Abstract
The Mdm2 proto-oncogene is amplified to high copy numbers in human sarcomas and is overexpressed in a wide variety of other human cancers. Because Mdm2 protein forms a complex with the p53 tumor suppressor protein and down-regulates p53 function, the oncogenic potential of Mdm2 is presumed to be p53-dependent. To model these conditions in mice, we have used the entire Mdm2 gene, under transcriptional control of its native promoter region, as a transgene to create mice that overexpress Mdm2. The transgenic mice are predisposed to spontaneous tumor formation, and the incidence of sarcomas observed in the Mdm2-transgenic mice in the presence or absence of functional p53 demonstrates that, in addition to Mdm2-mediated inactivation of p53, there exists a p53-independent role for Mdm2 in tumorigenesis.
The MDM2 proto-oncogene is amplified and overexpressed in a variety of human cancers. Approximately one-third of osteosarcomas show amplification and overexpression of MDM2, and MDM2 has been found to be overexpressed in numerous soft tissue sarcomas, including malignant fibrous histiocytomas, rhabdomyosarcomas, liposarcomas, and leiomyosarcomas (1–5). Amplification and overexpression of MDM2 also is observed in approximately 10% of glioblastomas and astrocytomas (6), and MDM2 overexpression has been documented in 53% of myeloid (7), 28% of B-cell lymphomas (8), and 40% of oral squamous cell carcinomas (9). The human MDM2 gene is located on chromosome 12 (q14.3–q15). This region contains many genes thought to be involved in control of cell growth, including GADD153—encoding a DNA-damage inducible C/EBP family member, SAS—encoding a putative trans-membrane protein involved in signal transduction, GLI—an oncogene encoding a zinc-finger transcription factor, and CDK4—encoding the cyclin-dependent kinase p34 protein. Many of the studies documenting MDM2 amplification in human cancers used Southern analysis to demonstrate an increase in the copy number of MDM2. This experimental approach does not address possible coamplification of MDM2 and other closely linked genes within the tumor. More recently, a number of studies have looked at possible coamplification of MDM2 and other linked genes in sarcomas and gliomas in greater detail (10–13). However, it remains unclear whether amplification of MDM2 is sufficient in and of itself to induce the broad range of tumor types and the numbers of tumors reported in the previous studies.
The Mdm2 gene was originally isolated from a mouse double minute chromosome that was present at a high copy number in a spontaneously transformed derivative of mouse 3T3 cells (14), and transfection and experimentally induced overexpression of Mdm2 has been found to immortalize rodent primary fibroblasts as well as induce a fully transformed phenotype in cultured cells (15). An explanation for the transforming capabilities of Mdm2 has been provided by reports indicating that Mdm2 forms a complex with the p53 tumor suppressor protein and inhibits p53-mediated transregulation of heterologous gene expression (16–18). Addition of exogenous Mdm2 can overcome p53-induced suppression of transformed cell growth (19) and can abrogate both p53-mediated, G1 phase cell cycle arrest and induction of apoptosis in cultured cells (20, 21). Complex formation occurs near the amino-terminal portion of both proteins, and Mdm2 has been proposed to inhibit p53 function by targeting p53 for proteolytic degradation (22, 23). Furthermore, p53 has been found to transactivate Mdm2 expression due to the presence of several p53-binding sites within the first intron of the Mdm2 gene (24, 25). Thus, complex formation between Mdm2 and p53 may serve to autoregulate Mdm2 expression as well as regulate p53 function (26).
