Abstract
FE65 is a multi-modular adaptor protein that binds the cytoplasmic tail of the β-amyloid precursor protein (APP). Genetic evidence suggests that APP is intimately involved in the pathogenesis of dementias of the Alzheimer type, neurodegenerative disorders that affect multiple cognitive domains, including learning and memory. Evidence from p97FE65-specific knockout mice (lacking the 97 kDa full-length FE65 protein, p97FE65) suggests an important role for FE65 in learning and memory. Interpretation of the learning and memory phenotype, however, is complicated by the up-regulation (compared to wild-type mice) of a novel 60 kDa FE65 isoform (p60FE65). Here, we report evidence that p60FE65 is translated from an alternative methionine, M261, on the p97FE65 transcript. Thus, p60FE65 has a shortened N-terminus, lacking part of the WW domain that is considered important for nuclear translocation and transactivation of gene expression. Consistently, p60FE65 exhibits an attenuated ability for APP-Gal4-mediated transcription as compared to p97FE65. Similar to p97FE65, however, both transfected and endogenous p60FE65 are able to translocate to the nucleus in cultured cells and in neurons. These results are consistent with earlier evidence from our laboratory that reduced FE65 nuclear signaling may contribute, in part, to the phenotypes observed in p97FE65 knockout mice.
Keywords: FE65 isoforms, alternative translation, transcription, nuclear localization, APP
INTRODUCTION
Dementias of the Alzheimer Type (DAT) are the most common forms of progressive memory and cognitive deterioration in the elderly. Mutations in three genes have been found to cause autosomal dominant early onset forms of DAT. These genes include the beta-amyloid precursor protein (APP), and presenilins 1 and 2 (Hardy 1997; Price and Sisodia 1998). Presenilins are associated with the γ–secretase activity that is involved in the proteolytic processing of APP. Cleavages of APP by α- or β- and γ-secretases lead to generation of secreted APP, β-amyloid peptides, APP intracellular domain (AICD), and various APP C-terminal fragments (Turner et al. 2003; Galvan et al. 2006; Saganich et al. 2006). Many functions have been described for APP and its proteolytic products that may impact several aspects of learning and memory processes.
FE65 is an adaptor protein that is abundantly expressed in neurons throughout the brain (Hu et al. 1999; Kesavapany et al. 2002). The full-length FE65 contains three protein-protein interaction domains (Fig. 1D). Its most C-terminal domain, PID2, binds to AICD, suggesting that the interaction may play a role in the pathogenesis of AD. Indeed, a minor FE65 allele in humans that codes for an FE65 isoform with weak binding to APP is associated with a reduced risk for development of very late onset DAT (Hu et al. 1998; Lambert et al. 2000; Hu et al. 2002). Given the observation that age is the major risk factor for DAT, it is intriguing that expression of the 97 kDa isoform of FE65 (p97FE65) decreases during aging in mice (Wang et al. 2004).
