels in the MCF-7-ΔN-FOXM1 cell line following cisplatin treatment (0.1 μM; 0-72 h), whilst in MCF-7 cells FOXM1 expression decreased rapidly after 24 h of cisplatin treatment (Figures 4B and 4C). These data suggest that the introduction of constitutive active ΔN-FOXM1 can protect breast cancer cells against cell death by enhancing cisplatin-induced DNA damage repair.
Figure 4. Overexpression of ΔN-FOXM1 is sufficient confer cisplatin resistance by enhancing DNA repair pathways in reducing DNA damage.
MCF-7 and MCF-7-ΔN-FOXM1 cells were treated with 0.1 μM of cisplatin for 0, 24, 48 and 72 h. A) Cell proliferation rates were determined by SRB assays, and MCF-7-CISR cells were also included as a control for cisplatin-resistant cells.The relative expression levels of FOXM1, BRCA2 and XRCC1 in MCF-7 and MCF-7-ΔN-FOXM1 cells were determined by B) Western blotting C) RTQ-PCR analysis. The expression levels of CDC25B and PLK1 proteins were also studied for comparison. RTQ-PCR results and means ± SD shown are from three independent experiments in triplicates.
FOXM1 can promote cisplatin resistance through DNA damage repair independent of BRCA2 and XRCC1 in MCF-7-CISR cells
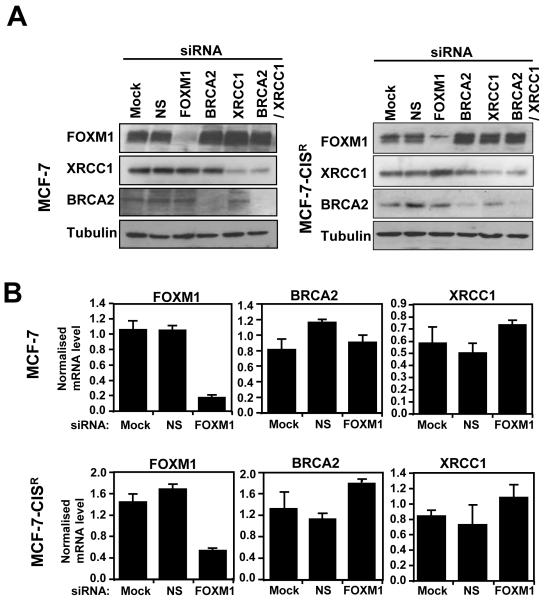
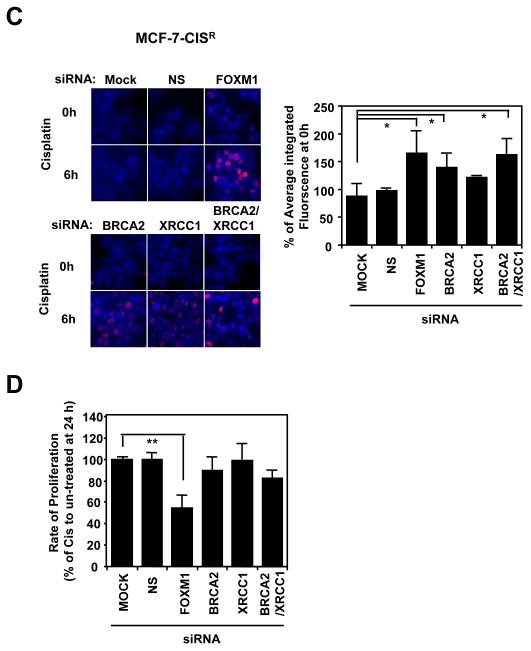
Having identified FOXM1 as a mediator of cisplatin resistance in breast cancer cells, we next examined whether BRCA2 and XRCC1 are the downstream targets of FOXM1 required in conferring cisplatin resistance in breast cancer cells. Surprisingly, the siRNA-mediated knockdown of FOXM1 did not result in a down-regulation of BRCA2 and XRCC1 expression in either the MCF-7 or MCF-7-CISR cell lines at the protein or mRNA level (Figure 5A and 5B), suggesting that FOXM1 is not the primary or sole regulator of BRCA2 and XRCC1 expression in these cells. The requirement of FOXM1, BRCA2, and XRCC1 expression for MCF-7-CISR cisplatin resistance was examined using siRNA-mediated knockdown of these genes. The knockdown of FOXM1, BRCA2 and BRCA2/XRCC1 in MCF-7-CISR cells increased the amount of DNA damage sustained by 1.5 to 2- fold (Figure 5C). Interestingly, following knockdown of FOXM1 the expression levels of BRCA2 and XRCC1 were maintained, and yet an increase in DNA damage was observed, suggesting that other FOXM1 downstream targets are involved. Moreover, SRB proliferation assay revealed that only the knockdown of FOXM1 was potent at re-sensitising MCF-7-CISR cells to cisplatin treatment and not the knockdown of BRCA2, XRCC1 or in combination (Figure 5D). This suggests that other unknown DNA repair targets or proliferative targets of FOXM1 could overcome the loss of one or two individual DNA repair. Thus, the inactivation of FOXM1 is essential for reversing cisplatin resistance and targeting FOXM1 could potentially be a better therapeutic strategy for overcoming cisplatin resistance, rather than just through the inactivation of DNA repair pathways.