We, and others, have used gene targeting in embryonic stem (ES) cells to create Mdm2-deficient mice (27, 28). The early embryonic lethal phenotype induced by Mdm2-deficiency is rescued by codeletion of functional p53, demonstrating that Mdm2 plays a critical role in development by regulating p53 function. Mice deficient for both Mdm2 and p53 undergo normal development are viable, and are fertile, suggesting that any functions possessed by Mdm2 aside from its ability to regulate p53 are dispensable for normal embryonic development. Mdm2/p53-deficient mice are susceptible to spontaneous tumorigenesis. A tumor susceptibility study performed by using these mice detected no difference between Mdm2/p53-deficient mice and p53-deficient mice in either the rate of tumor formation or in the spectrum of tumors indicating that physiologic levels of Mdm2 does not alter tumorigenesis when p53 is absent (29). We also have examined and compared the in vitro growth characteristics of p53-deficient and Mdm2/p53-deficient mouse fibroblasts (29). Embryonic fibroblasts deficient for both Mdm2 and p53 were indistinguishable from p53-deficient embryonic fibroblasts in their rate of proliferation and cell cycle characteristics. Our studies demonstrated that the presence or absence of Mdm2 had no effect on proliferation or cell cycling of p53-deficient cells and does not alter the development, viability, or tumorigenic potential of p53-null mice.
Several lines of evidence have been reported that suggest that Mdm2 may regulate normal and abnormal (neoplastic) cell growth not only by inhibiting p53 function, but through a p53-independent mechanism as well. In addition to an amino-terminal p53-binding domain, the primary structure of Mdm2 contains several other putative functional domains, including a nuclear localization signal, an acidic transcription activation domain, a central zinc finger element, and a carboxy-terminal zinc RING-finger motif (15, 30). The presence of domains suggests that Mdm2 may bind to DNA and affect transcription, though this has yet to be demonstrated. Alternatively, the RING-finger domain of Mdm2 has been shown to form a complex with the L5 ribosomal protein and its associated 5S ribosomal RNA (31). Human MDM2 has been reported to form a complex with the Retinoblastoma tumor suppressor protein and the E2F1 and DP1 transcription factors (32, 33). Transfection of MDM2 into a variety of cells was found to elevate expression of a reporter gene placed under transcriptional control of an E2F-responsive promoter in these studies, suggesting that MDM2-Rb complex formation and MDM2-E2F1/DP1 complex formation stimulates the expression of E2F-responsive genes. Complex formation was found to occur both in vitro and in cultured cells deficient for p53, suggesting that MDM2 might play a p53-independent role in promoting cell cycle progression from G1 to S phase.
Recently, massive overexpression of a full-length MDM2 cDNA in the mammary epithelium of transgenic mice was found to inhibit development of the mammary gland by inducing multiple rounds of S phase without completion of mitosis (34). The uncoupling of S phase from mitosis was seen in transgenic mice, which were either wild-type or deficient for p53, indicating a p53-independent role for Mdm2 in the regulation of DNA synthesis. Approximately 16% of the transgenic mice developed mammary carcinogenesis late in life, suggesting that overexpression of MDM2 may induce tumors in epithelial cells in vivo. However, it is uncertain from these experiments whether excess MDM2 promoted mammary tumorigenesis solely by inhibition of p53 function, or through a p53-independent pathway, because the tumor-bearing mice contained functional p53 genes.
Further evidence that Mdm2 may possess oncogenic potential in addition to its ability to inhibit p53 function when Mdm2 is overexpressed has been provided by several studies of human tumors that bear amplified MDM2 copy numbers and/or that overexpress MDM2. Although many of these human tumors retain wild-type p53, consistent with the notion that MDM2 overexpression serves as an alternate mechanism to render cells p53 deficient, several of the tumors harbored mutated p53 genes, a seemingly redundant genetic alteration if Mdm2 serves solely to inhibit p53 function (6, 9).
The results of our previous studies by using mice and embryonic fibroblasts deleted for Mdm2 indicated that the functional role of Mdm2 in development and in normal cell growth was solely to regulate p53 activity. However, these results do not exclude the possibility of a p53-independent role for Mdm2 in control of cell growth and neoplasia in cells that overexpress Mdm2. To determine whether amplification and overexpression of the Mdm2 gene is sufficient to promote tumor formation or whether tumorigenesis requires coamplification of genes that colocalize with Mdm2, we have created transgenic mice, which overexpress Mdm2 genes. Furthermore, we have analyzed the development and tumorigenic potential of these mice to determine whether Mdm2 overexpression is capable of inducing tumorigenesis in a p53-independent manner.
