Abstract
Interferon inducible protein kinase PKR is a component of innate immunity and mediates antiviral actions by recognizing pathogen associated molecular patterns (PAMPs). A well-known activator of PKR is long dsRNA, which can be produced during viral replication. Our recent results indicate that PKR can also be activated by short stem-loop RNA in a 5′-triphosphate-dependent fashion. A 5′-triphosphate is present primarily in foreign RNAs such as viral and bacterial transcripts, while a non-activating 5′-cap or 5′-monophosphate is present in most cellular RNAs. Recent studies indicate that internal RNA modifications and non-Watson-Crick motifs also repress PKR activation, and do so in an RNA structure-specific fashion. Interestingly, self-RNAs have more nucleoside modifications than non-self RNAs. Internal and 5′-end RNA modifications have repressive effects on other innate immune sensors as well, including TLR3, TLR7, TLR8, and RIG-I, suggesting that nucleoside modifications suppress innate immunity on a wide scale.
Keywords: modified RNA, self and non-self RNA, PKR, RNA structure, translation inhibition
Innate immunity is the first line of host defense against pathogen infection.1 Identification of innate immune-tripping pathogenic signals has been progressing rapidly. Innate antiviral defense is mediated through virus recognition and signaling by the host cell, resulting in the disruption of virus replication. Upon recognizing viruses by pathogen recognizing receptors (PRR), interferons (IFNs) are produced that are essential for immune defense against viruses and induce proteins such as PKR, OAS, and ADAR, which mediate antiviral actions.2,3 One critical issue is how does innate immunity distinguish self from pathogen. In innate immunity, motifs associated with pathogens include lipopolysaccharides (LPS), proteins, CpG-DNA, ssRNA and dsRNA. There are several excellent reviews available in the literature that elaborate on this subject.4–15 In this commentary, we describe some of our recent results and compare them to related studies, which add PKR to the list of innate immune sensors that are sensitive to RNA modifications. Overall, it appears that modifications in RNA provide a critical basis for PKR to distinguish self-RNAs from pathogenic RNAs.
Protein kinase PKR is an interferon-inducible protein and a key component of innate immunity. It consists of an N-terminal dsRNA-binding domain (dsRBD), which comprises two tandem copies of the dsRNA-binding motif (dsRBM) separated by a 20 amino acid linker, and a C-terminal catalytic kinase domain. It has been shown that long stretches of dsRNA (>33 base pairs) activate PKR to undergo autophosphorylation and phosphorylate its cellular substrate, translation initiation factor eIF2α at serine 51.16 As a consequence, eIF2 affinity for GTP exchange factor eIF2B increases up to 100-fold, leading to competitive inhibition of eIF2B and inhibition of translation initiation.17 We have recently found that short stem-loop (sSL) RNAs activate PKR in a 5′-triphosphate-dependent fashion and that such activation is largely abrogated by many natural modifications in RNA.18,19 These studies demonstrate that nucleoside modifications regulate PKR in an RNA structure-specific manner. In this article, we focus on PKR’s role in innate immunity, however it should be noted that PKR has also been implicated in numerous other cellular roles including regulating signal transduction and transcription, and controlling tumorigenesis and neurodegenerative diseases.20
5′-TRIPHOSPHATE OF RNA COMPRISES A PKR PAMP
Early studies used poly I:C as a model dsRNA to examine the enzymatic activity of PKR.21,22 Later analyses revealed that PKR is able to bind non-sequence specifically to dsRNA as short as 11 bp, with at least 33 bp required for activation.16 In addition to dsRNA, PKR activators include polyanionic molecules, several cellular proteins, and other cellular RNAs.20 In a search for novel activators, we identified a short dsRNA with single-stranded tails, the ‘ss-dsRNA’ motif, which activates PKR, with the length of the tail providing a critical determinant.23 This motif has an imperfect stem of 16 bp and is flanked by single-stranded tails. These RNAs were prepared by in vitro transcription, which provided a 5′-triphosphate. This raised the question of whether the 5′-triphosphate plays a role in PKR activation.