Figure 5. BRCA2 and XRCC1 are not the sole downstream targets of FOXM1 involved in cisplatin resistance.
MCF-7 and MCF-7-CISR cells were either untransfected (Mock), transfected with non-specific (NS) siRNA (100 nM) or siRNA smart pool against FOXM1 (100 nM), BRCA2 (100 nM), XRCC1 (100 nM) or BRCA2 plus XRCC1 (100 nM) for 24 h. A) The expression levels of FOXM1, BRCA2 and XRCC1 in MCF-7 and MCF-7-CISR cells were determined by Western blotting. B) RTQ-PCR analysis was performed to determine the relative FOXM1, BRCA2 and XRCC1 mRNA transcript levels. C) MCF-7-CISR cells were then treated with 0.1 μM of cisplatin for either 0h or 6h, and stained with γH2AX antibodies and DAPI. Images visualized by confocal microscopy and the average integrated fluorescence intensity is shown. Original magnification X 40. D) SRB assay was performed to gauge the changes in percentage in cell proliferation in the presence and absence of cisplatin treatment in MCF-7-CISR cells with different siRNA knockdown conditions. Cell proliferation results shown compared the proliferative rates of cisplatin-treated cells with the untreated cells of a given siRNA transfection. Data shown were derived from at least three independent experiments. The error bars show the standard deviation (mean ± SD). Statistical analysis was done using Student’s t tests. *, P ≤ 0.05; **, P ≤ 0.01, significant.
Thiostrepton can overcome cisplatin resistance in breast cancer cells through the down-regulation of FOXM1
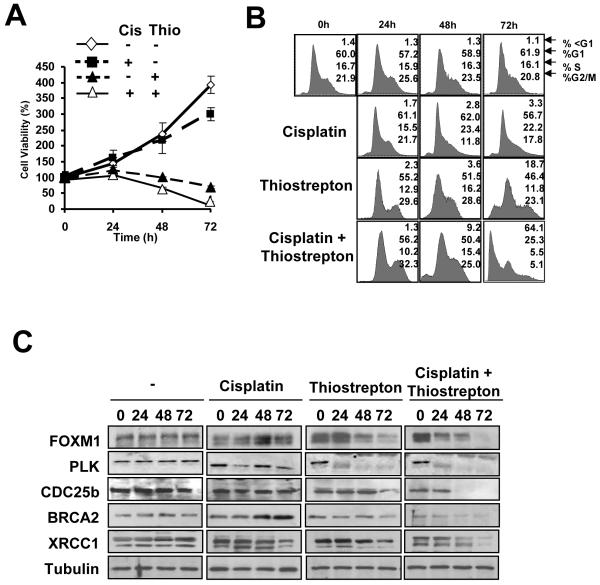
To test if FOXM1 inactivation is a viable strategy for overcoming cisplatin resistance, we studied the effects of MCF-7-CISR cells treated with the thiazole antibiotic thiostrepton, which has previously been showed to inhibit FOXM1 expression (33), alone (10 μM), and in combination with cisplatin (100 nM). SRB proliferative assays indicated that MCF-7-CISR cells treated with thiostrepton, or in combination with cisplatin showed a significant decrease in rate of cell proliferation (Figure 6A). Cell cycle analysis revealed that MCF-7-CISR cells treated with thiostrepton alone showed an 18.7 % cell death rate by 72 h, whilst in combination, cisplatin and thiostrepton showed synergy, exhibiting a cell death rate of 64.1% at 72 h in this experiment (Figure 6B). In MCF-7-CISR cells treated with thiostrepton or thiostrepton and cisplatin, the down-regulation of FOXM1 and its downstream targets CDC25B and PLK occurred at 48 h and 24 h following treatment respectively (Figure 6C). The shorter time needed for FOXM1 down-regulation in the co-treated cells may reflect the higher levels of cell death observed when both drugs were administered together. In conclusion, inhibition of FOXM1 is able to circumvent cisplatin resistance in breast cancer cells.