METHODS
Creation and Analysis of Mdm2-Transgenic Mice.
Cosmid 2A-43/pCV001 (gift from D. L. George, University of Pennsylvania School of Medicine, Philadelphia) containing the entire Mdm2 gene was linearized by SalI digestion and introduced into AB 2.1 ES cells by electroporation. Clones were isolated after G418 selection and expanded for Southern analysis to determine the copy number of transgene integrants and the number of insertion sites within the ES cell genome. Clones bearing high copy numbers of the transgene inserted at a single site within the host cell genome were analyzed by Northern analysis for relative levels of Mdm2 gene expression. Four separate clones bearing 2-fold, 6-fold, 9-fold, or 15-fold overexpression of Mdm2 were used for blastocyst injections to create transgenic mice. The resulting chimeric mice (129SvEvBrd × C57BL/6) were mated with C57BL/6 mice to establish an Mdm2-transgenic mouse line. Southern analysis by using a SacI–ScaI Mdm2 genomic probe fragment (27) was performed to identify transgenic offspring. Mdm2-transgenic mice were crossed with Mdm2-heterozygous mice (27), and with p53-heterozygous and p53-null mice (35) to create Mdm2-transgenic, Mdm2-null mice and Mdm2-transgenic, p53-null mice. Genotyping of the resulting offspring was performed as described previously (27). RNA was isolated from the tissues of transgenic and wild-type mice by using RNAzol B as described by the manufacturer (Tel-Test, Friendswood, TX). Northern analysis of Mdm2 message levels was performed by using full-length Mdm2 cDNA as a probe.
Histologic Analysis of Tumors in Mice.
During the spontaneous tumor study, mice were inspected every third day for morbidity and for obvious tumor formation. Necropsies were performed on moribund mice, and selected tissues were fixed in 10% buffered formalin phosphate and processed for paraffin embedding as described previously (35). Sections were prepared and stained with haematoxylin and eosin and examined under a microscope. All tumors were identified and classified in the absence of knowledge of their genotypes.
RESULTS
Creation of Mdm2-Transgenic Mice.
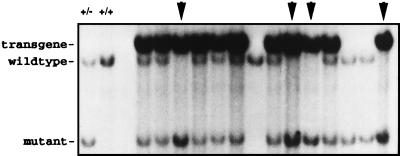
In an effort to closely mimic Mdm2 amplification events in human tumors, we elected to use as a transgene a cosmid construct, which contains the entire Mdm2 gene under transcriptional control of its native promoter region (15). Transfection of this cosmid into primary rodent fibroblasts has been reported previously to induce their transformation (15, 19). To confirm that the cosmid did not contain an “activating” mutation, we determined the structure of the mouse (129SvEvBrd strain) Mdm2 gene and compared the coding sequences of the Mdm2 gene with Mdm2 sequences contained on the cosmid clone (36). No differences were found in the coding sequences, indicating that the Mdm2 cosmid was structurally wild type. To generate transgenic mice, the cosmid was electroporated into AB2.1 ES cells, and G418-resistant clones were recovered and initially screened by Southern blot hybridization to identify clones that contained multiple copies of the Mdm2 transgene. These clones were subsequently examined for elevated levels of Mdm2 transcripts by Northern blot analysis. Multiple lines of ES cells were generated that overexpressed Mdm2, some by as much as 15-fold (data not shown). To establish these transgenes in the mouse germline, five different lines were microinjected into blastocysts. However, only one line of ES cells that displayed a 2-fold elevation in Mdm2 transcript levels was capable of producing chimeric mice after blastocyst injections. A line of Mdm2-transgenic mice was established by crossing these chimeric mice with wild-type mice. The Mdm2-transgenic mice were intercrossed with Mdm2m1/+ mice, which we generated previously (27) to yield Mdm2-heterozygous mice bearing the Mdm2 transgene. Intercrosses with these mice led to the recovery of Mdm2-null mice (Mdm2m1/Mdm2m1)—normally an early embryonic lethal genotype in the presence of wild-type p53, indicating that the transgene produces functional Mdm2 that was able to rescue the null allele (Fig. 1).