We tested the importance of the 5′-triphosphate in in vitro-transcribed ss-dsRNA by treating appropriate transcripts (tails of at least 9 nt) with calf intestinal phosphatase (CIP), which remove the 5′-triphosphate. Remarkably, CIP treatment of these ss-dsRNAs led to abrogation of PKR activation both in vitro and in cells. Chemically synthesized 5′-hydroxyl containing ss-dsRNA also showed no activation of PKR. Unlike ss-dsRNA, perfect dsRNA was found to not have a dependence on 5′-triphosphate, providing the first indication that modifications of RNA affect activation of PKR in an RNA structure-specific fashion. In parallel with the ss-dsRNA motif, transcripts of 47 and 110 nt with minimal secondary structures also revealed a dependence on 5′-triphosphate.
Cellular primary RNA transcripts have a 5′-triphosphate, but most processed self-RNAs do not. For example, pol I-transcripts (e.g. rRNAs) are processed to 5′-monophosphates, pol II-transcribed mRNAs to 7-methyl G caps, and pol III-transcribed tRNAs to 5′-monophosphates.24 In contrast to cellular RNAs, many mature viral and bacterial RNAs contain triphosphates at their 5′-end. We found that 5′-monophosphate, -diphosphate, and -7-methyl guanosine cap-containing ssRNA-47 and ssRNA-110 RNAs do not activate PKR, further implicating the 5′-triphosphate as the key 5′-end functional group in ssRNA needed to activate PKR.
It has been reported that RIG-I, an essential cytosolic viral RNA sensor, recognizes the 5′-triphosphate of RNA to induce interferon production as well.25,26 Cell line experiments showed that RIG-I signaling is not required for 5′-triphosphate-ssRNA activation of PKR, indicating that there are two strategies for recognizing a 5′-triphosphate.18 Similar to PKR displaying no 5′-triphosphate dependence for dsRNA, RIG-I can be activated in vitro by poly I:C, which contains little or no 5′-triphosphate, albeit weakly.27–29 This suggests that both PKR and RIG-I can be activated by 5′-triphosphate-ssRNAs and 5′-OH dsRNAs, although the molecular basis is likely to be different since PKR and RIG-I are not homologous (R. Toroney and P. C. Bevilacqua, unpublished observation).
Despite differences in sequence and structure of PKR and RIG-I, it is not surprising that these proteins recognize the same ligand. In the case of PKR, the catalytic ATP binds in the kinase domain through direct contacts of the phosphates with lysine residues, as well as through direct and water-mediated contacts of the two triphosphate-bound Mg2+ ions with acidic residues (unpublished observation). These interactions are common to the ATP binding sites of other protein kinases, polymerases, and restriction endonucleases.30,31 The electrostatic nature of these contacts suggests the possibility that binding of the 5′-triphosphate-Mg2+ complex may be achieved through a similar mixture of basic and acidic residues. Because both PKR and RIG-I contain ATP-binding sites, it is reasonable to suggest that these two proteins could bind a 5′-triphosphate in a chemically similar manner despite the overall lack of sequence and structural homology, although this may not hold.
INTERNAL RNA MODIFICATIONS DISGUISE SELF RNA
If some model structured RNAs activate PKR, then one might ask why structured self-RNAs, abundantly available in the host cell, do not activate PKR. One possibility is that cellular RNAs encode information to avoid activating PKR. If so, one mechanism by which self-RNA might avoid activating PKR is through nucleoside modifications. RNAs are modified post-transcriptionally as part of their maturation process. To date, over 100 naturally occurring modified nucleosides have been identified.32,33 Most of the modifications involve methylation of the nucleobase or 2′-O-methyl formation in the ribose. The heaviest modified RNAs are tRNAs. In addition, mammalian rRNAs contain more modifications than bacterial rRNA. Mammalian mRNAs possess 5-methylcytidine, 6-methyladenosine (m6A), inosine, 2′-O-methyl ribose, and a 5′-terminal cap structure of 7-methylguanosine, while bacterial mRNAs contain no nucleoside modifications. Some viral RNAs such as adenovirus, herpes simplex virus, influenza, respiratory syncytial virus, and simian virus have been shown to contain modifications, but it is not clear at present what roles these modifications play in viruses.5
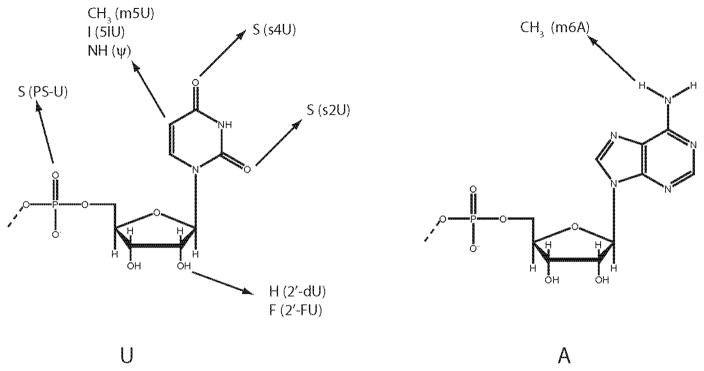