Figure 6. Thiostrepton can overcome cisplatin resistance in MCF-7-CISR breast cancer cells.
MCF-7-CISR cells were treated with DMSO (vehicle control), 0.1μM cisplatin, 10 μM thiostrepton or a combination of 0.1 μM cisplatin and 10 μM thiostrepton for 72 h. A) SRB proliferation assays were performed on these cells and the percentage of viable cells at each time point is shown. B) Cells were fixed at 0, 24, 48, and 72 h after treatment, and cell cycle phase distribution was analyzed by flow cytometry after propidium iodide staining. Percentage of cells in each phase of the cell cycle (sub-G1, G1, S, and G2/M) is indicated. Representative data from three independent experiments are shown. C) Cell lysates were prepared at the times indicated, and the expression of FOXM1, CDC25B, XRCC1, BRCA2, and β-tubulin analyzed by Western blotting.
Discussion
In the present study we have demonstrated for the first time that FOXM1 possesses a crucial role in cisplatin resistance in breast cancer cells through enhancing DNA-damage repair pathways. Several observations suggest that FOXM1 expression is an important determinant of cisplatin sensitivity and resistance. Firstly, the basal levels of FOXM1 protein and mRNA were higher in the cisplatin resistant MCF-7-CISR cells relative to the parental MCF-7 cells. Following cisplatin treatment, FOXM1 was down-regulated in the sensitive MCF-7 cells while the resistant MCF-7-CISR cells, there was an up-regulating of both FOXM1 mRNA and protein expression levels. Moreover, expression of the constitutively active ΔN-FOXM1 was sufficient to confer resistance to the cisplatin-sensitive MCF-7 breast cancer cells whereas the depletion of FOXM1 through siRNA knockdown reversed cisplatin resistance in MCF-7-CISR breast cancer cells.
FOXM1 has previously been reported to regulate the expression of the DNA repair genes BRCA2 and XRCC1 (25). However, despite the fact that BRCA2 and XRCC1 levels are elevated in the MCF-7-CISR and MCF-7-ΔN-FOXM1 expressing cells, evidence suggests that FOXM1 is not the sole regulator of BRCA2 and XRCC1. For instance, both BRCA2 and XRCC1 expression was not down-regulated upon FOXM1 silencing in both MCF-7 and MCF-7-CISR breast cancer cells. Additionally, we have cloned a BRCA2 gene promoter whose activity is repressible by cisplatin but is not responsive to FOXM1 transactivation (Supplemental Figure S3). This promoter is different from a previously published BRCA2 promoter (25), which locates further downstream the BRCA2 gene (Supplemental Figure S3). Furthermore, despite the fact the expression levels of BRCA2 and XRCC1 were higher in the MCF-7-ΔN-FOXM1 cells compared with the parental cells, transient transfection of MCF-7 with ΔN-FOXM1 failed to up-regulate either BRCA2 or XRCC1 expression (Supplemental Figure S4). All the supplementary evidence implies that additional regulators are needed, though FOXM1 plays a part in their activation.
Furthermore, it was also interesting to observe that although both BRCA2 and XRCC1 knockdown sensitized MCF-7-CISR elevated the amount of cisplatin-induced DNA damage and SRB proliferation assay also revealed that whilst BRCA2 and XRCC1 knockout did not show an appreciable anti-proliferative effect, but the knockdown FOXM1 significantly reduced the proliferative rate of MCF-7-CISR cells in response to cisplatin. These findings also suggest that besides DNA repair, other possible roles for FOXM1 such as the promotion of cell cycle progression or inhibition of cell cycle checkpoints and apoptosis, may also contribute to cisplatin resistance.