Figure 1.
Southern analysis of the offspring of Mdm2-transgenic, Mdm2 heterozygous mice crossed with Mdm2 heterozygous mice. Genomic DNA was isolated from tail biopsies, digested with EcoRI, and run on a 1% agarose gel. Southern analysis by using a Mdm2 genomic probe confirmed the presence of the transgene and the genotype of the mice. Arrows indicate those mice which are Mdm2-deficient and which are rescued by the presence of the transgene. Controls in first two lanes are DNA isolated from an Mdm2-heterozygous (+/−) mouse and from a wild-type (+/+) mouse
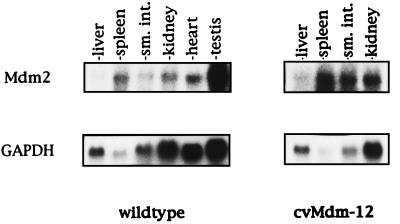
To determine the level of Mdm2 expression in various tissues, Northern blot analysis of total RNA isolated from liver, spleen, intestine, and kidney of wild-type and transgenic Mdm2 adult mice was performed by using full-length Mdm2 cDNA as a probe (Fig. 2). Human glyceraldehyde-3-phosphate dehydrogenase cDNA was used as a control for RNA levels and integrity. Phosphorimaging analysis of the Northern blots revealed that the Mdm2-transgenic mice contain an average 4-fold increase in the level of Mdm2 in various tissues relative to nontransgenic mice.
Figure 2.
Northern analysis of RNA isolated from wild-type and Mdm2-transgenic mouse tissues. Northern analysis of total RNA was performed by using an Mdm2 full-length cDNA probe. Human glyceraldehyde-3-phosphate dehydrogenase was probed to provide a phosphorimaging control for RNA load and integrity.
Tumorigenesis in Mdm2-Transgenic Mice.
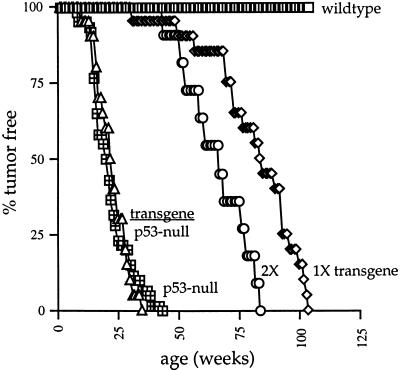
We have examined the tumor susceptibility of mice bearing the Mdm2 transgene. Twenty-two Mdm2-transgenic mice were monitored for the appearance of tumors. The rate of tumorigenesis in this population was compared with the rates of tumorigenesis in p53-null mice and in wild-type mice. The results (Fig. 3) demonstrated that overexpression of Mdm2 was sufficient to induce spontaneous tumor formation in mice, albeit at a rate much slower than that observed in p53-null mice. One-half of the Mdm2-transgenic mice succumbed to neoplasia by 84 weeks, compared with 20 weeks for one-half of the p53-null cohort.
Figure 3.
Tumorigenesis in Mdm2-transgenic mice. Twenty Mdm2-transgenic mice (diamond), 11 homozygous (2×) transgenic mice (circle), and 20 Mdm2-transgenic/p53-null mice (triangle) were assayed for tumor formation. The rates of tumorigenesis in these populations was compared with the rates of tumorigenesis in 42 p53-null mice (hatched square) and 30 wild-type mice (open square).
The Mdm2-transgenic mice were intercrossed to generate mice that were homozygous for the transgene, and these mice were monitored for tumorigenesis. Tumor formation was accelerated in these mice relative to the hemizygous Mdm2-transgenic mice but remained slower than the rate of tumorigenesis in p53-null mice. One-half of the homozygous transgenic mice (2×) developed neoplasia by 61 weeks of age. These data demonstrate that the rate of tumor formation depends on the level of Mdm2 expression, but it does not reveal whether Mdm2 overexpression is capable of inducing tumors by some mechanism aside from inhibition of p53 activity.