In a recent report, we described the effects of RNA modifications on PKR activation.19 Systematic analysis was performed on the effects of naturally occurring nucleobase, sugar, and phosphodiester modifications on activation of PKR by ssRNA-47 and dsRNA-47. In addition, effects of GU wobble base pairing on PKR activation were examined. Modifications were incorporated by replacing the unmodified NTP with the corresponding modified NTP in the T7 transcription reaction. Modified nucleosides used in the study are provided in Figure 1 and effects of the modifications are summarized in Table 1. Effects of the nucleoside modifications in 5′-triphosphate-containing ssRNA-47 on PKR were analyzed. Our results indicate that minor grove modified nucleosides (s2U and 2′-dU), major grove modified nucleosides (s4U, Ψ, m5U, I5U, and m6A), and Watson-Crick base-pairing-modified nucleosides (s2U, and s4U) abrogate activation of PKR in ssRNA-47. Remarkably, ssRNA-47 with the minor groove-modifying nucleoside 2′FU, retains the ability to activate PKR (Table 1). The opposing effects of 2′-FU and 2′-dU in ssRNA-47 support a role for hydrogen bonding between PKR and the ribose portion of ssRNA. Further mechanistic and structural studies are needed to understand the molecular basis of PKR regulation by modified ssRNA.
Figure 1.
Schematic of modified nucleosides. Unmodified uridine (U) and adenosine (A) are depicted. Position of change in atom or functional group is shown.
TABLE 1.
EFFECTS OF RNA MODIFICATIONS ON PKR ACTIVATION
‘+++’, “++”, “+”, and “−” indicate 50–100%, 25–50%, 10–25%, and less than 10% of PKR activation levels relative to unmodified ssRNA-47 (column 2) or dsRNA-47 (column 3), respectively.
The details of the modifications employed in the study can be found in ref. 19
With the exception of ‘2-FU’, minor groove modifications abrogate PKR activation by both ssRNA-47 and dsRNA-47.
Major groove modifications in ssRNA-47 abrogated PKR activation, with the exception of ‘PS-U’, whereas all major groove modifications in dsRNA-47 had virtually no effect with the exception of ‘s4U’, which also perturbs the W-C face.
When we tested the same modifications in dsRNA (with 47 bp), a striking difference was observed for some modifications. Modifications to the minor groove (s2U, 2′-dU and 2′FU) and Watson-Crick base pairing face (s2U and s4U) in dsRNA led to similar effects on PKR activation as in ssRNA-47. However, modifications to the major groove, which abrogated PKR activation by ssRNA, had either no effect on activation by dsRNA (m5U and m6A), or just a slight effect (Ψ– and I5U).19 Overall, we conclude that internal nucleoside modifications, like 5′-end specific modifications, modulate PKR activation in an RNA structure-specific context.
Observation that dsRNA substituted with 2′-dU does not activate PKR, while dsRNA substituted with 2′-FU does is consistent with a crystal structure of a dsRNA-dsRBM complex,34 which showed that RNA-protein interactions are primarily through water-bridged interactions to 2′-OHs present in the minor groove of dsRNA. This suggests that 2′-FU-substituted dsRNA activates PKR by maintaining a water-bridged hydrogen bonding network.
Abrogation of PKR activation by modified ssRNA and dsRNA is not due to an inability to bind, as our results show that modified RNAs bind PKR with approximately equal affinity as unmodified RNA. In fact, non-activator modified dsRNAs act as potent inhibitors of PKR.19 This suggests a nonequivalence of binding to and activation of PKR by modified RNAs. Similar behavior has been observed by Kawahara et. al. in that unmodified and edited pre-miR-151 both bind tightly to Dicer-TRBP, but only unedited RNA is cleaved.35
We also examined activation of PKR by dsRNA with GU wobble base pairs, which are building blocks in RNA structure that have been found in virtually every class of functional RNA. Moderate levels of GU wobble insertion abrogated PKR activation, suggesting that wobble base pairing may also serve to discriminate self-RNA and pathogenic RNA. GU wobble base pairs embedded within A-form RNA helices make the major groove wide and shallow, offering a possible structural basis for this effect.36,37 Interestingly, IU base pairs, which occur through editing of AU to IU in dsRNA by ADAR enzymes, also result in GU-like wobbled structures.