These observations may have implications in the development of a treatment regime for cisplatin resistance patients, suggesting it would be more efficient to target a “master” oncogene such as FOXM1, rather than targeting a subset of DNA repair machinery, where potential compensatory mechanisms could be present to hamper the treatment. Moreover, the inhibition of FOXM1 and co-treatment with DNA-damaging agents may be hypothesized to enhance therapeutic response. In order to test this hypothesis, we employed the specific FOXM1 inhibitor thiostrepton (33). Our results indicate that thiostrepton synergised with cisplatin to reverse acquired cisplatin resistance in breast cancer cells, and caused a substantial increase in the amount of cisplatin-induced cell death. This probably reflects the role of FOXM1 in several aspects of physiological processes including proliferation, cell cycle transition and DNA repair, and thus a reduction in the ability of cisplatin resistant cells evade the cytotoxic effects of cisplatin. As a consequence, we have also demonstrated for the first time that the employment of a FOXM1 inhibitor like thiostrepton, in conjunction with chemotherapy, could be provide a mechanism for reversing the phenomenon of wide-spread chemoresistance in breast cancer patients. Moreover, the fact that thiostrepton exclusively targets cancer cells and not non-malignant cells can further enhance the specificity of cisplatin in combinatorial treatments (33).
The mechanism by which FOXM1 activity and/or expression is up-regulated in MCF-7-CISR cells requires further investigation. Treatment with DNA damaging agents, including γ-irradiation, etoposide and UV, has been reported to increase CHK2-induced phosphorylation of FOXM1, potentially stabilizing the protein and leading to the transcriptional up-regulation of downstream DNA repair genes (25). However, several previous studies suggest that it is unlikely that CHK2 phosphorylation of FOXM1 is relevant in acquired cisplatin resistant breast cancer cells. For example, clinical assessment of a selective CHK2 inhibitor VRX0466617 has shown that it can sensitise cancer cells to apoptosis following exposure to ionizing radiation but not cisplatin (36). Moreover, loss or down-regulation of CHK2 expression has been shown to contribute to cisplatin resistance in ovarian cancer and lung cancer (37, 38). Thus, it is possible that additional mechanisms are responsible for the enhanced levels of FOXM1 in MCF-7-CISR cells. Indeed, our studies have showed that resistant cells up-regulated FOXM1 expression at the mRNA level in response to cisplatin. This adds a new dimension to the FOXM1 signalling network whereby following DNA damage FOXM1 activity and expression may be modulated differentially depending on cellular background. In consequence, it will also be interesting to unravel further the exact molecular mechanisms by which FOXM1 mRNA expression levels are regulated in response to cisplatin.
In summary, FOXM1 is a critical mediator of cisplatin sensitivity and resistance in breast cancer cells. Therefore, FOXM1 can be a useful marker for predicting and monitoring cisplatin response. Through the inhibition of FOXM1, it is possible that acquired cisplatin resistance can be reversed, and FOXM1 could be a new therapeutic target in acquired cisplatin resistant breast cancer.
Supplementary Material
Acknowledgments
We thank Evie Maifoshie for help in cloning the human BRCA2 promoter, and Julian Gronau and Dennis Zhang in the quantification of fluorescence intensity in confocal microscopy.
Grant support: Medical Research Council (J. M. Kwok and E.W-F. Lam), Cancer Research UK (S. S. Myatt, R. C. Coombes, E.W-F. Lam), Breast Cancer Campaign (E.W-F. Lam, J. Millour), Biotechnology and Biological Sciences Research Council (B. Peck and E.W-F. Lam).
References
- 1.Kelland L. The resurgence of platinum-based cancer chemotherapy. Nat Rev Cancer. 2007;7:573–84. doi: 10.1038/nrc2167. [DOI] [PubMed] [Google Scholar]