To determine whether overexpression of Mdm2 promotes tumorigenesis through a p53-independent pathway, we generated Mdm2-transgenic, p53-null mice by conventional breedings. Twenty-one Mdm2-transgenic, p53-null mice were used in a tumor assay and monitored for their tumor susceptibility. The rate of tumor formation in this colony was statistically indistinguishable from the rate of tumorigenesis in p53-null mice (Fig. 3).
The spectrum of tumor types formed in the Mdm2-transgenic mice and in the Mdm2-transgenic, p53-null mice was determined by histopathology. This analysis revealed far more sarcomas in Mdm2-transgenic mice (regardless of their p53 status), compared with p53-null mice lacking the Mdm2-transgene (Table 1). Thirty-eight percent of all tumors observed in Mdm2-transgenic mice were sarcomas, whereas sarcomas comprised only 9% of all tumors obtained in nontransgenic, p53-null mice (Fisher’s exact test, P < 0.01).
Table 1.
Spectrum of tumors occurring in Mdm2-transgenic mice
| Mdm2-transgenic micea | Mdm2 transgenic/p53-null miceb | p53-null micec |
|---|---|---|
| 14 Malignant lymphoma | 14 Malignant lymphoma | 35 Malignant lymphoma |
| 9 Hemangiosarcoma | 8 Hemangiosarcoma | 2 Osteosarcoma |
| 1 Rhabdomyosarcoma | 2 Osteosarcoma | 1 Rhabdomyosarcoma |
| 1 Adenocarcinoma | 1 Squamous cell carcinoma | 1 Undifferentiated sarcoma |
| 1 Pituitary adenoma | 1 Medulloblastoma | 3 Carcinomas |
| 10/26 Tumors = sarcomas (38%) | 10/26 Tumors = sarcomas (38%) | 4/42 Tumors = sarcomas (9%) |
Tumors isolated from 22 Mdm2-transgenic, p53 +/+ mice, 21 Mdm2-transgenic, p53 −/− mice were classified by histopathology. Numbers in parentheses represent the percentage of all tumors that are sarcomas.
Approximately 80% of p53-null mice succumb to malignant lymphomas, which are chiefly thymic in origin. A high percentage of Mdm2-transgenic mice (64%) or Mdm2-transgenic, p53-null mice (67%) also present with lymphoma. Thus the large increase in the percentage of total tumors that are sarcomas (shown in Table 1), and the large increase in the percentage of transgenic mice that present with sarcoma (45% of Mdm2-transgenic mice and 48% of Mdm2-transgenic, p53-null mice compared with 9% of nontransgenic, p53-null mice), does not reflect a large decrease in the total number of mice bearing a lymphoma but rather reflects the fact that many of the transgenic mice exhibit an additional tumor, typically a hemangiosarcoma. This type of sarcoma accounts for approximately one-third of all tumors observed in the Mdm2-transgenic mice.
DISCUSSION
We have generated Mdm2-transgenic mice to determine whether Mdm2 overexpression induces spontaneous tumorigenesis, and whether the oncogenic activity of Mdm2 overexpression results solely from its ability to inhibit p53 function. Transgenic mice bearing an amplified copy number of Mdm2 were constructed by electroporation of the entire Mdm2 gene into embryonic stem cells and microinjection of transfected ES cells into blastocysts to generate germ line chimeras. Five separate ES cell clones were used in these experiments. However, only one clone with modest levels of Mdm2 overexpression was capable of generating chimeras with a high degree of ES cell contribution and germ line transmission of the Mdm2 transgene, despite multiple repeated attempts with the other ES cell lines that contained far greater levels of Mdm2-expression. Our inability to create mice that greatly overexpress Mdm2 in a nontissue confined manner is consistent with the experience of several other labs, and suggests that overexpression of Mdm2 during development may induce embryonic lethality. Although our study uses only one line of transgenic mice, the fact that the penetrance of the tumor phenotype was 100% in the Mdm2-transgenic, p53 wild-type mice coupled with a zero incidence of tumorigenesis in nontransgenic, sibling mice, indicates that the tumorigenic phenotype of the mice results from the transgene and not from an unrelated mutation arising in the transfected ES cells.
