Abstract
Background
Deconjugation of ubiquitin and/or ubiquitin-like modified protein substrates is essential to modulate protein-protein interactions and, thus, signaling processes in cells. Although deubiquitylating (deubiquitinating) enzymes (DUBs) play a key role in this process, however, their function and regulation remain insufficiently understood. The "loss-of-function" phenotype studies can provide important information to elucidate the gene function, and zebrafish is an excellent model for this goal.
Results
From an in silico genome-wide search, we found more than 90 putative DUBs encoded in the zebrafish genome belonging to six different subclasses. Out of them, 85 from five classical subclasses have been tested with morpholino (MO) knockdown experiments and 57 of them were found to be important in early development of zebrafish. These DUB morphants resulted in a complex and pleiotropic phenotype that, regardless of gene target, always affected the notochord. Based on the huC neuronal marker expression, we grouped them into five sets (groups I to V). Group I DUBs (otud7b, uchl3 and bap1) appear to be involved in the Notch signaling pathway based on the neuronal hyperplasia, while group IV DUBs (otud4, usp5, usp15 and usp25) play a critical role in dorsoventral patterning through the BMP pathway.
Conclusion
We have identified an exhaustive list of genes in the zebrafish genome belonging to the five established classes of DUBs. Additionally, we performed the corresponding MO knockdown experiments in zebrafish as well as functional studies for a subset of the predicted DUB genes. The screen results in this work will stimulate functional follow-up studies of potential DUB genes using the zebrafish model system.
Background
During the past 15 years, protein modification by ubiquitin (UBQ) and ubiquitin-like (UBL) molecules has been suggested to be an important mechanism in regulating numerous critical cellular processes, such as signal transduction, transcriptional control, protein degradation, epigenetic modification and intracellular localization. UBQ is a 76 amino-acid protein that is covalently linked to lysine residues of target proteins in a multi-step process. The process of UBQ conjugation involves three basic classes of enzymes. UBQ-activating enzymes (E1s) activate the C terminus of UBQ, resulting in the formation of an ATP-dependent thio-ester that is linked to the E1's active site - cysteine residue. Afterwards, UBQ is transferred to the active site - cysteine residue - of an E2 conjugating enzyme. The third class of enzymes, the UBQ-protein ligases (E3s) catalyze the final reaction (the UBQ-conjugation process), in which the UBQ is attached to the lysine residue of the protein substrate (reviewed in [1,2]). As a result of the ubiquitylation (ubiquitination) process, the target protein can be modified by a single UBQ (mono-ubiquitylation) or several UBQs (poly-ubiquitylation), which determines the tagging outcome [3].
Deconjugation of UBQ- and/or UBL-modified protein substrates is essential to recycle both the substrate proteins and the UBQ or UBL modifiers; thus, this process influences protein-protein interactions, signaling processes and the pool of free UBQ/UBL within the cell. The deconjugation process is managed by deubiquitylating enzymes (DUBs). The DUBs are in a heterogeneous group of cysteine or metalloproteases [4], which can cleave the scissile peptide bond between the last residue of UBQ (Gly 76) and target proteins. Since their discovery in 1990s, DUBs have been emerging as key regulators in many basic cellular processes. For example, they modulate the ubiquitin-proteasome system (reviewed in [5,6]). Recently, numerous studies have shown that DUBs are linked to diseases such as cancer. CYDL is a tumor suppressor [7-11]; UCHL1 and USP24 are involved in Parkinson's disease [12-16]; ATXN3 influences ataxias [17,18].
Most extensively, DUBs have been studied in the human genome [4]. The classical DUBs can be divided into five subclasses based on their UBQ-protease domains. These are (i) UBQ-specific proteases (USPs), (ii) otubain proteases (OTUs), (iii) UBQ C-terminal hydrolases (UCHs), (iv) Machado-Joseph disease proteases (MJDs) and (v) JAMM motif proteases (JAMMs). Nijman et al. have identified 95 DUBs, out of which the overwhelming majority (58) belongs to the USP class. Whereas the DUBs in the JAMM family are metalloproteases, the other four groups belong to the cysteine protease class. Additionally, two more subclasses of DUBs have been suggested based on evidence from sequence homology [19]. The so-called PPPDE group involves papain-like proteases and the WLMs (Wss1p-like proteases) are metalloproteases. Both are hypothesized to play a role in removal of UBQ or UBL from substrate proteins. Experimentally, DUBs can be identified with several approaches such as using hemagglutinin-tagged (HA) UBQ-derived probes [20] or biotin-labeled UBQ aldehyde probes [21] for labeling. These methods had been used to identify several members of the USP, UCH and OTU families [6]. Alternatively, extension of families of known or suspected representatives of DUB classes is possible with the in silico collection of families of significantly similar sequences within the homology concept.
It must be noted that the understanding of the DUBs' functions remains very limited despite considerable research efforts. Apparently, the problems are associated with the large number of various DUBs (some of which might compensate for others in parallel pathways) and with their position in the subcellular gene networks where a single DUB can affect several pathways via its protein substrates. One possibility to get new insight into the problem is to involve new model organisms into the analysis and to rely on their specific methodological advantages. The zebrafish (Danio rerio) system with its morpholino (MO) technology is superbly suited for easily studying the loss-of-function phenotype in vivo, important information necessary to elucidate the genes' function [22,23]. In this work, we use the zebrafish model to identify the MO knockdown phenotypes of potential DUB genes that we have identified with in silico methods from the genome sequence. The respective experiments have been carried out for 85 putative DUBs. 57 out of the 85 genes were found to be required for normal development and their morphants have exhibited complex phenotypes always with inclusion of notochord deformations. In order to narrow down the functions in development for some of the genes, we chose a neuronal marker, huC, for grouping the DUBs into subgroups that provide hints for specific pathway involvement. This first loss-of-function study of DUB genes in zebrafish provides a valuable resource for further DUB studies in vertebrates.
Results
Zebrafish has more than 90 putative DUBs encoded in its genome
DUB-subclass-specific Hidden Markov model and BLAST/PSI-BLAST searches (as described in the Methods section) have been applied for identifying DUBs in the proteome of zebrafish. It should be noted that the first genome-wide search has been started in February 2008 and was regularly repeated (last time, in April 2009). Depending on the zebrafish proteome and the non-redundant protein database, the numbers of hits were slightly different and not all putative DUB sequences were known at the planning stage of experimental work. Therefore, we conclude that our current list of potential DUBs in zebrafish might not be completely exhaustive; yet, it appears comprehensive at the time of writing. For the ubiquitin-specific protease (USP) class containing a UCH (PF0043) domain, we determined 51 proteins. 15 zebrafish proteins resembling OTU-like cysteine proteases (OTUs) have an OTU (PF02338) domain. Another three proteins have the features of Machado-Joseph disease proteases (MJDs) with Josephine domain (PF02099). There are four zebrafish ubiquitin C-terminal hydrolase proteins (UCHs) sharing a Peptidase_C12 (PF1088) domain. The DUB metalloproteases are represented by 14 proteins carrying a Mov34/MPN/PAD-1 family (PF01398) domain (JAMMs). Another four proteins belong to the PPPDE class (PF05903). In total, this results in 91 candidate targets. In Additional file 1, the names and accession numbers of all 91 hits are shown.
The sequence domain architectures of all 91 DUB hits are also shown in Additional file 1. The combination of UBQ-protease domains with domains and motifs from the ubiquitylation pathway (such as ubiquitin-associated UBA, ubiquitin UBQ and ubiquitin-interacting motif, UIM) is not a real surprise, whereas the association with nucleic-acid binding domains (Myb_DNA_binding, PRO8 and Tudor) and protein-binding domains (SWIRM and MATH) would make sense in the context of the involvement of DUB proteins in the regulation of gene expression. The PRPF8 (pre-mRNA-processing-splicing factor 8) protein, a member of the JAMM protein family, is known to be one of the largest and most highly conserved nuclear protein that occupies a central position in the catalytic core of the spliceosome [24]. Another member of the JAMM protein family, MYSM1, is a histone H2A deubiquitinase that regulates transcription by coordinating histone acetylation and deubiquitylation, and destabilizing the association of linker histone H1 with nucleosomes [25]. We searched the scientific literature extensively for evidence for DUB functions and the summary of known DUBs is shown in Additional file 2.
We think that both the human and zebrafish DUB lists are not complete at present and revisions of the genome build or the proteome lists will lead to some correction. As a general trend, the total numbers as well as the numbers of DUBs in the subclasses are quite comparable between both organisms [4]. For the overwhelming majority (about four fifths), there are also clear orthologues in fish and humans (these genes carry the names of their human counterparts in Additional file 1); yet, there are also a few cases of missing equivalents in either organism or organism-specific duplications and it is not clear at present whether genome revisions will resolve these discrepancies. So far, it appears too early to draw biological conclusions out of differences in the DUB lists.
57 DUB genes are required for zebrafish early development
At the time of starting the MO experiments, our list comprised 85 gene entries. For most of them, we found records in GenBank and/or the Ensembl genome database (Additional file 3) suitable for straightforward MO design. It is a common and general practice to design the MOs that target the ATG translation sites. Translation-blocking MO interferes with the protein translation process of the target gene. However, ATG translation start site information was missing for 11 sequences and we used a workaround as described in the Methods section. For these genes, we designed MOs targeting splicing of exon-intron junction that is on or in the vicinity of conserved domains (marked in bold italics in Additional file 3).