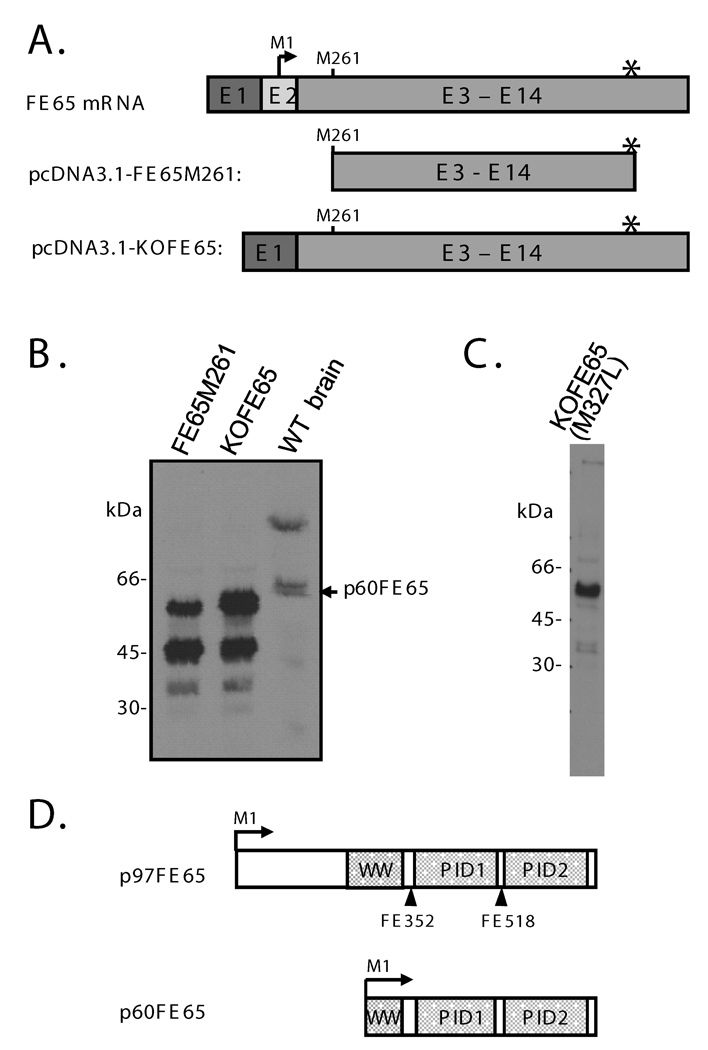
Fig. 1. The p60FE65 isoform of FE65 is likely translated from M261 within the WW domain of the p97FE65 transcript.
A: Schematic diagrams of the full-length FE65 mRNA, and the corresponding cDNA cloned into pcDNA3.1-FE65M261 or pcDNA3.1-KOFE65 construct. The full-length FE65 mRNA is shown for reference. Note that exon 2 (E2) is absent in the KOFE65 construct. The bent arrow and asterisk represent the translation initiation site and the stop codon utilized by p97FE65, respectively. B: Western analysis of lysates from mouse brain and from COS cells that were transfected with pcDNA3.1-FE65M260 or pcDNA3.1-KOFE65 using antibody FE518. C: Western analysis of lysates from COS cells that were transfected with pcDNA3.1-KOFE65(M327L). Note the absence of the 45 kDa protein band that was generated from pcDNA3.1-FE65M261 and pcDNA3.1-KOFE65 (shown in B). The remaining band (~60 kd), expressed from all three constructs, represents FE65 proteins translated from M261. D: Diagram of the domain structures of p97FE65 and p60FE65. Bent arrows symbolize the respective translation initiation sites. Straight arrows indicate the epitope positions of antibodies FE352 and FE518.
The two N-terminal domains of FE65, WW and PID1, bind to several other proteins, including neural cytoskeletal Mena (Ermekova et al. 1997), non-receptor tyrosine kinase c-Abl (Zambrano et al. 2001), histone acetyltransferase Tip60 (Cao and Sudhof 2001), transcription factor CP2/LSF/LBP1 (Zambrano et al. 1998), and a member of the low-density lipoprotein receptor-related protein family, LRP1 (Trommsdorff et al. 1998). The functional significance of these interactions will require additional research, but may include modulation of APP processing (Sabo et al. 1999; Pietrzik et al. 2004; Hu et al. 2005), plasma membrane dynamics (including neurite outgrowth and neuronal positioning) (Sabo et al. 2001 and 2003; Guenette et al. 2006; Ikin et al. 2007), and transcriptional activation (Cao and Sudhof 2001; Yang et al. 2006).