Mdm2 is normally expressed in low levels in all tissues during development and in the adult mouse (15, 37), and absence of Mdm2 is a lethal condition in the presence of functional p53 (27, 28). The Mdm2-transgenic mice displayed an approximate four-fold elevation in Mdm2 transcript levels in the various tissues examined, and rescue of Mdm2-null, p53 wild-type mice by the presence of the transgene indicates that the transgene is appropriately regulated during development and produces Mdm2 products which are capable of inhibiting p53 activity.
The Mdm2-transgenic mice contain 25 copies of the Mdm2 transgene inserted into a single site within the genome. The 100% incidence of tumorigenesis in these transgenic mice suggests that human tumors bearing amplified copy numbers of MDM2 did not require coamplification of other genes localizing to chromosome 12q14-q15 to undergo transformation. The 25% increase in the rate of tumorigenesis seen in the mice homozygous for the transgene insertion relative to hemizygous, Mdm2-transgenic mice suggest that the rate of tumorigenesis correlates with the level of Mdm2 expression.
Although the transgenic mice were susceptible to spontaneous tumorigenesis, the rate of tumor formation was much slower than that seen in p53-null mice. This is likely to be due to the relatively modest increase in the level of Mdm2 expression in the transgenic mice. The levels of Mdm2 overexpression observed in human tumor samples or in cell lines derived from such tumors range from 5–50 fold (1, 7). The level of Mdm2-overexpression in the transgenic mice may also account for the inability of the transgene to elevate the rapid rate of tumorigenesis seen in p53-null mice. However, the increase in the incidence of sarcomas in the Mdm2-transgenic, p53-null mice relative to p53-null mice must be due to a p53-independent mechanism mediated by Mdm2, since p53 is absent in these mice. Thus, the results of this experiment provides in vivo evidence that Mdm2 overexpression can promote tumorigenesis in a p53-independent manner, at least in tissues of mesenchymal origin. The increase in sarcomas in the Mdm2-transgenic mice is consistent with the observation that 30–40% of human sarcomas contain MDM2 amplification. Interestingly, although the incidence of sarcomas in the transgenic mice was not altered by changes in p53 status, the rate of sarcoma formation was accelerated when p53 was deleted.
Human and mouse Mdm2 loci generate multiple transcripts which differ in their coding capacities (15, 38). A number of these transcripts encode Mdm2 products which lack sequences required for binding to p53 and which fail to inhibit p53-mediated transactivation of a p53-responsive promoter. Alternately spliced forms of human MDM2 which lack the p53-binding domain have been found more frequently in poorly differentiated, late stage ovarian and bladder carcinomas (39). Transfection of cDNAs encoding these various MDM2 isoforms induced transformation of NIH 3T3 cells, even though the MDM2 proteins were incapable of complexing with p53. Preliminary analysis of sarcomas harvested from the Mdm2-transgenic mice suggests that these tumors likewise contain alternatively spliced isoforms of Mdm2 message, a number of which lack sequences encoding the p53-binding site near the amino terminus of the Mdm2 protein. Further analysis of these transcripts will likely help resolve the significance of these transcripts to sarcoma formation in the mice.
Elucidation of the p53-independent mechanism used by Mdm2 in cellular transformation may have direct clinical relevance. Many laboratories are presently investigating the feasibility of small molecule intervention to inhibit Mdm2-p53 binding as a means of treating cancers which exhibit high levels of Mdm2 expression. If Mdm2 promotes tumorigenesis solely by inhibiting p53, then restoration of p53 function via inhibition of Mdm2-p53 binding may likely be of therapeutic value. However, if overexpression of Mdm2 promotes tumorigenesis by other pathways as well, then inhibition of Mdm2-p53 complex formation may not be sufficient to reverse the neoplasia.