Among all 85 MOs tested, 26 MOs (31%) led to no detectable morphological phenotypes, while two MOs (2%) resulted in cell death in early developmental stages (cell arrest after 30% epiboly). The remaining 57 MOs (67%) gave different observable phenotypes, including abnormal development in head, brain, eyes, body axis, notochord, precardial region, yolk and tail. Most of the MOs did not specially affect one region but several. The notochord was always affected in the MO knockdown experiments. The results are summarized in Additional file 4. In this table, brief phenotypic descriptions and images of the morphants are shown.
Classifying DUB genes into five subgroups according to their huC expression level and pattern
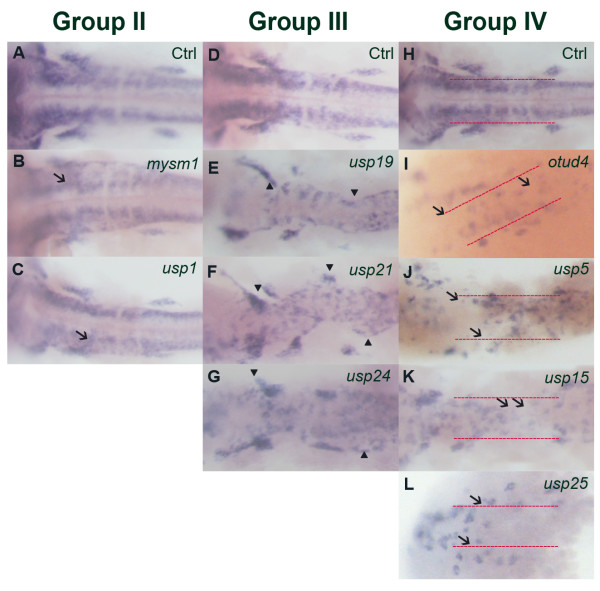
As we mentioned above, most of the DUB genes gave pleiotropic phenotypes. It appears difficult to classify the diverse phenotypes based on their varied morphology. Therefore, we applied additional in situ staining of the neuronal huC marker. We chose huC as the primary marker because of its strong and easily recognizable staining patterns in zebrafish early development. In addition, neuronal development is always a hot topic and research-focusing point. In the screen, its distinctive distribution patterns allowed classifying 83 out of 85 DUB genes into five groups (except for zranb1a and usp46 (2%) that resulted in cell death). The largest group includes 47 members (group V, 55%), which shows no change in huC expression level or patterning. Within this group, all the members exhibit the same neuronal pattern as the control. Neurons are formed in a well-organized manner (data not shown). Group I is the only group that has an increased huC expression (increase in both expression level and number of neurons) and/or enlarged patterning and is composed of three members (4%). Overgrown hindbrain neurons were found among them (Figure 1A-D). Groups II to IV are characterized by a decrease in huC expression with different patterns. Group II shows decreased huC expression without changed patterning and comprises 16 members (19%). Group III exhibits decreased huC expression with slightly destructive patterning effect and has 13 members (15%). They have abnormal shape of hindbrain neurons, such as curved neuron pattern and uneven cranial ganglia length. Finally, group IV features a decrease of huC expression together with severely destructed patterning and it is made up of 4 members (5%). All the members in this group cannot form organized neuronal patterns. Besides, individual small clusters of neurons were observed. Moreover, all the cranial ganglia structures were lost in this group. Selected results among them are shown in Figure 2. Since groups I and IV showed distinct huC in situ results, we next focused on these two groups for general signaling and developmental studies. The classification is summarized in Additional file 4.
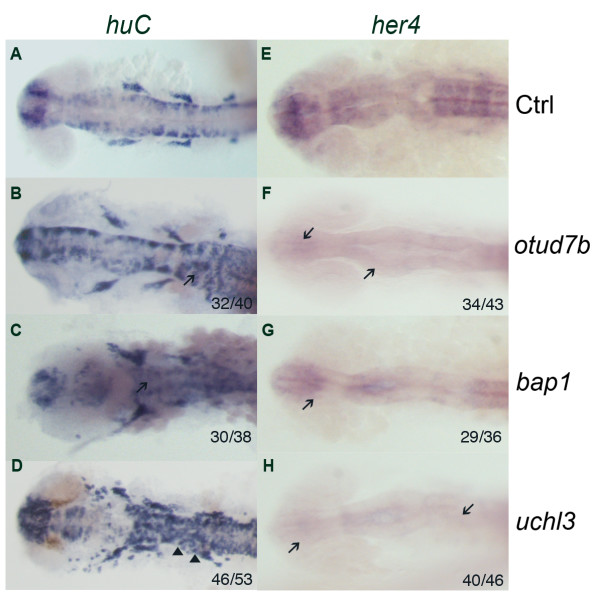
Figure 1.
huC and her4 expression in morphants of group I zebrafish DUBs at 24 hpf. huC and her4 expression at 24 hpf, dorsal view. Increase in huC expression was found in morphants of group I DUBs when compared to control (A-D). There are 3 members in this group, which named as otud7b, bap1 and uchl3. The arrows mark the increased huC expression area (B-C), while the arrowheads mark the denser expression pattern (D). uchl3 morphants showed the most severe effect with both stronger and denser huC expression. her4 is a downstream target gene in the Notch signaling pathway, increase of neurons may be due to the decrease of the Notch activation. Decrease in her4 expression was found in morphants of all group I DUBs when compared to control (E): otud7b (F), bap1 (G) and uchl3 (H). Arrows show the absence of her4 expression in the morphants. The number of embryos with the presented phenotype is shown in the right bottom corner of each panel.
Figure 2.
Comparison of huC expression in morphants of groups II, III and IV zebrafish DUBs at 24 hpf. huC expression at 24 hpf, dorsal view. Decreased huC expression was found in morphants of these 3 groups. Group II has 16 members that showed a typical decreased huC expression (arrows). Group III has 13 members that showed the decreased huC expression with destructive patterning. Lastly, group IV has 4 members that had the most severe decreased and destructive huC expression. All 4 members named otud4, usp5, usp15 and usp25 could not form a normal neuronal pattern in the head region. Arrowheads mark the abnormal patterns in morphants of group III; while arrows indicate the absence of neurons in morphants of group IV.
otud7b, bap1 and uchl3 (group I) may be involved in Notch signaling
The knockdown of group I DUB genes resulted in increased huC expression (Figure 1A-D, Additional file 5), indicating an increase in neurons. Neurogenesis is the process of forming neurons in the early development. Neuronal death during the development is a normal process happened in the normal development [26]. One of the well known midline cell fate signaling is the Notch pathway [27,28]. Inactivation of Notch pathway leads to a failure of the lateral inhibition, which results in premature differentiation of neural progenitors and permits neuron mass production.
Activation of Notch leads to her4 expression [29]. Morphants of all group I DUB genes (otud7b, bap1 and uchl3) showed neuronal hyperplasia, indicating a compromise in Notch-dependent lateral inhibition during development (Figure 1A-D). Consistently, all their morphants showed decreased her4 expression in descending order of otud7b, bap1 and uchl3 (Figure 1E-H, Additional file 5), indicating that their corresponding genes may be involved in the Notch signaling pathway.
otud4, usp5, usp15 and usp25 (group IV) play critical roles in dorsoventral patterning
From the huC expression, we noticed that neurons of group IV morphants are not well organized, implying that their dorsoventral patterning might be affected in the early stages. From the phenotypic studies at 24 hpf, the group IV morphants have dorsalized phenotypes that ranged from C1 to C5 (Additional file 6), where C5 shows the strongest dorsalized phenotype [30,31]. Their phenotypes became milder afterwards (Additional file 4), which might be due to the decreasing MO blocking effect at later stages and/or negative feedback regulation on gene expression [32]. We collected the MO-injected embryos for in situ staining at about 50%-60% epiboly and 10-somite stages. Several molecular markers were used to examine the mesoderm, ectoderm and dorsolateral regions.
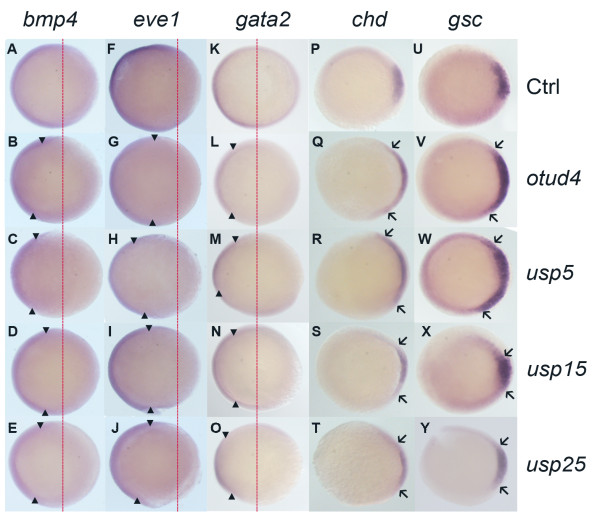
Three different markers of ventral territories (bmp4, eve1 and gata2) and two dorsal ectoderm and mesoderm markers (chd and gsc) were used. Bone morphogenetic protein 4 (Bmp4) is important to the dorsoventral patterning of the mesoderm. The failure of its expression in dorsal blastomeres causes the ventralization of the embryo [33-35]. eve1 is a zebrafish homeobox gene similar to even-skipped in Drosophila [36]. It is strongly expressed in the ventrolateral marginal cells. gata2 is a hematopoietic transcription factor gene [37] that is expressed in a pattern similar to eve1. It is a marker for ventral ectoderm and hematopoietic cells in ventral mesoderm. All three markers showed similar results in the group IV morphants. Their transcripts in the ventral half of the marginal and the animal zone are expressed in a more restricted area than controls (Figure 3A-O, Additional file 7). For the dorsal patterning, we used chordin (chd) and goosecoid (gsc). Chordin can antagonize and repress BMP4 and is normally dorsally restricted [38]. Similarly, gsc expression is also restricted to dorsal region [39]. Expression of chd and gsc in the morphants was expanded into lateral domains (Figure 3P-Y). To summarize, the ventral fates were affected and restricted in a smaller area while the dorsal region was expanded during gastrula.