We conclude the portion of this article specifically on PKR by noting that it is likely that other mechanisms besides molecular signals encoded in the RNA itself serve to assist PKR in accurately discriminating between self and non-self RNA. One possibility is that RNA-binding proteins compete with PKR for cellular RNA. It appears that the vast majority of cellular RNAs are protein-bound.38 For example, the canonical RNA-recognition motif (RRM), is a common motif found in ssRNA-binding proteins such as nucleolin, a nuclear protein that binds pre-rRNA, and p14, a component of the spliceosome. Endogenous dsRNAs, such as miRNAs and their precursurs, are bound by other dsRBD-containing proteins such as ADARs and Dicer, presumably inhibiting binding by PKR.39 Much of the RNA in the cell is even bound by multiple proteins, such as rRNA in the ribosome and mRNA in the messenger ribonucleoprotein particle (mRNP), an RNA-protein complex which consists of numerous proteins that accompany the RNA from the nucleus out to the cytoplasm.40 As PKR is found in the nucleus as well as the cytoplasm,41 binding of RNAs by these and other proteins may be an essential checkpoint in preventing PKR activation by self-RNA.
COMPARISON OF EFFECTS OF RNA MODIFICATIONS: PKR, TLRs, AND RIG-I
Recently Kariko and colleagues42 showed that monocyte-derived dendritic cells transfected with human mitochondrial RNA, which has just one modification, secreted TNF-α (a proinflammatory cytokine secreted as part of the RNA-sensing TLR-initiated immune response)43,44 at similar levels as cells transfected with total RNA obtained from E. coli. In contrast, transfections with RNAs from other compartments of the cell, which tend to have more modifications, showed little or no production of TNF-α. The authors concluded that mammalian self-RNA might be masked by naturally occurring nucleoside modifications.
To understand the effect of nucleoside modifications on TLRs, they conducted experiments on in vitro-transcribed modified RNAs in HEK293 cells overexpressing TLR3, TLR7, and TLR8, measuring IL-8 production. Interestingly, production of IL-8 was suppressed by several different types of modified nucleotides. In parallel, Hornung et. al.26 showed that 5′-triphosphate-dependent activation of RIG-I is suppressed by certain nucleoside modifications, namely Ψ, s2U, and 2′-O-Me-U. Pseudouridine and s2U also suppress PKR, as does introduction of 2′-dU (2′-O-Me-U was not tested with PKR). The effects of other nucleoside modifications on RIG-I activation remain to be tested and compared to PKR.
In general, the finding that stimulation of innate immunity is suppressed by RNA modifications is similar to that observed with PKR.19,26,42 Nonetheless, it is quite likely that the molecular basis for the effects is different with each protein since TLRs and RIG-I do not have dsRBMs and in general have little sequence homology. Interestingly, TLR3, TLR7, and TLR8 are present in the endosomal membrane, with RIG-I and PKR primarily in the cytosol. TLR3 is also expressed on the surface of cells. Taken together, these studies suggest that introduction of modifications into siRNA that serve to mimic self-RNA may avoid unwanted immunostimulatory effects,19,26,42,45 while introduction of modifications such as 2′-FU may lead to immunostimulation (isRNAs), which may be desirable in some applications.11,46 In addition, these data suggest that incorporation of certain modifications into RNA may produce better immunotherapy with RNA-based vaccines or an RNA-based adjuvant.
CONCLUDING REMARKS
A major issue in innate immunity is the specific recognition of molecular signals emanating from pathogens. Some of these signals are encoded in RNA. Primary transcripts with a 5′-triphosphate and no internal modifications activate an immune response. These features are disguised in most endogenous transcripts through 5′-end modifications and internal modifications and non-Watson-Crick motifs. A series of RNA sensors exist in the innate immune system that discriminate self and non-self RNA on the basis of nucleoside modifications at 5′-end and internal regions of the RNA, although the molecular basis is likely to be different for each sensor. It appears that endosomal and cytosolic viral recognition pathways complement each other. Further discovery and characterization of RNA PAMPs will aid in understanding innate immunity and thereby provide the opportunity to make new and better RNA-based therapeutics.
Acknowledgments
This work was supported by NIH Grant R01-58709. We thank Katalin Kariko and Drew Weissman for helpful comments.