Acknowledgments
We thank Mary Ellen Perry for comments on the manuscript, Donna L. George for kindly providing the Mdm2 cosmid used in creating the transgenic mice, and Eva Regel for technical assistance. This work was supported, in part, by a National Cancer Institute Grant CA54897 (to L.D.), and by Grant IRG-93-033-05-IRG from the American Cancer Society (to S.J.). A.B. is an investigator with the Howard Hughes Medical Institute.
ABBREVIATION
- ES
embryonic stem
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Oliner J D, Kinzler K W, Meltzer P S, George D L, Vogelstein B. Nature (London) 1992;358:80–83. doi: 10.1038/358080a0. [DOI] [PubMed] [Google Scholar]
- 2.Leach F S, Tokino T, Meltzer P, Burrell M, Oliner J D, Smith S, Hill D E, Sidransky D, Kinzler K W, Vogelstein B. Cancer Res. 1993;53:2231–2234. [PubMed] [Google Scholar]
- 3.Nakayama T, Toguchida J, Wadayama B, Kanoe H, Kotoura Y, Sasaki M. Int J Cancer. 1995;64:342–346. doi: 10.1002/ijc.2910640511. [DOI] [PubMed] [Google Scholar]
- 4.Cordon-Cardo C, Latres E, Drobnjak M, Oliva M, Pollack D, Woodruff J M, Marachel V, Chen J, Brennan M F, Levine A. Cancer Res. 1994;54:794–799. [PubMed] [Google Scholar]
- 5.Miller C W, Aslo A, Won A, Tan M, Lampkin B, Koeffler H P. J Cancer Res Clin Oncol. 1996;122:559–565. doi: 10.1007/BF01213553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reifenberger G, Liu L, Ichimura K, Schmidt E E, Collins V P. Cancer Res. 1993;53:2736–2739. [PubMed] [Google Scholar]
- 7.Bueso-Ramos C E, Yang Y, deLeon E, McCown P, Stass S A, Albitar M. Blood. 1993;82:2617–2623. [PubMed] [Google Scholar]
- 8.Watanabe T, Hotta T, Ichikawa A, Kinoshita T, Nagai H, Uchida T, Murate T, Saito H. Blood. 1994;84:3158–3165. [PubMed] [Google Scholar]
- 9.Matsumura T, Yoshihama Y, Kimura T, Shintani S, Alcade R E. Oncology. 1996;53:308–312. doi: 10.1159/000227578. [DOI] [PubMed] [Google Scholar]
- 10.Forus A, Florenes V A, Maelandsmo G M, Meltzer P S, Fodstad O, Myklebost O. Cell Growth Diff. 1993;4:1065–1070. [PubMed] [Google Scholar]
- 11.Khatib Z A, Matsushime H, Valentine M, Shapiro D N, Sherr C J, Look A T. Cancer Res. 1993;53:5535–5541. [PubMed] [Google Scholar]
- 12.Reifenberger G, Ichimura K, Reifenberger J, Elkahloun A G, Meltzer P S, Collins V P. Cancer Res. 1996;56:5141–5145. [PubMed] [Google Scholar]
- 13.Fischer U, Meltzer P, Meese E. Hum Genet. 1996;98:625–628. doi: 10.1007/s004390050271. [DOI] [PubMed] [Google Scholar]
- 14.Cahilly-Snyder L, Yang-Feng T, Francke U, George G L. Somatic Cell Mol Genet. 1987;13:235–244. doi: 10.1007/BF01535205. [DOI] [PubMed] [Google Scholar]
- 15.Fakharzadeh S S, Trusko S P, George D L. EMBO J. 1991;10:1565–1569. doi: 10.1002/j.1460-2075.1991.tb07676.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Momand J, Zambetti G P, Olson D C, George D, Levine A J. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- 17.Barak Y T, Oren M. EMBO J. 1992;11:2115–2121. doi: 10.1002/j.1460-2075.1992.tb05270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hoppe-Seyler F, Butz K. J Virol. 1993;67:3111–3117. doi: 10.1128/jvi.67.6.3111-3117.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay C A. Mol Cell Biol. 1993;13:301–306. doi: 10.1128/mcb.13.1.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Wu X, Levine A J. Mol Cell Biol. 1996;16:2445–2452. doi: 10.1128/mcb.16.5.2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Haupt Y, Barak Y, Oren M. EMBO J. 1996;15:1596–1606. [PMC free article] [PubMed] [Google Scholar]