Results from our laboratory suggest that FE65 plays a direct role in learning and memory. Isoform-specific p97FE65 null mice (lacking p97FE65) exhibit impairments in passive avoidance and Morris water maze behavioral tests, with features of slow memory formation and dramatically reduced ability to replace old memory associations with new associations (Wang et al. 2004). These phenotypes likely reflect suboptimal synaptic plasticity. Interpretation of the learning/memory deficits found in the p97FE65 null mice, however, is complicated by the up-regulation (compared to wild-type mice) of a previously undescribed 60-kDa FE65 isoform, p60FE65, that retains the APP-binding ability. Therefore, the impaired learning and memory observed in the p97FE65 null mice might potentially be due to the loss of p97FE65, the up-regulation of p60FE65, or the combination of both. The relative roles of the FE65 isoforms in learning and memory and in disorders of memory, such as DAT, remain to be determined.
In search of a Notch-like transcriptional function for AICD (Schroeter et al. 1998; Struhl and Adachi 1998), Cao and Sudhof (2001) were among the first to show that p97FE65 is required for transactivation of APP-GAL4 mediated reporter gene expression. The WW domain appears to be particularly important for this function (Cao and Sudhof 2001; Minopoli et al. 2001). Our recent evidence, however, indicates that FE65-Gal4 by itself is able to trigger robust reporter activities, and that APP may not be absolutely required for that nuclear signaling pathway (Yang et al. 2006). This assertion is supported by results from Hebert et al (Hebert et al. 2006), who showed that AICD co-expression has little effect on the ability of FE65 to transactivate various promoters. The FE65 nuclear signaling pathway may be important for learning and memory, given the learning/memory deficits observed in our p97FE65-deficient mice (Wang et al. 2004).
To begin the initial exploration of this hypothesis, we cloned the p60FE65 isoform and compared its capacity to transactivate the APP-Gal4 assay to that of p97FE65. In this paper, we show evidence that isoform p60FE65 up-regulated in p97FE65 null mice is translated from an alternative translation initiation site, M261, on the p97FE65 transcript. Thus, p60FE65 lacks part of the WW domain that is important for FE65 transcriptional function (Cao and Sudhof 2001; Minopoli et al. 2001). Consistently, p60FE65 exhibits an attenuated ability for Gal4-mediated transcription as compared to p97FE65. This deficiency is unlikely due to hindered access to the nucleus, as immunostaining results indicate that p60FE65, like p97FE65, is present within the nucleus of cultured cells as well as neurons in vivo. These results are consistent with our hypothesis that impaired FE65 nuclear signaling contributes to the learning/memory-deficits observed in our p97FE65 null mice. Much additional research is required, however, to elucidate detailed mechanisms underlying these deficits.
EXPERIMENTAL PROCEDURES
Ethics Statement
All animal procedures were approved by the Institutional Animal Care and Use Committee at the University of Washington and the National Institute of Health Guide for the Care and Use of Laboratory animals.
Plasmids