ABBREVIATIONS
- ADAR
adenosine deaminase that acts on RNA
- LPS
lipopolysaccharide
- MDA5
melanoma differentiation-associated gene-5
- PKR
RNA activated protein kinase
- PAMP
pathogen-associated molecular pattern
- PRR
pathogen recognizing receptors
- RIG-I
retinoic acid inducible gene-I
- TLR
toll-like receptor
References
- 1.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–64. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 2.Samuel CE. Antiviral actions of interferons. Clin Microbiol Rev. 2001;14:778–809. doi: 10.1128/CMR.14.4.778-809.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat Rev Drug Discov. 2007;6:975–90. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85:435–45. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 5.Kariko K, Weissman D. Naturally occurring nucleoside modifications suppress the immunostimulatory activity of RNA: implication for therapeutic RNA development. Curr Opin Drug Discov Devel. 2007;10:523–32. [PubMed] [Google Scholar]
- 6.Chi H, Flavell RA. Innate recognition of non-self nucleic acids. Genome Biol. 2008;9:211. doi: 10.1186/gb-2008-9-3-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Randall RE, Goodbourn S. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J Gen Virol. 2008;89:1–47. doi: 10.1099/vir.0.83391-0. [DOI] [PubMed] [Google Scholar]
- 8.Bowie AG, Fitzgerald KA. RIG-I: tri-ing to discriminate between self and non-self RNA. Trends Immunol. 2007;28:147–50. doi: 10.1016/j.it.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 9.Saito T, Gale M., Jr Principles of intracellular viral recognition. Curr Opin Immunol. 2007;19:17–23. doi: 10.1016/j.coi.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 10.Lamphier MS, Sirois CM, Verma A, Golenbock DT, Latz E. TLR9 and the recognition of self and non-self nucleic acids. Ann N Y Acad Sci. 2006;1082:31–43. doi: 10.1196/annals.1348.005. [DOI] [PubMed] [Google Scholar]
- 11.Schlee M, Hornung V, Hartmann G. siRNA and isRNA: two edges of one sword. Mol Ther. 2006;14:463–70. doi: 10.1016/j.ymthe.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 12.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that cooperate in innate immunity. Trends Immunol. 2006;27:352–7. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 13.Sioud M. Innate sensing of self and non-self RNAs by Toll-like receptors. Trends Mol Med. 2006;12:167–76. doi: 10.1016/j.molmed.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–7. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 15.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 16.Manche L, Green SR, Schmedt C, Mathews MB. Interactions between double-stranded RNA regulators and the protein kinase DAI. Mol Cell Biol. 1992;12:5238–48. doi: 10.1128/mcb.12.11.5238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Williams BR. PKR; a sentinel kinase for cellular stress. Oncogene. 1999;18:6112–20. doi: 10.1038/sj.onc.1203127. [DOI] [PubMed] [Google Scholar]
- 18.Nallagatla SR, Hwang J, Toroney R, Zheng X, Cameron CE, Bevilacqua PC. 5′-triphosphate-dependent activation of PKR by RNAs with short stem-loops. Science. 2007;318:1455–8. doi: 10.1126/science.1147347. [DOI] [PubMed] [Google Scholar]
- 19.Nallagatla SR, Bevilacqua PC. Nucleoside modifications modulate activation of the protein kinase PKR in an RNA structure-specific manner. RNA. 2008;14:1201–13. doi: 10.1261/rna.1007408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sadler AJ, Williams BR. Structure and function of the protein kinase R. Curr Top Microbiol Immunol. 2007;316:253–92. doi: 10.1007/978-3-540-71329-6_13. [DOI] [PubMed] [Google Scholar]
- 21.Farrell PJ, Sen GC, Dubois MF, Ratner L, Slattery E, Lengyel P. Interferon action: two distinct pathways for inhibition of protein synthesis by double-stranded RNA. Proc Natl Acad Sci U S A. 1978;75:5893–7. doi: 10.1073/pnas.75.12.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hovanessian AG. On the discovery of interferon-inducible, double-stranded RNA activated enzymes: the 2′–5′oligoadenylate synthetases and the protein kinase PKR. Cytokine Growth Factor Rev. 2007;18:351–61. doi: 10.1016/j.cytogfr.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 23.Zheng X, Bevilacqua PC. Activation of the protein kinase PKR by short double-stranded RNAs with single-stranded tails. RNA. 2004;10:1934–45. doi: 10.1261/rna.7150804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schlee M, Barchet W, Hornung V, Hartmann G. Beyond double-stranded RNA-type I IFN induction by 3pRNA and other viral nucleic acids. Curr Top Microbiol Immunol. 2007;316:207–30. doi: 10.1007/978-3-540-71329-6_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichlmair A, Schulz O, Tan CP, Naslund TI, Liljestrom P, Weber F, Reis e Sousa C. RIG-I-mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314:997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 26.Hornung V, Ellegast J, Kim S, Brzozka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann KK, Schlee M, Endres S, Hartmann G. 5′-Triphosphate RNA is the ligand for RIG-I. Science. 2006;314:994–7. doi: 10.1126/science.1132505. [DOI] [PubMed] [Google Scholar]