- 22.Kubbutat M H G, Jones S N, Vousden K H. Nature (London) 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- 23.Haupt Y, Maya R, Kazaz A, Oren M. Nature (London) 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- 24.Barak Y T, Juven R, Haffner R, Oren M. EMBO J. 1993;12:461–468. doi: 10.1002/j.1460-2075.1993.tb05678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juven T, Barak Y, Zauberman A, George D L, Oren M. Oncogene. 1993;8:3411–3416. [PubMed] [Google Scholar]
- 26.Wu X, Bayle H, Olson D, Levine A J. Genes Dev. 1993;7:1126–1132. doi: 10.1101/gad.7.7a.1126. [DOI] [PubMed] [Google Scholar]
- 27.Jones S N, Roe A E, Donehower L A, Bradley A. Nature (London) 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- 28.Montes de Oca Luna R, Wagner D S, Lozano G. Nature (London) 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- 29.Jones S N, Sands A, Hancock A R, Vogel H, Donehower L A, Linke S P, Wahl G M, Bradley A. Proc Natl Acad Sci USA. 1996;93:14106–14111. doi: 10.1073/pnas.93.24.14106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Boddy M N, Freemont P S, Borden K L B. Trends Biochem Sci. 1994;19:198–199. doi: 10.1016/0968-0004(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 31.Elenbaas B, Dobbelstein M, Roth J, Shenk T, Levine A J. Mol Med. 1996;2:439–451. [PMC free article] [PubMed] [Google Scholar]
- 32.Xiao Z-X, Chen J, Levine A J, Modjtahedi N, Xing J, Sellers W R, Livingston D M. Nature (London) 1995;375:694–698. doi: 10.1038/375694a0. [DOI] [PubMed] [Google Scholar]
- 33.Martin K, Trouche D, Hagemeier C, Sorensen T S, La Thangue N B, Kouzarides T. Nature (London) 1995;375:691–694. doi: 10.1038/375691a0. [DOI] [PubMed] [Google Scholar]
- 34.Lundgren K, Montes de Oca Luna R, McNeill B, Emerick E P, Spencer B, Barfield C R, Lozano G, Rosenberg M P, Finlay C A. Genes Dev. 1997;11:714–725. doi: 10.1101/gad.11.6.714. [DOI] [PubMed] [Google Scholar]
- 35.Harvey M, McArthur M J, Montgomery C A, Jr, Butel J S, Bradley A, Donehower L A. Nat Genet. 1993;5:225–229. doi: 10.1038/ng1193-225. [DOI] [PubMed] [Google Scholar]
- 36.Jones S N, Ansari-Lari M A, Hancock A R, Jones W J, Gibbs R A, Donehower L A, Bradley A. Gene. 1996;175:209–213. doi: 10.1016/0378-1119(96)00151-5. [DOI] [PubMed] [Google Scholar]
- 37.Montes De Oca Luna R, Tabor A D, Eberspaecher H, Hulboy D L, Worth L L, Colman M S, Finlay C A, Lozano G. Genomics. 1996;33:352–357. doi: 10.1006/geno.1996.0210. [DOI] [PubMed] [Google Scholar]
- 38.Haines D S, Landers J E, Engle L J, George D L. Mol Cell Biol. 1994;14:1171–1178. doi: 10.1128/mcb.14.2.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sigalas I, Calvert H, Anderson J J, Neal D E, Lunec J. Nat Med. 1996;2:912–917. doi: 10.1038/nm0896-912. [DOI] [PubMed] [Google Scholar]