pcDNA3.1-p97FE65 (containing mouse FE65 cDNA starting from the 1st methionine (M1) and ending at the stop codon) and pcDNA3.1-FE65M261 (containing FE65 cDNA starting from the fifth methionine (M261) and ending at the stop codon) were generated from pCAGGS-FE65 (Wang et al. 2004) by PCR subcloning. FE65 cDNA was amplified using forward (5’-AGTTCAAGCTTATGTCTGTTCCATCATCCCTGAGC-3’ for pcDNA3.1-p97FE65 or 5’-AGTTCAAGCTTATGAGGGTACAGGACACCTCAG-3’ for pcDNA3.1-FE65M261) and reverse (5’-GTTCTCGAGCGGCCGCGTCATGGGGTCTGGGATCCCAGAC-3’) primers containing a HindIII or a stop codon and a BamHI site at the 5’ ends, respectively. The PCR products were cloned into the HindIII and BamHI sites of pcDNA3.1. pcDNA3.1-KOFE65 was made by reverse transcribing FE65 mRNA isolated from the cerebral cortex of a p97FE65 null mouse (Wang et al. 2004), and inserting the amplified cDNA product into a pcDNA3.1 vector. In brief, the FE65 transcript, which lacks exon 2, was reverse-transcribed into cDNA using an FE65-specific primer (5’-AGTGATATCAGTACAGCGGAGG-3’) and Superscript II (Invitrogen, Carlsbad, CA). The resultant single-stranded FE65 cDNA (exons 1–14, minus exon 2) was PCR amplified using forward (5’-AGTTCAAGCTTATGTTGTGATGGAGAAGCTGCG-3’) and reverse (5’-GTTCTCGAGCGGCCGCGGAGGTGCCCGCAGAACAG-3’) primers containing a HindIII or a NotI restriction site at the 5’ end, respectively. The PCR product was cloned into the HindIII and NotI sites of pcDNA3.1. pcDNA3.1-KOFE65(M327L) was generated by site-directed mutagenesis from pcDNA3.1-KOFE65 with complementary oligonucleotides 5’-CCAGTGAGGAGGCCCCACTCGAGTTGGGACTGAAGGA-3’ (sense strand). pMst-app-Gal4DB was generously provided by Dr. Thomas C. Sudhof (University of Texas Southwestern Medical Center). pMst-Gal4DB was generated by PCR amplifying Gal4DB cDNA using pMst-app-Gal4DB as a template and forward (5’-ATCTTTGCTAGCGATGAAGCTACTGTCTTCTATCGAACAAGC-3’) and reverse (5’-TCGCAGTCGACTCACGATACAGTCAACTGTCTTTGACC-3’) primers containing a NheI or a SalI site at the 5’ end, respectively. The PCR product was cloned into the NheI and SalI sites of the pMst vector.
Cell cultures and transfections
A transformed human embryonic kidney cell line, HEK293E, a transformed African green monkey kidney cell line, COS-7, and a human brain neuroblastoma cell line, SK-N-SH, were cultured as previously described (Hu et al. 2005). For DNA transfection assays, cells at 50–75% confluence were typically transfected with 3µg total plasmids per well of 6-well plates. Transfection was mediated by 15µl of 1µg/µl polyethylenimines (Polysciences, Inc., Warrington, PA) (Hu et al. 2005). For immunocytochemistry, cells were transfected with 0.8µg plasmids and 4µl polyethylenimines per well in 24-well plates.
Western analysis
Mouse whole brains were lysed as previously described (Wang et al. 2004). Cultured cells were typically harvested 48 hours after transfection in the same lysis buffer as used for whole brains. Alternatively, cells were lysed in luciferase reporter lysis buffer (BD biotechnology, Palo Alto, CA). Western blotting to detect FE65 was performed essentially as described, using 1:20,000 diluted FE518 antibody (Hu et al. 2002).
Reporter assays
Cells were transfected with equal amounts of pFRluc (Stratagene, La Jolla, CA), pcDNA3.1-LacZ, pMst-Gal4DB or pMst-app-Gal4DB, and either pcDNA3.1-p97FE65 or pcDNA3.1-KOFE65(M327L). Cells were harvested 48 hours after transfection. Luciferase and β-galactosidase activities were determined using a Luciferase reporter assay kit (BD biotechnology, Palo Alto, CA) and a β-galactosidase enzyme assay system (Promega, Madison, WI), respectively, according to the manufacturers’ instructions. All samples were examined in triplicate and reported results are representative of at least two independent experiments.