- 27.Yoneyama M, Kikuchi M, Natsukawa T, Shinobu N, Imaizumi T, Miyagishi M, Taira K, Akira S, Fujita T. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5:730–7. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 28.Gitlin L, Barchet W, Gilfillan S, Cella M, Beutler B, Flavell RA, Diamond MS, Colonna M. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc Natl Acad Sci U S A. 2006;103:8459–64. doi: 10.1073/pnas.0603082103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Yamaguchi O, Otsu K, Tsujimura T, Koh CS, Reis e Sousa C, Matsuura Y, Fujita T, Akira S. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441:101–5. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 30.Johnson LN, Noble ME, Owen DJ. Active and inactive protein kinases: structural basis for regulation. Cell. 1996;85:149–58. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 31.Berg JM, Tymoczko JL, Stryer L. Biochemistry. New York: W. H. Freeman and Company; 2007. [Google Scholar]
- 32.Grosjean H. Modification and editing of RNA: historical overview and important facts to remember. Topics in Current Genetics: Fine Tuning of RNA Functions by Modifications and Editing. Berlin Heidelberg: Springer-Verlag. 2005;12:1–22. [Google Scholar]
- 33.Rozenski J, Crain PF, McCloskey JA. The RNA Modification Database: 1999 update. Nucleic Acids Res. 1999;27:196–7. doi: 10.1093/nar/27.1.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ryter JM, Schultz SC. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17:7505–13. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of the microRNA-151 precursor blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007;8:763–9. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weeks KM, Crothers DM. Major groove accessibility of RNA. Science. 1993;261:1574–7. doi: 10.1126/science.7690496. [DOI] [PubMed] [Google Scholar]
- 37.Varani G, McClain WH. The G x U wobble base pair. A fundamental building block of RNA structure crucial to RNA function in diverse biological systems. EMBO Rep. 2000;1:18–23. doi: 10.1093/embo-reports/kvd001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hall KB. RNA-protein interactions. Curr Opin Struct Biol. 2002;12:283–8. doi: 10.1016/s0959-440x(02)00323-8. [DOI] [PubMed] [Google Scholar]
- 39.Tian B, Bevilacqua PC, Diegelman-Parente A, Mathews MB. The double-stranded-RNA-binding motif: interference and much more. Nat Rev Mol Cell Biol. 2004;5:1013–23. doi: 10.1038/nrm1528. [DOI] [PubMed] [Google Scholar]
- 40.Le Hir H, Gatfield D, Izaurralde E, Moore MJ. The exon-exon junction complex provides a binding platform for factors involved in mRNA export and nonsense-mediated mRNA decay. EMBO J. 2001;20:4987–97. doi: 10.1093/emboj/20.17.4987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeffrey IW, Kadereit S, Meurs EF, Metzger T, Bachmann M, Schwemmle M, Hovanessian AG, Clemens MJ. Nuclear localization of the interferon-inducible protein kinase PKR in human cells and transfected mouse cells. Exp Cell Res. 1995;218:17–27. doi: 10.1006/excr.1995.1126. [DOI] [PubMed] [Google Scholar]
- 42.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–75. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 43.Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, McNally E, Pickart C, Ma A. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol. 2004;5:1052–60. doi: 10.1038/ni1110. [DOI] [PubMed] [Google Scholar]
- 44.Perkins ND. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol. 2007;8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 45.Judge AD, Bola G, Lee AC, MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 46.Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S, Noronha A, Manoharan M, Akira S, de Fougerolles A, Endres S, Hartmann G. Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med. 2005;11:263–70. doi: 10.1038/nm1191. [DOI] [PubMed] [Google Scholar]