Figure 3.
Comparison of ventral (bmp4, eve1, gata2) and dorsal (chd, gsc) markers expression in morphants of group IV zebrafish DUBs at 50-60% epiboly. Ventral markers (bmp4, eve1 and gata2) and dorsal markers (chd and gsc) expression of group IV morphants at 50-60% epiboly stage, animal pole views, dorsal towards the right. otud4 (B, G, L, Q, V), usp5 (C, H, M, R, W) and usp25 (E, J, O, T, Y) morphants showed narrower expression pattern for ventral markers (A-O) but wider expression pattern of dorsal markers (P-Y). usp15 showed a similar expression pattern (D, I, N, S, X) with control (A, F, K, P, U). Red dot lines indicate the normal expression margin of ventral markers in wild-type embryos.
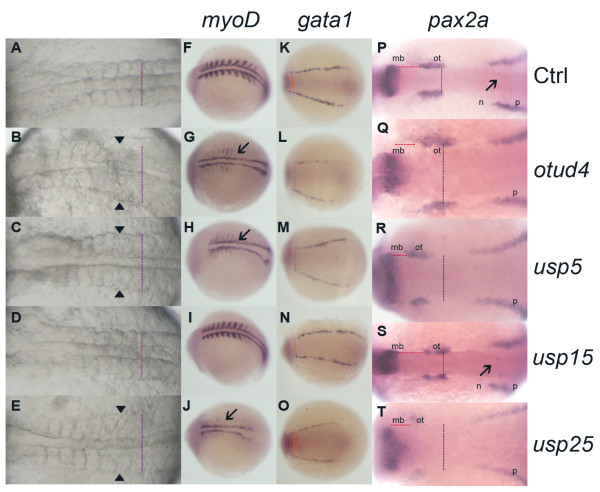
The shift of various marker genes (bmp4, chd, eve1, gata2 and gsc) would result in phenotypic changes in the later developmental stages. In order to confirm that the dorsalized effects are maintained after gastrulation, we chose some other in situ markers for the studies at 10-somite stage: myoD is used for dorsal mesoderm and somite muscle staining [40,41]; gata1 is ventrally expressed in presumptive hematopoietic cells in two lateral stripes [37,42]; and pax2a is used for marking the presumptive neural region [43]. From the in situ results, we found that myoD and pax2a expression patterns were expanded in group IV morphants, indicating a dorsalized phenotype [41,44]. myoD expression in otud4, usp5 and usp25 morphants was weaker but wider. The number of stained somatic muscles was decreased. usp25 morphants showed the most severe result while usp15 showed a similar myoD pattern to wild-type (Figure 4F-J). For gata1 staining, widening of the two lateral stripes of presumptive hematopoietic cells was found in otud4, usp5 and usp25 morphants. Same as the results of other probes, usp15 morphants showed a similar gata1 expression pattern as wild-type (Figure 4K-O). Similarly, in pax2a in situ staining, usp15 morphants showed a similar expression pattern as wild-type. In otud4, usp5 and usp25 morphants, distance between two otic vesicles was increased, which featured dorsalization. In addition, usp25 morphants also showed a decreased pax2a expression in otic vesicles (Figure 4P-T). Taken together, the abnormal dorsoventral patterning in morphants of otud4, usp5, usp15 and usp25 genes started from gastrula and continued in later developmental stages. In addition, while usp25 morphants always showed a more severe dorsalized pattern, usp15 always showed the least.
Figure 4.
Comparison of myoD, gata1 and pax2a expression in morphants of group IV zebrafish DUBs at 10-somite stage. Morphology of group IV morphants at 10-somite stage (A-E). Lateral expansion of somite muscles were indicated (arrowheads and purple dot lines) in otud4 (B), usp5 (C) and usp25 (E) morphants. usp15 (D) morphants showed a similar phenotype as control (A). myoD, gata1 and pax2a expression at 10-somite stage, dorsal view (F-T). Orange dot lines represent the distance between two lateral stripes; blue dot lines indicate the distance between two optic vesicles; while red dot lines show the distance between midbrain and otic vesicle. mb, midbrain; ot, otic vesicle; n, neuronal and p, pronephric precursor expression domains. otud4, usp5 and usp25 morphants showed features of dorsalization: lateral expression of myoD (G-J, marked with arrows), pax2a (Q-T) and widening of the gata1 (L-O) mesoderm distance expression. usp15 (I, N, S) showed a similar expression to control (F, K, P). All are head to the left.
The group IV DUB genes are likely involved in the BMP pathway
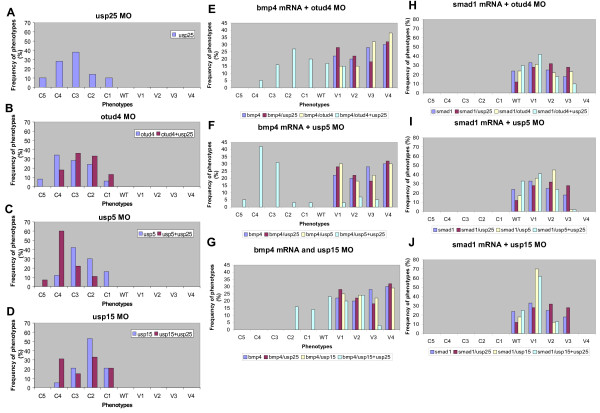
The BMP signaling pathway is well known for its essentiality in many developmental processes [45]. In zebrafish, it plays a main role in determining the ventral cell fates [46]. Briefly, the BMP signaling pathway starts from the BMP receptors activation. The receptors are mediated by BMPs and will active the Smad proteins (R-Smad) through phosphorylation. The Smad protein will then bind to a common mediator Smad (Co-Smad) to form complexes, which will finally be translocated into the nucleus and act as transcriptional regulators. Mutations of ventral bmp gene will result in the dorsalization of the embryo. swirl (swr) and snailhouse (snh) [30] are two well known mutants that have mutations in bmp2b [31] and bmp7 genes [47,48], respectively. To gain further insights into where group IV DUBs function in the BMP pathway, we preformed co-injection of group IV MOs and selected mRNA of BMP signaling genes. Single mRNA injections of bmp4 and smad1 resulted in ventralized phenotypes as previously described [31,49]. When embryos were injected with bmp4 mRNA and group IV MO, we observed ventralized phenotypes (Figure 5E-G). The results seemed to disagree with our hypothesis, since Bmp4 is at the uppermost position in the BMP pathway. However, because DUBs are enzymes, their functions might be compensated or replaced by others. Thus, we preformed co-injection of bmp4 mRNA with different combination of group IV DUB MOs. Based on the single MO injection result, we chose the most effective usp25 MO as a basic component and all the MO concentrations were halved as single injection. Except for the mild effect of usp15+usp25 double knockdown, both otud4+usp25 and usp5+usp25 MOs significantly enhanced the dorsalized phenotypes in embryos co-injected with bmp4 mRNA (Figure 5E-G). For co-injection of smad1 mRNA and group IV MOs, ventralized phenotypes were found (Figure 5H-J). Specific injection amounts were tabulated in Table 1. In the table, we introduced the Dorsal-Ventral (DV) value, which was calculated by the frequencies of phenotypes times a fixed value (5 for C5, 4 for C4, -4 for V4 and -3 for V3 phenotypes, etc.). The final DV value indicated the average result of the co-injection experiments. To summarize, the combination MOs could not rescue ventralized phenotypes caused by smad1 mRNA injection (negative DV value), while maintained their dorsalized phenotypes when co-injected with the bmp4 mRNA (positive DV value except the mild effect of usp15+usp25). These data suggested that the group IV DUBs act downstream of Bmp4 and upstream of Smad1 in dorsoventral patterning. The substrate of the particular E3 ligase in the BMP pathway can be deubiquitylated more efficiently by two DUBs. Collectively, all the group IV DUB genes are involved in the BMP pathway and can affect the early dorsoventral patterning in the zebrafish. The proposed working model is depicted in Figure 6A-G. We classified the phenotypes based on Additional file 6 and further confirmed by in situ staining using different molecular markers at different stages (Additional file 8).
Figure 5.
Frequency of dorsalized and ventralized phenotypes after knockdown of group IV DUB genes and overexpression of Bmp signaling genes. Panels A to D showed the frequency of dorsalized phenotypes after knockdown of group IV DUB genes. Noted that severe dorsalized phenotypes were found after double-knockdown of usp5+usp25 or usp15+usp25 (C, D). Panels E to G showed the phenotypes' frequencies after overexpression of bmp4 and group IV DUB genes knockdown. Noted that there were phenotypic shift from ventralization to dorsalization after double knockdown of two DUB genes, with more obvious changes in otud4+usp25 (E) and usp5+usp25 (F) double knockdown. Panels H to J showed the results of the smad1 overexpression and double knockdown of group IV DUB genes. Noted that the ventralized effects were maintained in the double knockdown of group IV DUB genes. Collectively, the results suggested that group IV DUB genes are involved in the BMP pathway and function between Bmp4 and Smad1. Noted that there were three different Y axis scales in the figure (A-D; E-G; H-J). The number of injected embryos was showed in Table 1.