Immunohistochemistry and Immunocytochemistry
Whole brains were removed, halved sagittally, and immersion fixed in 4% paraformaldehyde overnight at 4°C. The brains were then placed in 30% sucrose overnight at 4°C before cryosectioning. 20µm sagittal sections were cut and collected on glass slides. Sections were pretreated with 50mM ammonium chloride for 30 min before blocking with 10% filtered fetal bovine serum/0.25% triton X-100 for 1hr. Sections were co-incubated with monoclonal Anti-MAP2, clone AP-20, ascites fluid (1:500) (Sigma, St. Louis, MO) and polyclonal FE65 antibodies (5µg/ml of either FE352 or FE518) (Wang et al. 2004) at 4°C for approximately 24 hours, followed by fluorescent labeled secondary antibodies (10µg/ml of Alexa-fluor 488 donkey anti-rabbit IgG and Alexa-fluor 594 donkey anti-mouse IgG (Invitrogen, Carlsbad, CA) for 2 hr at room temperature. Sections were then incubated with 0.1% Sudan Black B (Sigma, St. Louis, MO) in 70% methanol for 10 min at room temperature. Sections were coverslipped with DAPI mounting medium (Vector Laboratories, Burlingame, CA) to label nuclei. Images were collected using an Eclipse E600 microscope (Nikon, Melville, NY) equipped with a Retigia EX cooled CCD camera and QCapture software (Qimaging, Surrey, BC, Canada), or were captured in some cases using a Deltavision Restoration Microscopy System (Applied Precision; Issaquah, WA) in which out-of-focus flare introduced into the image at different optical sections is reversed by computer deconvolution. Image qunatitation was analyzed with Softworx suite (Applied Precision, Inc.; Issaqah, WA). Images were assembled using Photoshop (Adobe). Immunocytochemistry was performed as previously described (Hu et al. 2005). Transfected COS cells or non-transfected SK-N-SH cells were incubated with an FE65-specific antibody, FE518 (1 µg/ml) (Hu et al. 2002) or FE352 (5µg/ml) (Wang et al., 2004), at room temperature for 1 hr, followed by Alexa-Fluor-568-conjugated secondary antibody (10 µg/ml) (Molecular Probes, Eugene, OR).
RESULTS
Evidence that the p60FE65 isoform is translated from M261 within the WW domain of the p97FE65 transcript
We recently identified a previously unrecognized 60 kDa FE65 protein isoform (p60FE65) that was up-regulated in our p97FE65 null mice generated by conventional knockout targeting on exon 2 (Wang et al. 2004). The exon carrying the translation initiation site for p97FE65 was deleted during RNA splicing in the knockout mice (Wang et al. 2004). An FE65 antibody (FE27) specific to the N-terminus of the full-length FE65 confirmed the knockout of p97FE65 and failed to detect p60FE65 in Western blots, whereas two other antibodies (FE352 and FE518) specific to more C-terminal epitopes identified the 60 kDa bands from both p97FE65 null and wild-type mouse brains (Wang et al. 2004) (Fig. 1D). We therefore propose that p60FE65 is translated from an alternative in-frame methionine downstream of exon 2 on the p97FE65 transcript. Methionine 261 (M261), located within the WW domain (amino acids 254–289 in p97FE65) immediately following the first signature tryptophan (W260), is the first available methionine downstream of exon 2 and in-frame with the p97FE65 initiation site. In order to determine whether M261 might be the translation initiation site for p60FE65, we generated an N-terminally truncated FE65 expression construct that lacks the p97FE65 cDNA sequence upstream of the M261 codon (pcDNA3.1-FE65M261) (Fig. 1A). Western blotting of lysates from COS cells overexpressing FE65M261 proteins revealed the presence of a ~60 kDa protein band (Fig. 1B), suggesting that p60FE65 is indeed translated from M261 within the WW domain of the p97FE65 transcript. Although the protein band ran slightly faster on SDS-PAGE than the endogenous p60FE65 detected in p97FE65 wild-type brain lysates, we also observed that overexpressed p97FE65 proteins run slightly faster than endogenous p97FE65 when the protein gel was allowed to run long enough (data not shown). The size discrepancies may reflect a post-translational modification(s) that occurs in brain cells in vivo but not in cultured cells. We also noticed that in addition to p60FE65, the FE65M261 construct also generated smaller proteins (~45 kDa) (Fig. 1B) similar in size to FE65 proteins initiated at Methionine 327 (M327) (data not shown; see below). We have occasionally observed a band of approximately this size on Western blots of brain lysates. Collectively, the results suggest that both methionines, M261 and M327, can serve as functional translation initiation sites in vitro and perhaps in vivo.