- 2.Siddik ZH. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–79. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 3.Chang IY, Kim MH, Kim HB, et al. Small interfering RNA-induced suppression of ERCC1 enhances sensitivity of human cancer cells to cisplatin. Biochem Biophys Res Commun. 2005;327:225–33. doi: 10.1016/j.bbrc.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Dabholkar M, Bostick-Bruton F, Weber C, Bohr VA, Egwuagu C, Reed E. ERCC1 and ERCC2 expression in malignant tissues from ovarian cancer patients. J Natl Cancer Inst. 1992;84:1512–7. doi: 10.1093/jnci/84.19.1512. [DOI] [PubMed] [Google Scholar]
- 5.Ferry KV, Hamilton TC, Johnson SW. Increased nucleotide excision repair in cisplatin-resistant ovarian cancer cells: role of ERCC1-XPF. Biochem Pharmacol. 2000;60:1305–13. doi: 10.1016/s0006-2952(00)00441-x. [DOI] [PubMed] [Google Scholar]
- 6.Nowosielska A, Marinus MG. Cisplatin induces DNA double-strand break formation in Escherichia coli dam mutants. DNA Repair (Amst) 2005;4:773–81. doi: 10.1016/j.dnarep.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 7.Hurley J, Doliny P, Reis I, et al. Docetaxel, cisplatin, and trastuzumab as primary systemic therapy for human epidermal growth factor receptor 2-positive locally advanced breast cancer. J Clin Oncol. 2006;24:1831–8. doi: 10.1200/JCO.2005.02.8886. [DOI] [PubMed] [Google Scholar]
- 8.Lee YJ, Doliny P, Gomez-Fernandez C, Powell J, Reis I, Hurley J. Docetaxel and cisplatin as primary chemotherapy for treatment of locally advanced breast cancers. Clin Breast Cancer. 2004;5:371–6. doi: 10.3816/cbc.2004.n.044. [DOI] [PubMed] [Google Scholar]
- 9.Pegram MD, Pienkowski T, Northfelt DW, et al. Results of two open-label, multicenter phase II studies of docetaxel, platinum salts, and trastuzumab in HER2-positive advanced breast cancer. J Natl Cancer Inst. 2004;96:759–69. doi: 10.1093/jnci/djh133. [DOI] [PubMed] [Google Scholar]
- 10.Sirohi B, Arnedos M, Popat S, et al. Platinum-based chemotherapy in triple-negative breast cancer. Ann Oncol. 2008;19:1847–52. doi: 10.1093/annonc/mdn395. [DOI] [PubMed] [Google Scholar]
- 11.Stewart DJ. Mechanisms of resistance to cisplatin and carboplatin. Crit Rev Oncol Hematol. 2007;63:12–31. doi: 10.1016/j.critrevonc.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 12.Eckstein N, Servan K, Girard L, et al. Epidermal growth factor receptor pathway analysis identifies amphiregulin as a key factor for cisplatin resistance of human breast cancer cells. J Biol Chem. 2008;283:739–50. doi: 10.1074/jbc.M706287200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yde CW, Issinger OG. Enhancing cisplatin sensitivity in MCF-7 human breast cancer cells by down-regulation of Bcl-2 and cyclin D1. Int J Oncol. 2006;29:1397–404. [PubMed] [Google Scholar]
- 14.Hong Y, Yang J, Wu W, et al. Knockdown of BCL2L12 leads to cisplatin resistance in MDA-MB-231 breast cancer cells. Biochim Biophys Acta. 2008;1782:649–57. doi: 10.1016/j.bbadis.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 15.Chu F, Barkinge J, Hawkins S, Gudi R, Salgia R, Kanteti PV. Expression of Siva-1 protein or its putative amphipathic helical region enhances cisplatin-induced apoptosis in breast cancer cells: effect of elevated levels of BCL-2. Cancer Res. 2005;65:5301–9. doi: 10.1158/0008-5472.CAN-04-3270. [DOI] [PubMed] [Google Scholar]
- 16.Bu Y, Lu C, Bian C, et al. Knockdown of Dicer in MCF-7 human breast carcinoma cells results in G1 arrest and increased sensitivity to cisplatin. Oncol Rep. 2009;21:13–7. [PubMed] [Google Scholar]
- 17.Korver W, Roose J, Heinen K, et al. The human TRIDENT/HFH-11/FKHL16 gene: structure, localization, and promoter characterization. Genomics. 1997;46:435–42. doi: 10.1006/geno.1997.5065. [DOI] [PubMed] [Google Scholar]
- 18.Petrovic V, Costa RH, Lau LF, Raychaudhuri P, Tyner AL. FoxM1 regulates growth factor-induced expression of kinase-interacting stathmin (KIS) to promote cell cycle progression. J Biol Chem. 2008;283:453–60. doi: 10.1074/jbc.M705792200. [DOI] [PubMed] [Google Scholar]
- 19.Wang IC, Chen YJ, Hughes D, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25:10875–94. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung TW, Lin SS, Tsang AC, et al. Over-expression of FoxM1 stimulates cyclin B1 expression. FEBS Lett. 2001;507:59–66. doi: 10.1016/s0014-5793(01)02915-5. [DOI] [PubMed] [Google Scholar]