Table 1.
Dorsal-ventral value of RNA and/or MO injection studies
| RNA/MO | pg/em | pmol | n ep | n | DV value |
|---|---|---|---|---|---|
| -/otud4 | 0.5 | 3 | 104 | 3.14 | |
| -/usp5 | 2.0 | 3 | 101 | 2.50 | |
| -/usp15 | 0.5 | 3 | 127 | 2.10 | |
| -/usp25 | 2.0 | 3 | 185 | 3.14 | |
| -/otud4+usp25 | 0.25/1.0 | 2 | 100 | 2.59 | |
| -/usp5+usp25 | 1.0/1.0 | 2 | 110 | 3.63 | |
| -/usp15+usp25 | 0.25/1.0 | 2 | 78 | 2.56 | |
| bmp4/- | 2.5 | 3 | 98 | -2.66 | |
| bmp4/otud4 | 2.5 | 0.5 | 3 | 71 | -2.93 |
| bmp4/usp5 | 2.5 | 2.0 | 3 | 65 | -2.52 |
| bmp4/usp15 | 2.5 | 0.5 | 3 | 78 | -2.55 |
| bmp4/usp25 | 2.5 | 2.0 | 3 | 82 | -2.54 |
| bmp4/otud4+usp25 | 2.5 | 0.25/1.0 | 2 | 93 | 1.27 |
| bmp4/usp5+usp25 | 2.5 | 1.0/1.0 | 2 | 118 | 2.63 |
| bmp4/usp15+usp25 | 2.5 | 0.25/1.0 | 2 | 132 | -0.31 |
| smad1/- | 800 | 3 | 96 | -1.37 | |
| smad1/otud4 | 800 | 0.5 | 3 | 103 | -1.44 |
| smad1/usp5 | 800 | 2.0 | 3 | 111 | -1.32 |
| smad1/usp15 | 800 | 0.5 | 3 | 104 | -0.94 |
| smad1/usp25 | 800 | 2.0 | 3 | 98 | -1.76 |
| smad1/otud4+usp25 | 800 | 0.25/1.0 | 2 | 68 | -1.08 |
| smad1/usp5+usp25 | 800 | 1.0/1.0 | 2 | 71 | -0.95 |
| smad1/usp15+usp25 | 800 | 0.25/1.0 | 2 | 75 | -0.88 |
Two mRNA (bmp4 and smad1) were co-injected with the group IV MOs (otud4, usp5, usp15 and usp25). The dorsal-ventral values were calculated as follows: Frequency of phenotypes * [C5: 5; C4: 4; C3: 3; C2: 2; C1: 1; Wt: 0; V1: -1; V2: -2; V3: -3; V4: -4]. The highest DV value is 5, which indicates 100% C5 phenotypes, while lowest is -4, which represents 100% V4 phenotypes. Negative values (ventralized phenotypes) were shown in bold.
Abbreviations: n ep, number of experiments; n, number of scored embryos; DV value, dorsal-ventral value.
Figure 6.
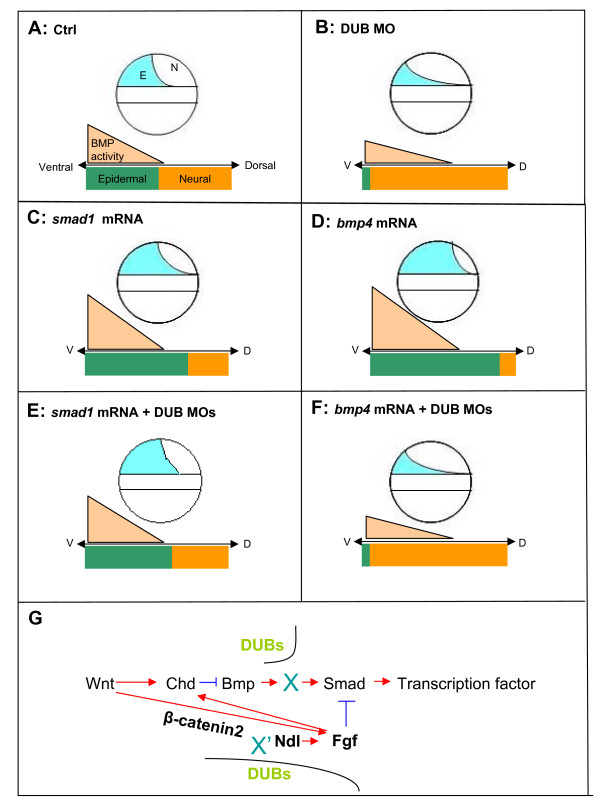
Proposed model of group IV DUB genes in the BMP pathway. Group IV DUB genes (otud4, usp5, usp15 and usp25) are involved in the BMP pathway. Panel A shows the normal wild type embryo with usual BMP activity. Panel B shows that single group IV MO knockdown results in dorsalized phenotypes. Panel C shows that overexpression of smad1 leads to ventralized phenotypes (V1-V3). Panel D shows that bmp4 overexpression results in severe ventralized phenotypes (V3, V4). Panel E shows that co-injection of combination of group IV MOs and smad1 mRNA results in mild ventralized phenotypes. Panel F shows that co-injection of combination group IV MOs and bmp4 mRNA causes severe dorsalized phenotypes as in panel B. Double knockdown of group IV MOs with bmp4 or smad1 overexpression causes different results. Overexpression of bmp4 could not rescue the dorsalized phenotypes that caused by double MOs knockdown, while smad1 could. This implies that group IV DUBs function between Bmp4 and Smad1 in the BMP pathway. Since co-injection experiments could not result in absolute dorsalized or ventralized phenotypes, which suggests that the DUB genes might interact with other dorsoventral signaling pathways. The proposed working model is shown in G. Taken together with the real-time results (Additional file 9), the group IV MO will increase the mRNA expression of different signaling genes (bolded), which results in stimulating Chd and inhibiting Smad. Collectively, all the effects will inhibit the BMP pathway and cause dorsalized effect. We propose that group IV DUBs will act on the substance X that is directly involved in the BMP pathway. Besides, they may interact with another common substrate X' that stimulates different signaling genes and generates secondary dorsal-ventral effects in the zebrafish early development.
Discussion
In this paper, we describe our identification of candidate DUB genes in zebrafish and the results of the "loss-of-function" screen based on the morpholino technology; thus, vastly extending the available biological information about DUB gene function and creating a resource for their further functional study. To our knowledge, "loss-of-function" studies of DUBs in any kinds of organism have been carried out only for very few genes due to the considerable cost associated with raising genetic knockout- and/or knockdown-modulated animals. Zebrafish is a model organism that is well suited to fill this gap. It is well known for its fast life cycle and the transparency of its embryos. Recently, the techniques of using MO knockdown and mRNA overexpression by microinjection further advance the versatility of using zebrafish as a model system. Developmental processes are always an interesting area for studying gene functions. Thus, we used the zebrafish to perform the DUB genes "loss-of-function" screen to understand their developmental functions.
More than 90 DUB candidates in zebrafish were found
To begin with, we started an in silico screen aimed at identifying potential DUB genes from the zebrafish genome. Although both the genome and the proteome of zebrafish are subject to some changes and corrections in the future, the available sequence data is sufficient for deriving the overwhelming part of the complete genes of DUB candidates. For six of the described seven classes of DUBs (USP, OTU, UCH, MJD, JAMM, PPPDE), we found 91 candidates. As expected, there are no candidates for the WLM group. Both in total number and in respect to the distribution among subclasses, zebrafish and human are not very different. For most (ca. 80%) of the candidates, there are obvious orthologues in both organisms although there are occasional organism-specific duplications or missing DUBs (Table 2). The sequence domain architecture analysis of the DUBs showed co-occurrence with domains characteristic for UBQ/UBL pathways as well as for gene expression, chromatin and cytoskeletal re-structuring processes; thus, it is likely that DUBs influence the respective biomolecular mechanisms (Additional file 1).
Table 2.
Comparison of DUB genes in human and zebrafish
| DUB Family Name | Extra DUB Genes in Zebrafish | Missing DUB Genes (Protein) in Zebrafish | Subtotal in Zebrafish | Subtotal in Human |
|---|---|---|---|---|
| USP | cyldb | USP6 | 51 | 58 |
| usp2b | USP9X | |||
| usp9 | USP9Y | |||
| usp12b | USP17 | |||
| usp53 | USP26 | |||
| usp54b | USP29 | |||
| unclassified (1) | USP35 | |||
| unclassified (2) | USP41 | |||
| unclassified (3) | USP50 | |||
| USP51 | ||||
| USP52 | ||||
| USP55 | ||||
| #59 (DUB3) | ||||
| #60 | ||||
| #61 (TL132-l) | ||||
| #62 (TL132) | ||||
| OTU | otud1l | OTUD6A | 15 | 14 |
| otud3 | OTUBQ2 | |||
| otud5b | OTUD1 | |||
| zranb1b | ||||
| UCH | 4 | 4 | ||
| MJD | ATXN3L | 3 | 5 | |
| JOS3 | ||||
| JAMM | eif3hb | #89 (IFP38) | 14 | 14 |
| stambpb | #91 | |||
| PPPDE | pppde1 | 4 | n.a. | |
| loc794838 | ||||
| pppde2a | ||||
| pppde2b | ||||
Through the in silico screen, 91 putative DUB genes were found in zebrafish, which were classified into 6 families. Details were summarized in the table. For the first 5 families (USP, OTU, UCH, MJD and JAMM), gene and protein names in human were based on Nijman et al., 2005 [4]. The human homologues of the PPPDE family were not studied in this work. n.a. stands for "not analyzed".
DUB genes play different roles in the developmental processes
Out of 85 putative zebrafish DUB genes that were subjected to a MO test, 57 DUB morphants show pleiotropic phenotypes throughout the body. From the results, we infer that DUB genes play different roles in the early zebrafish developmental processes, ranging from the dorsoventral patterning during the gastrulation to the fine-tuning of neurons and fin phenotypes. Most of the DUB genes appear multi-functional: they affect not only one and single developmental process. Taking Cyld as an example, there are published papers using the Cyld knockout mice for the studies. They have different effects on spermatogenesis and osteoclastogenesis [50,51]. Deubiquitylation is an important process for cells to maintain the cellular level of UBQ. Different cellular processes require ubiquitylation. E3 ligase is an enzyme that is involved in the last step of ubiquitylation. E3 ligases like BIR repeat-containing Ubiquitin-conjugating enzyme (BRUCE) and Mind bomb (Mib) perform different functions. BRUCE can regulate the apoptosis and cytokinesis [52,53], while Mib can facilitate the internalization of cell surface Delta in signal-sending cell and promote Notch activation in signal-receiving cells [54,55]. The number of UBQ E3 ligases is far greater than that of DUB enzymes, which implies that, as a trend, one DUB enzyme could deubiquitylate the substrates of several E3 ligases [4]. This fact may explain why our DUB-morphants did not show any unique and specific phenotypes. The resulting normal phenotype in some morphants may be due to the non-specific or/and low dosage of the MOs. Besides, we could not exclude the possibility that some other DUBs could compensate the deubiquitylation process of the knockdown DUB gene. Thus, those 26 DUB genes that did not give phenotypes could be regarded as the "non-essential" DUB genes, while those 59 (including two that are required for survival) DUB genes caused abnormal phenotypes are critical for development.
Notch signaling and the group I DUB genes
In the vertebrate neuronal development, formation of excess number of neurons during the neurogenesis will decay through apoptosis to ensure the formation of correct neurons' connection and pattern [56]. Neuronal death is a normal process happens during development [26]. Thus, it is abnormal that there are an increased number of neurons in the developmental process. The selection of the neuronal cell death and/or survival is mainly based on various neurotrophic factors and their related signaling pathway [57-59]. Notch signaling is well known for its control of cell fate and regulation of pattern formation [60]. Notch is a transmembrane receptor that is involved in the Notch signaling pathway, which was first identified in the mutant fly with "notches" in the wing [61]. Reports had shown that Notch signaling restricts neural differentiation by inhibiting the expression of proneural genes and its deficiency results in neurogenic phenotypes [62,63]. Our huC in situ screen identified three group I DUB genes, whose morphants exhibit an increase of neurons, indicating a premature differentiation - a possible consequence of a failure in the Notch-dependent lateral inhibition. Within these morphants, we further confirmed that the expression level of one of the Notch signaling downstream target gene, her4, was decreased. These results suggested that group I DUB genes affected Notch signaling directly or indirectly.
BMP signaling and the group IV DUB genes
In the huC screen, there were four DUB genes whose morphants had severely destructive neuronal patterning. One of the reasons to cause this could be the early defect in dorsoventral patterning. We tested different dorsal and ventral molecular markers at different time points and the results suggested that these morphants are dorsalized. One of the well known pathways involved in the dorsoventral patterning is the BMP pathway, which is required to specify the ventral cell fate [46]. We further underwent the co-injections experiments of mRNA of BMP signaling genes and the DUB MOs to further determine whether these DUBs are involved in the BMP pathway. When we co-injected smad1 mRNA and group IV DUB MOs into embryos, the dorsalized phenotypes caused by MO injection alone was rescued, indicating that group IV DUB genes are involved in the BMP pathway. Surprisingly, bmp4 mRNA injection with single group IV MO also resulted in ventralized phenotypes, which suggested that their functions might be compensated by others. Thus, we decided to make a combination of MOs among the group IV DUBs. After double knockdown with bmp4 overexpression, results showed that there was a shift from V3-V4 phenotypes to dorsalized phenotypes. It is notable that all the experiments could not completely transform the overexpression (ventralized) or knockdown (dorsalized) effects. This observation suggests that group IV DUBs may not simply be involved in the BMP pathway, but also involved in other dorsoventral pathways such as Nodal, Fgf, and Wnt. Reports have shown that these pathways can interact with each others [64-66]. We tested different signaling genes mRNA expressions by the real-time PCR. Consistently, results showed that the group IV MO could affect the signaling genes expression slightly but significantly in Nodal, Fgf and Wnt pathways. Details were shown in Additional file 9. It is not surprising that DUBs are involved and function differently in more than one pathway. Many cellular processes involve ubiquitylation, which suggests that one DUB may deubiquitylate more than one substrate. Here, we only showed the knockdown of group IV DUB genes will change the mRNA expression of different signaling genes; however, we did not know exactly how they are regulated and what is the mechanistic details between group IV DUB genes and them. Further biochemical studies have to be conducted in order to fully understand the nature of the DUBs.
Conclusion
From the in silico screen, we have identified 91 zebrafish DUB candidate genes belonging to six families. Out of the 85 targets that were subjected to a MO knockdown test: two were found to cause cell death and 57 resulted in pleiotropic developmental phenotypes; the remaining 26 did not show a detectable phenotype. Based on the expression level and pattern changes of huC, the 57 cases are classified into five groups. Further functional analyses of group I DUB genes suggest that three DUB genes (uchl3, otud7b and bap1) are the candidate genes to be functionally associated with the Notch pathway. Group IV DUB genes (otud4, usp5, usp15 and usp25) appear to be involved in the BMP pathway. Besides, our results suggest that substrates of the E3 ligase in BMP pathway are deubiquitylated more efficiently by two DUBs. To conclude, this paper provides a basic frame for the functional studies of DUB genes and acts as a screening step for researchers to pick up their specific DUB genes for follow-up studies based on the phenotypes. We understand that the screen might not be fully complete and that further research and in-depth studies are required. By releasing the genetic and functional information at this stage, the research efficiency with respect to zebrafish DUB genes will be accelerated.
Methods
Fish strains and maintenance
The wild-type strain used in the screen was AB line. They were raised and staged as described [67]. Throughout the experiments, the embryos were incubated at 25°C. All experimental procedures on zebrafish embryos were approved by the Biological Research Centre, A*STAR, Singapore (BRC IACUC No. 080390).
Identification of potential DUB genes in the zebrafish genome
For 5 types of DUB domains, PFAM [68] entries do exist and the corresponding hidden Markov models (HMMs) [69] were used for searches against the zebrafish proteome from ENSEMBL genome build database (originally with version v48 from February 2008 and, repeatedly, with later versions up to and including v53 from April 2009). The accessions of these five entries are: (i) UBQ-specific proteases (USPs) with the UBQ C-terminal hydrolase domain (UCH, PF0043), (ii) otubain proteases (OTUs) with the OTU-like cysteine protease domain (OTU, PF02338), (iii) UBQ C-terminal hydrolases (UCHs) with the UBQ C-terminal hydrolase family 1 domain (Peptidase_C12, PF1088), (iv) Machado-Joseph disease proteases (MJDs) with the Josephine domain (PF02099) and (v) JAMM motif metalloproteases (JAMMs) with the Mov34/MPN/PAD-1 family domain (PF01398).
Subsequently, the hits were blasted [70] against the zebrafish subset of the non-redundant protein sequence database for finding additional hits among sequences that were not included into the zebrafish genome build. Finally, blast searches against the human subset of the nrdb identified the closest human homologous proteins. Similar procedures were carried out for the PPPDE group and the WLMs (Wss1p-like proteases) group, where we relied on the published alignments [19] and on the PFAM entries PF08325 (WLM) and PF05903 (PPPDE). It should be noted that, depending on the zebrafish genome release and on the inclusion/removal of sequences into/from the non-redundant protein database, the in silico search results varied to some degree over time; the results presented here are an integration of the repeated searches.
Morpholino (MO) sequence site selection and design
For each putative DUB gene found in the genome-wide in silico search, an individual morpholino was designed. First, we tried to find the translation initiation site (TIS) by mapping the protein sequence onto the genome or transcripts with BLAST http://blast.ncbi.nlm.nih.gov/Blast.cgi[70]. For proteins with obviously missing or incorrect N-termini, we inspected the respective genomic data and tried to find a TIS in agreement with sequences from other organisms. If this was not possible (and for sequences whose MOs for the TIS did not match the minimal MO quality criteria (see below)), we searched for exon/intron boundaries as a possible morpholino sequence site with the goal that the MO targeting site was located on or 5' to the UBQ protease domains.
Once the initiation site was selected, all potential upstream sequences for MO target oligos were processed with AMOD [71]. Antisense MOs were selected based on the guidelines from Gene Tools (reviewed by [23]). Basically, the MOs should have a GC content about 40-60%, less than 37% G content, and without any consecutive tri- or tetra-G nucleotide sequences. In addition, the selected MOs should minimize self or pair sequence homology. Furthermore, MOs that target 5'-UTR were generated for some selected DUB genes to test their specificity. Designed MOs are tabulated (Additional file 3).