- 21.Madureira PA, Varshochi R, Constantinidou D, et al. The Forkhead box M1 protein regulates the transcription of the estrogen receptor alpha in breast cancer cells. J Biol Chem. 2006;281:25167–76. doi: 10.1074/jbc.M603906200. [DOI] [PubMed] [Google Scholar]
- 22.Wierstra I, Alves J. FOXM1c and Sp1 transactivate the P1 and P2 promoters of human c-myc synergistically. Biochem Biophys Res Commun. 2007;352:61–8. doi: 10.1016/j.bbrc.2006.10.151. [DOI] [PubMed] [Google Scholar]
- 23.Wang Z, Banerjee S, Kong D, Li Y, Sarkar FH. Down-regulation of Forkhead Box M1 transcription factor leads to the inhibition of invasion and angiogenesis of pancreatic cancer cells. Cancer Res. 2007;67:8293–300. doi: 10.1158/0008-5472.CAN-07-1265. [DOI] [PubMed] [Google Scholar]
- 24.Dai B, Kang SH, Gong W, et al. Aberrant FoxM1B expression increases matrix metalloproteinase-2 transcription and enhances the invasion of glioma cells. Oncogene. 2007;26:6212–9. doi: 10.1038/sj.onc.1210443. [DOI] [PubMed] [Google Scholar]
- 25.Tan Y, Raychaudhuri P, Costa RH. Chk2 mediates stabilization of the FoxM1 transcription factor to stimulate expression of DNA repair genes. Mol Cell Biol. 2007;27:1007–16. doi: 10.1128/MCB.01068-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–9. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 27.Francis RE, Myatt SS, Krol J, et al. FoxM1 is a downstream target and marker of HER2 overexpression in breast cancer. Int J Oncol. 2009;35:57–68. doi: 10.3892/ijo_00000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bektas N, Haaf A, Veeck J, et al. Tight correlation between expression of the Forkhead transcription factor FOXM1 and HER2 in human breast cancer. BMC Cancer. 2008;8:42. doi: 10.1186/1471-2407-8-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goode EL, Ulrich CM, Potter JD. Polymorphisms in DNA repair genes and associations with cancer risk. Cancer Epidemiol Biomarkers Prev. 2002;11:1513–30. [PubMed] [Google Scholar]
- 30.Turner N, Tutt A, Ashworth A. Hallmarks of ‘BRCAness’ in sporadic cancers. Nat Rev Cancer. 2004;4:814–9. doi: 10.1038/nrc1457. [DOI] [PubMed] [Google Scholar]
- 31.Sakai W, Swisher EM, Karlan BY, et al. Secondary mutations as a mechanism of cisplatin resistance in BRCA2-mutated cancers. Nature. 2008;451:1116–20. doi: 10.1038/nature06633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swisher EM, Sakai W, Karlan BY, Wurz K, Urban N, Taniguchi T. Secondary BRCA1 mutations in BRCA1-mutated ovarian carcinomas with platinum resistance. Cancer Res. 2008;68:2581–6. doi: 10.1158/0008-5472.CAN-08-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kwok JM, Myatt SS, Marson CM, Coombes RC, Constantinidou D, Lam EW. Thiostrepton selectively targets breast cancer cells through inhibition of forkhead box M1 expression. Mol Cancer Ther. 2008;7:2022–32. doi: 10.1158/1535-7163.MCT-08-0188. [DOI] [PubMed] [Google Scholar]
- 34.Huang X, Okafuji M, Traganos F, Luther E, Holden E, Darzynkiewicz Z. Assessment of histone H2AX phosphorylation induced by DNA topoisomerase I and II inhibitors topotecan and mitoxantrone and by the DNA cross-linking agent cisplatin. Cytometry A. 2004;58:99–110. doi: 10.1002/cyto.a.20018. [DOI] [PubMed] [Google Scholar]
- 35.Brozovic A, Damrot J, Tsaryk R, et al. Cisplatin sensitivity is related to late DNA damage processing and checkpoint control rather than to the early DNA damage response. Mutat Res. 2009;670:32–41. doi: 10.1016/j.mrfmmm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Carlessi L, Buscemi G, Larson G, Hong Z, Wu JZ, Delia D. Biochemical and cellular characterization of VRX0466617, a novel and selective inhibitor for the checkpoint kinase Chk2. Mol Cancer Ther. 2007;6:935–44. doi: 10.1158/1535-7163.MCT-06-0567. [DOI] [PubMed] [Google Scholar]
- 37.Zhang P, Wang J, Gao W, Yuan BZ, Rogers J, Reed E. CHK2 kinase expression is down-regulated due to promoter methylation in non-small cell lung cancer. Mol Cancer. 2004;3:14. doi: 10.1186/1476-4598-3-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang P, Gao W, Li H, Reed E, Chen F. Inducible degradation of checkpoint kinase 2 links to cisplatin-induced resistance in ovarian cancer cells. Biochem Biophys Res Commun. 2005;328:567–72. doi: 10.1016/j.bbrc.2005.01.007. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.