Morpholino specificity
Since the screen is a high-throughput knockdown analysis by using MO. Mistakes or non-specific knockdown might happen. However, we have tried our best to confirm the phenotypes by injecting the MOs and examined the phenotypes by at least two researchers. Besides, for those group I genes, we have designed a second MO that targets 5'-UTR region to further verify the knockdown phenotypes. In addition, we also used splicing MOs, which theoretically lead to un-spliced fragments encoding a stop codon within, and 5mis-match MOs of group IV DUBs to confirm that our knockdown results were specific and efficient. Sequences of MOs were listed in Additional file 3, and the result was tabulated in Additional file 10. Besides, the RT-PCR checks of splicing MOs were shown in Additional file 5.
Expression construct generation and mRNA synthesis
bmp4 and smad1 were amplified with primers (for bmp4: 5'-CCGGATCCATGATTCCTGGTAATCGAATGCTGA-3' and 5'-CCCTCGAGTTAGCGGCAGCCACACCCCT-3'; for smad1: 5'-CCGAATTCATGAATGTCACCTCACTCTTTTCCT-3' and 5'-CCCTCGAGCTAGGACACTGAAGAAATGGGGTT-3') containing BamHI and XhoI; EcoRI and XhoI restriction sites, respectively, from full-length cDNA using Pfu DNA polymerase (Stratagene, La Jolla, CA) and ligated into pCS2+ to generate pCS2+bmp4 and pCS2+-smad1 expression constructs. All constructs cloned in the pCS2+ vectors were linearized by NotI. Capped RNA was prepared with the SP6 Message Machine kit (Ambion, Austin, TX) and finally dissolved in DEPC-treated water.
Morpholino (MO) and mRNA injection
All MOs were purchased from Gene Tools, LLC (Philomath, OR), re-suspended in DEPC-treated water to make a 5 mM stock and stored at -20°C. Diluted MOs and/or mRNA (amount specified in Table 1 and Additional file 4) were injected into one- or two-cell stage embryos. Embryos from four different pairs of fish were used for each MO and/or mRNA injection.
Screening procedure
Injected embryos (including control) were mainly scored at two time points (1 and 3 days postfertilization (dpf)), corresponding to the pharyngula period and the hatching period, respectively [67]. The screening examination was mainly based on the method described previously [22]. Briefly, embryos at early stages were examined for abnormalities in the shape and morphology of those early developing tissues or organs like eyes, brain, notochord, spinal cord and somites. Afterwards, in the latter stage, pigmentation and phenotypic changes in cardiovascular system and the fins were also examined. All phenotypes were observed in a dominant feature of injected embryos at a non-toxic dose by at least two screeners. Besides, the MO-induced effects were not accompanied with those non-specific effects caused by overdose MO treatment (unpublished observations). In addition, we also fixed embryos at 50-60% epiboly, 10-somite stage and 24 hpf for whole mount in situ hybridization (WISH).
Whole mount in situ hybridization (WISH)
Plasmids that were used to make antisense mRNA probes have been published previously: bmp4 [72], chd [73], eve1 [36], gata1 [37], gata2 [37], gsc [39], her4 [29], huC [74], myoD [40], pax2a [43]. Single in situ hybridization was performed as described [75].
RNA extraction, reverse transcription and quantitative real-time PCR
Embryos at 10-somite and prim-5 stages were collected. Their total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA). Purified sample RNA with a ratio of 1.8-2.0 at A260/A280 was used. Briefly, 0.5 μg of total RNA extracted was reversely transcribed by iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). In case of real-time RT-PCR, PCRs were conducted using LightCycler 480 real-time PCR detection system using SYBR Green I Master (Roche, Basel, Switzerland). The copy number of the transcripts for each sample was calculated in reference to the parallel amplifications of known concentrations of the respective cloned PCR fragments. Standard curves were constructed and the amplification efficiencies were about 0.9-0.95. The data were then normalized using the expression levels of β-actin mRNA. The occurrence of primer-dimers and secondary products was inspected using melting curve analysis. Our data indicated that the amplification was specific. There was only one PCR product amplified for individual set of primers. Control amplification was done either without RT or without RNA. In the case of RT-PCR (underlined), embryos at prim-5 stage were used. PCRs were conducted by DNA Engine Dyad Peltier Thermal Cycler (Bio-Rad) using PCR Core Kit (Roche). Primers for β-actin (F: AGATCTGGCATCACACCTTC, R: TCACCAGAGTCCATCACGAT), β-catenin2 (F: AGGATCTGGACAACCAGGTG, R: GCACCATCACTGCAGCTTTA), fgf8 (F: GCAAGAAAAATGGTCTGGGA, R: TGCGTTTAGTCCGTCTGTTG), her4 (F: TGGCTCAAGAGTTCGTCAAG, R: AGTGGTCTGAGGATTGTCCA), huC (F: TCGAGTCCTGCAAATTGGTC, R: GTGAGGTGATGATCCTTCCA), lefty1 (F: CGCAAATTCACAAGAGGGAT, R: TCTCGGGGATTCTTGATGTC), nodal1 (F: GAGTGTGAGAGAAGCCCCTG, R: AGGTTCACTTCCACCACCAG), otud4 (F: GCGGTGCTTTGTCATTTACA, R: CTCTGAGAACGATCTTCTGG), sprouty2 (F: AGCAATGAGTACACGGAGGG, R: CACCTGCATTTCCCAAAAGT), usp5 (F: ATTTCGCTGCACCTTTGGTG, R: TGTTGGTCCTTCTTGATGTGG), usp15 (F: CATGCAGTGCGCGAGCGAAG, R: CTGAGAGCAGGCCGCTGTTG), usp25 (F: TTCATGCAGGAGCTTAGGCA, R: GCCAGGAAACGTCCATAAAA) and wnt8a (F: TGGTCGACTTGCTGTCAAAG, R: TCCATGTAGTCCCATGCTGA).
Statistical analysis
All data are represented as the mean ± SEM. Statistical significance is tested by Student's t-test. Groups were considered significantly different if P < 0.05.
List of abbreviations used
24 hpf: 24 hours after fertilization; BMP: bone morphogenetic protein; BRUCE: BIR repeat-containing Ubiquitin-conjugating enzyme; chd: chordin; DUBs: deubiquitylating enzymes; gsc: goosecoid; HA: hemagglutinin-tagged; JAMM: JAMM motif proteases; MIB: Mind bomb; MJD: Machado-Joseph disease protease; MO: morpholino; OTU: otubain protease; TGF-β: transforming growth factor β; TIS: translation initiation site; UBL: Ubiquitin-like; UBQ: Ubiquitin; UCH: Ubiquitin C-terminal hydrolase; UIM: Ubiquitin-interacting motif; USP: Ubiquitin-specific protease; WISH: whole mount in situ hybridization; WLMs: Wsslp-like proteases.
Authors' contributions
WKFT, BE, FE and YJJ planned and designed the experiments. BE and FE contributed in the bioinformatics analysis of DUB genes. WKFT designed the MO and mRNA. WKFT, SHKH and QN performed the experiments. WKFT, BE, FE and YJJ wrote the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Sequence Domain Architecture of zebrafish DUBs.
Reported DUB functions between 2005-2009.
DUB genes in zebrafish. The table lists the basic information of their names, Ensembl access/GenBank numbers and their MO sequences.
Early developmental phenotypes of zebrafish after DUBs MO knockdowns.
This figure shows the PCR products of group IV DUBs after injecting splicing MOs and the expression levels of huC and her4 in morphants of group I and selected group II DUB genes by using RT-PCR.
This figure shows the morphologies of dorsalized (C1-C5) and ventralized (V1-V4) embryos after microinjections at 24-30 hpf.
This figure presents the lateral view of in situ hybridization data of ventralized markers (bmp4, eve1, gata2) at 50-60% epiboly morphants.
This figure shows the results of in situ hybridization experiments, using gsc, pax2a + myoD probe at 50-60% epiboly and prim-5 stage.
This figure shows the mRNA expression level of different signaling genes (β-catenin2, fgf8, lefty1, nodal1, sprouty2 and wnt8a) at 10-somite stage of group IV DUB morphants.
Frequency of dorsalized phenotypes of group IV MOs injection studies.
Contributor Information
William KF Tse, Email: kftse@imcb.a-star.edu.sg.
Birgit Eisenhaber, Email: beisenhaber@etc.a-star.edu.sg.
Steven HK Ho, Email: hkho@imcb.a-star.edu.sg.
Qimei Ng, Email: qimei_85@yahoo.com.sg.
Frank Eisenhaber, Email: franke@bii.a-star.edu.sg.
Yun-Jin Jiang, Email: yjjiang@imcb.a-star.edu.sg.
Acknowledgements
We thank HL Ang, AWO Cheong and KL Poon for providing information and plasmids of in situ probes. We also thank the members of Jiang lab for helpful comments and discussions in the experiments. We thank the staff of Zebrafish Facility in IMCB for their excellent maintenance of fish stocks. This work is supported by the Agency of Science, Technology and Research (A*STAR), Singapore.
References
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Pickart CM. Ubiquitin enters the new millennium. Mol Cell. 2001;8(3):499–504. doi: 10.1016/S1097-2765(01)00347-1. [DOI] [PubMed] [Google Scholar]
- Hicke L. A new ticket for entry into budding vesicles-ubiquitin. Cell. 2001;106(5):527–530. doi: 10.1016/S0092-8674(01)00485-8. [DOI] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, Bernards R. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123(5):773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Amerik AY, Hochstrasser M. Mechanism and function of deubiquitinating enzymes. Biochim Biophys Acta. 2004;1695(1-3):189–207. doi: 10.1016/j.bbamcr.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Love KR, Catic A, Schlieker C, Ploegh HL. Mechanisms, biology and inhibitors of deubiquitinating enzymes. Nat Chem Biol. 2007;3(11):697–705. doi: 10.1038/nchembio.2007.43. [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature. 2003;424(6950):797–801. doi: 10.1038/nature01811. [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature. 2003;424(6950):801–805. doi: 10.1038/nature01802. [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature. 2003;424(6950):793–796. doi: 10.1038/nature01803. [DOI] [PubMed] [Google Scholar]
- Reiley W, Zhang M, Sun SC. Negative regulation of JNK signaling by the tumor suppressor CYLD. J Biol Chem. 2004;279(53):55161–55167. doi: 10.1074/jbc.M411049200. [DOI] [PubMed] [Google Scholar]
- Ikeda F, Dikic I. CYLD in ubiquitin signaling and tumor pathogenesis. Cell. 2006;125(4):643–645. doi: 10.1016/j.cell.2006.05.003. [DOI] [PubMed] [Google Scholar]
- Leroy E, Boyer R, Auburger G, Leube B, Ulm G, Mezey E, Harta G, Brownstein MJ, Jonnalagada S, Chernova T. The ubiquitin pathway in Parkinson's disease. Nature. 1998;395(6701):451–452. doi: 10.1038/26652. [DOI] [PubMed] [Google Scholar]
- Liu Y, Fallon L, Lashuel HA, Liu Z, Lansbury PT Jr. The UCH-L1 gene encodes two opposing enzymatic activities that affect alpha-synuclein degradation and Parkinson's disease susceptibility. Cell. 2002;111(2):209–218. doi: 10.1016/S0092-8674(02)01012-7. [DOI] [PubMed] [Google Scholar]
- Saigoh K, Wang YL, Suh JG, Yamanishi T, Sakai Y, Kiyosawa H, Harada T, Ichihara N, Wakana S, Kikuchi T. Intragenic deletion in the gene encoding ubiquitin carboxy-terminal hydrolase in gad mice. Nat Genet. 1999;23(1):47–51. doi: 10.1038/12647. [DOI] [PubMed] [Google Scholar]
- Oliveira SA, Li YJ, Noureddine MA, Zuchner S, Qin X, Pericak-Vance MA, Vance JM. Identification of risk and age-at-onset genes on chromosome 1p in Parkinson disease. Am J Hum Genet. 2005;77(2):252–264. doi: 10.1086/432588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Schrodi S, Rowland C, Tacey K, Catanese J, Grupe A. Genetic evidence for ubiquitin-specific proteases USP24 and USP40 as candidate genes for late-onset Parkinson disease. Hum Mutat. 2006;27(10):1017–1023. doi: 10.1002/humu.20382. [DOI] [PubMed] [Google Scholar]
- Burnett B, Li F, Pittman RN. The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet. 2003;12(23):3195–3205. doi: 10.1093/hmg/ddg344. [DOI] [PubMed] [Google Scholar]
- Scheel H, Tomiuk S, Hofmann K. Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet. 2003;12(21):2845–2852. doi: 10.1093/hmg/ddg297. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Novel predicted peptidases with a potential role in the ubiquitin signaling pathway. Cell Cycle. 2004;3(11):1440–1450. doi: 10.4161/cc.3.11.1206. [DOI] [PubMed] [Google Scholar]
- Borodovsky A, Ovaa H, Kolli N, Gan-Erdene T, Wilkinson KD, Ploegh HL, Kessler BM. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem Biol. 2002;9(10):1149–1159. doi: 10.1016/S1074-5521(02)00248-X. [DOI] [PubMed] [Google Scholar]
- Balakirev MY, Tcherniuk SO, Jaquinod M, Chroboczek J. Otubains: a new family of cysteine proteases in the ubiquitin pathway. EMBO Rep. 2003;4(5):517–522. doi: 10.1038/sj.embor.embor824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang Y-J, Heisenberg C-P. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development. 1996;123:1–36. doi: 10.1242/dev.123.1.1. [DOI] [PubMed] [Google Scholar]
- Eisen JS, Smith JC. Controlling morpholino experiments: don't stop making antisense. Development. 2008;135(10):1735–1743. doi: 10.1242/dev.001115. [DOI] [PubMed] [Google Scholar]
- Grainger RJ, Beggs JD. Prp8 protein: at the heart of the spliceosome. RNA. 2005;11(5):533–557. doi: 10.1261/rna.2220705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu P, Zhou W, Wang J, Puc J, Ohgi KA, Erdjument-Bromage H, Tempst P, Glass CK, Rosenfeld MG. A histone H2A deubiquitinase complex coordinating histone acetylation and H1 dissociation in transcriptional regulation. Mol Cell. 2007;27(4):609–621. doi: 10.1016/j.molcel.2007.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ER, Schwartz JH, Jessell TM. Principles of Neural Science, 4/e. New York: McGraw-Hill; 2000. [Google Scholar]
- Latimer AJ, Dong X, Markov Y, Appel B. Delta-Notch signaling induces hypochord development in zebrafish. Development. 2002;129(11):2555–2563. doi: 10.1242/dev.129.11.2555. [DOI] [PubMed] [Google Scholar]
- Jülich D, Lim CH, Round J, Nicolaije C, Schroeder J, Davies A, Geisler R, Consortium TS, Lewis J, Jiang Y-J. beamter/deltaC and the role of Notch ligands in the zebrafish somite segmentation, hindbrain neurogenesis and hypochord differentiation. Dev Biol. 2005;286(2):391–404. doi: 10.1016/j.ydbio.2005.06.040. [DOI] [PubMed] [Google Scholar]
- Takke C, Dornseifer P, von Weizsäcker E, Campos-Ortega JA. her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development. 1999;126(9):1811–1821. doi: 10.1242/dev.126.9.1811. [DOI] [PubMed] [Google Scholar]
- Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, Brand M, van Eeden FJ, Furutani-Seiki M, Granato M, Haffter P, Heisenberg C-P. Genes establishing dorsoventral pattern formation in the zebrafish embryo: the ventral specifying genes. Development. 1996;123:81–93. doi: 10.1242/dev.123.1.81. [DOI] [PubMed] [Google Scholar]
- Kishimoto Y, Lee KH, Zon L, Hammerschmidt M, Schulte-Merker S. The molecular nature of zebrafish swirl: BMP2 function is essential during early dorsoventral patterning. Development. 1997;124(22):4457–4466. doi: 10.1242/dev.124.22.4457. [DOI] [PubMed] [Google Scholar]
- Topczewska JM, Topczewski J, Shostak A, Kume T, Solnica-Krezel L, Hogan BL. The winged helix transcription factor Foxc1a is essential for somitogenesis in zebrafish. Genes Dev. 2001;15(18):2483–2493. doi: 10.1101/gad.907401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köster M, Plessow S, Clement JH, Lorenz A, Tiedemann H, Knöchel W. Bone morphogenetic protein 4 (BMP-4), a member of the TGF-beta family, in early embryos of Xenopus laevis: analysis of mesoderm inducing activity. Mech Dev. 1991;33(3):191–199. doi: 10.1016/0925-4773(91)90027-4. [DOI] [PubMed] [Google Scholar]
- Dale L, Howes G, Price BM, Smith JC. Bone morphogenetic protein 4: a ventralizing factor in early Xenopus development. Development. 1992;115(2):573–585. doi: 10.1242/dev.115.2.573. [DOI] [PubMed] [Google Scholar]
- Jones CM, Lyons KM, Lapan PM, Wright CV, Hogan BL. DVR-4 (bone morphogenetic protein-4) as a posterior-ventralizing factor in Xenopus mesoderm induction. Development. 1992;115(2):639–647. doi: 10.1242/dev.115.2.639. [DOI] [PubMed] [Google Scholar]
- Joly JS, Joly C, Schulte-Merker S, Boulekbache H, Condamine H. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development. 1993;119(4):1261–1275. doi: 10.1242/dev.119.4.1261. [DOI] [PubMed] [Google Scholar]
- Detrich HW, Kieran MW, Chan FY, Barone LM, Yee K, Rundstadler JA, Pratt S, Ransom D, Zon LI. Intraembryonic hematopoietic cell migration during vertebrate development. Proc Natl Acad Sci USA. 1995;92(23):10713–10717. doi: 10.1073/pnas.92.23.10713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y, Lu B, Steinbeisser H, De Robertis EM. Regulation of neural induction by the Chd and Bmp-4 antagonistic patterning signals in Xenopus. Nature. 1995;376(6538):333–336. doi: 10.1038/376333a0. [DOI] [PubMed] [Google Scholar]
- Stachel SE, Grunwald DJ, Myers PZ. Lithium perturbation and goosecoid expression identify a dorsal specification pathway in the pregastrula zebrafish. Development. 1993;117(4):1261–1274. doi: 10.1242/dev.117.4.1261. [DOI] [PubMed] [Google Scholar]
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Grunwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122(1):271–280. doi: 10.1242/dev.122.1.271. [DOI] [PubMed] [Google Scholar]
- Little SC, Mullins MC. Twisted gastrulation promotes BMP signaling in zebrafish dorsal-ventral axial patterning. Development. 2004;131(23):5825–5835. doi: 10.1242/dev.01464. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Warga RM, Schilling TF. Origin and organization of the zebrafish fate map. Development. 1990;108(4):581–594. doi: 10.1242/dev.108.4.581. [DOI] [PubMed] [Google Scholar]
- Krauss S, Johansen T, Korzh V, Fjose A. Expression of the zebrafish paired box gene pax[zf-b] during early neurogenesis. Development. 1991;113(4):1193–1206. doi: 10.1242/dev.113.4.1193. [DOI] [PubMed] [Google Scholar]
- Xie J, Fisher S. Twisted gastrulation enhances BMP signaling through chordin dependent and independent mechanisms. Development. 2005;132(2):383–391. doi: 10.1242/dev.01577. [DOI] [PubMed] [Google Scholar]
- Hogan BL. Bone morphogenetic proteins: multifunctional regulators of vertebrate development. Genes Dev. 1996;10(13):1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- Holley SA, Ferguson EL. Fish are like flies are like frogs: conservation of dorsal-ventral patterning mechanisms. Bioessays. 1997;19(4):281–284. doi: 10.1002/bies.950190404. [DOI] [PubMed] [Google Scholar]
- Dick A, Hild M, Bauer H, Imai Y, Maifeld H, Schier AF, Talbot WS, Bouwmeester T, Hammerschmidt M. Essential role of Bmp7 (snailhouse) and its prodomain in dorsoventral patterning of the zebrafish embryo. Development. 2000;127(2):343–354. doi: 10.1242/dev.127.2.343. [DOI] [PubMed] [Google Scholar]
- Schmid B, Furthauer M, Connors SA, Trout J, Thisse B, Thisse C, Mullins MC. Equivalent genetic roles for bmp7/snailhouse and bmp2b/swirl in dorsoventral pattern formation. Development. 2000;127(5):957–967. doi: 10.1242/dev.127.5.957. [DOI] [PubMed] [Google Scholar]
- Dick A, Meier A, Hammerschmidt M. Smad1 and Smad5 have distinct roles during dorsoventral patterning of the zebrafish embryo. Dev Dyn. 1999;216(3):285–298. doi: 10.1002/(SICI)1097-0177(199911)216:3<285::AID-DVDY7>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- Wright A, Reiley WW, Chang M, Jin W, Lee AJ, Zhang M, Sun SC. Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell. 2007;13(5):705–716. doi: 10.1016/j.devcel.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Jin W, Chang M, Paul EM, Babu G, Lee AJ, Reiley W, Wright A, Zhang M, You J, Sun SC. Deubiquitinating enzyme CYLD negatively regulates RANK signaling and osteoclastogenesis in mice. J Clin Invest. 2008;118(5):1858–1866. doi: 10.1172/JCI34257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke T, Pohl C, Pyrowolakis G, Jentsch S. Dual role of BRUCE as an antiapoptotic IAP and a chimeric E2/E3 ubiquitin ligase. Mol Cell. 2004;14(6):801–811. doi: 10.1016/j.molcel.2004.05.018. [DOI] [PubMed] [Google Scholar]
- Pohl C, Jentsch S. Final stages of cytokinesis and midbody ring formation are controlled by BRUCE. Cell. 2008;132(5):832–845. doi: 10.1016/j.cell.2008.01.012. [DOI] [PubMed] [Google Scholar]
- Le Borgne R, Bardin A, Schweisguth F. The roles of receptor and ligand endocytosis in regulating Notch signaling. Development. 2005;132(8):1751–1762. doi: 10.1242/dev.01789. [DOI] [PubMed] [Google Scholar]
- Chitnis A. Why is Delta endocytosis required for effective activation of Notch? Dev Dyn. 2006;235(4):886–894. doi: 10.1002/dvdy.20683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger V, Levi-Montalcini R. Proliferation, differentiation and degeneration in the spinal ganglia of the chick embryo under normal and experimental conditions. J Exp Zool. 1949;111(3):457–501. doi: 10.1002/jez.1401110308. [DOI] [PubMed] [Google Scholar]
- Klein R. Role of neurotrophins in mouse neuronal development. FASEB J. 1994;8(10):738–744. doi: 10.1096/fasebj.8.10.8050673. [DOI] [PubMed] [Google Scholar]
- Peruzzi F, Gordon J, Darbinian N, Amini S. Tat-induced deregulation of neuronal differentiation and survival by nerve growth factor pathway. J Neurovirol. 2002;8(Suppl 2):91–96. doi: 10.1080/13550280290167885. [DOI] [PubMed] [Google Scholar]
- Althaus HH, Klöppner S, Klopfleisch S, Schmitz M. Oligodendroglial cells and neurotrophins: a polyphonic cantata in major and minor. J Mol Neurosci. 2008;35(1):65–79. doi: 10.1007/s12031-008-9053-y. [DOI] [PubMed] [Google Scholar]
- Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004;131(5):965–973. doi: 10.1242/dev.01074. [DOI] [PubMed] [Google Scholar]
- Morgan TH. The theory of the gene. Am Nat. 1917;51:513–544. doi: 10.1086/279629. [DOI] [Google Scholar]
- Chitnis AB. The role of Notch in lateral inhibition and cell fate specification. Mol Cell Neurosci. 1995;6(4):311–321. doi: 10.1006/mcne.1995.1024. [DOI] [PubMed] [Google Scholar]
- Parks AL, Huppert SS, Muskavitch MAT. The dynamics of neurogenic signalling underlying bristle development in Drosophila melanogaster. Mech Dev. 1997;63(1):61–74. doi: 10.1016/S0925-4773(97)00675-8. [DOI] [PubMed] [Google Scholar]
- Mizoguchi T, Izawa T, Kuroiwa A, Kikuchi Y. Fgf signaling negatively regulates Nodal-dependent endoderm induction in zebrafish. Dev Biol. 2006;300(2):612–622. doi: 10.1016/j.ydbio.2006.08.073. [DOI] [PubMed] [Google Scholar]
- Poulain M, Furthauer M, Thisse B, Thisse C, Lepage T. Zebrafish endoderm formation is regulated by combinatorial Nodal, FGF and BMP signalling. Development. 2006;133(11):2189–2200. doi: 10.1242/dev.02387. [DOI] [PubMed] [Google Scholar]
- Varga M, Maegawa S, Bellipanni G, Weinberg ES. Chordin expression, mediated by Nodal and FGF signaling, is restricted by redundant function of two beta-catenins in the zebrafish embryo. Mech Dev. 2007;124(9-10):775–791. doi: 10.1016/j.mod.2007.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. Stages of embryonic development of the zebrafish. Dev Dyn. 1995;203(3):253–310. doi: 10.1002/aja.1002030302. [DOI] [PubMed] [Google Scholar]
- Sammut SJ, Finn RD, Bateman A. Pfam 10 years on: 10,000 families and still growing. Brief Bioinform. 2008;9(3):210–219. doi: 10.1093/bib/bbn010. [DOI] [PubMed] [Google Scholar]
- Eddy SR. What is a hidden Markov model? Nat Biotechnol. 2004;22(10):1315–1316. doi: 10.1038/nbt1004-1315. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee EW, Shim KJ, Pickart MA, Ekker SC, Ellis LB. AMOD: a morpholino oligonucleotide selection tool. Nucleic Acids Res. 2005. pp. W506–511. [DOI] [PMC free article] [PubMed]
- Chin AJ, Chen J-N, Weinberg ES. Bone morphogenetic protein-4 expression characterizes inductive boundaries in organs of developing zebrafish. Dev Genes Evol. 1997;207:107–114. doi: 10.1007/s004270050097. [DOI] [PubMed] [Google Scholar]
- Miller-Bertoglio VE, Fisher S, Sanchez A, Mullins MC, Halpern ME. Differential regulation of chordin expression domains in mutant zebrafish. Dev Biol. 1997;192(2):537–550. doi: 10.1006/dbio.1997.8788. [DOI] [PubMed] [Google Scholar]
- Kim CH, Ueshima E, Muraoka O, Tanaka H, Yeo SY, Huh TL, Miki N. Zebrafish elav/HuC homologue as a very early neuronal marker. Neurosci Lett. 1996;216(2):109–112. doi: 10.1016/0304-3940(96)13021-4. [DOI] [PubMed] [Google Scholar]
- Ma M, Jiang Y-J. Jagged2a-Notch signaling mediates cell fate choice in zebrafish pronephric duct. PLoS Genet. 2007;3(1):e18. doi: 10.1371/journal.pgen.0030018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence Domain Architecture of zebrafish DUBs.
Reported DUB functions between 2005-2009.
DUB genes in zebrafish. The table lists the basic information of their names, Ensembl access/GenBank numbers and their MO sequences.
Early developmental phenotypes of zebrafish after DUBs MO knockdowns.
This figure shows the PCR products of group IV DUBs after injecting splicing MOs and the expression levels of huC and her4 in morphants of group I and selected group II DUB genes by using RT-PCR.
This figure shows the morphologies of dorsalized (C1-C5) and ventralized (V1-V4) embryos after microinjections at 24-30 hpf.
This figure presents the lateral view of in situ hybridization data of ventralized markers (bmp4, eve1, gata2) at 50-60% epiboly morphants.
This figure shows the results of in situ hybridization experiments, using gsc, pax2a + myoD probe at 50-60% epiboly and prim-5 stage.
This figure shows the mRNA expression level of different signaling genes (β-catenin2, fgf8, lefty1, nodal1, sprouty2 and wnt8a) at 10-somite stage of group IV DUB morphants.
Frequency of dorsalized phenotypes of group IV MOs injection studies.