To further confirm that p60FE65 is a product of the p97FE65 transcript, an FE65 construct (pcDNA3.1-KOFE65) was directly cloned from the full-length FE65 cDNA (minus exon 2; see supplemental information) that was reverse transcribed from p97FE65 null mouse brain RNA (Fig. 1A). All proteins expressed from this construct migrated at the same positions in parallel with those expressed from pcDNA3.1-FE65M261 (Fig. 1B), including 60 kDa, 45 kDa, and a faint band at ~37 kDa. Compared to pcDNA3.1-FE65M261, pcDNA3.1-KOFE65 generated relatively greater amounts of the ~60 kDa proteins, suggesting that the sequences upstream of M261 may positively regulate protein translation from M261. An M327L mutation in the pcDNA3.1-KOFE65 construct prevented the expression of the 45 kDa proteins (Fig. 1C), confirming the assumption that M327 is used as an alternative translation initiation site. The pcDNA3.1-KOFE65(M327L) construct was therefore used for all subsequent functional analysis requiring the expression of p60FE65, as the 60 kDa protein is the main focus of this study. These results indicate that M261 serves as a primary translation initiation site when the upstream methionines are deleted (e.g., in the case of p97FE65 null mice), or serves as an alternative translation initiation site (e.g., in the case of FE65 wild-type mice). The resulted p60FE65 lacks the N-terminal third, approximately, of the p97FE65 sequence, including the first seven amino acids of the WW domain, but retains the two PID domains (Fig. 1D).
The p60FE65 isoform weakly transactivates APP-Gal4 nuclear signaling
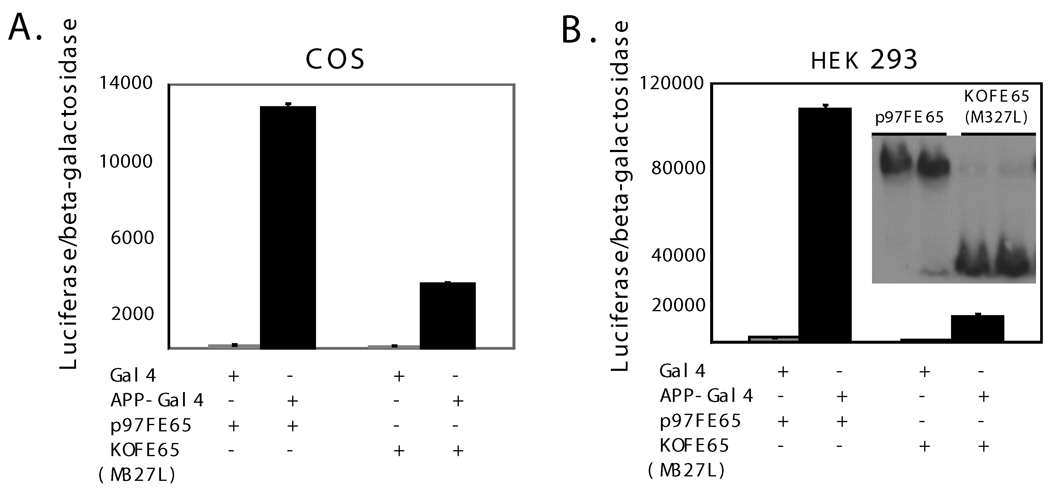
Because the WW domain is thought to be important for FE65 nuclear signaling (Cao and Sudhof, 2001), the nuclear function of p60FE65 is therefore likely to be affected. To test this hypothesis, an APP-Gal4DB reporter system (Cao and Sudhof, 2001) was used. Our previous evidence indicated that FE65 is the key transcriptional activator in FE65-Gal4DB (not requiring co-transfection of APP) or APP-Gal4DB (requiring co-transfection of FE65) reporter systems. The two systems work in a similar fashion; the APP moiety in APP-Gal4DB mainly serves as a linker to connect FE65 to Gal4DB (Yang et al. 2006). The APP-Gal4DB and luciferase reporter constructs were co-transfected with pcDNA3.1-p97FE65 or pcDNA3.1-KOFE65(M327L) in COS and HEK293 cell cultures. When compared to p97FE65, p60FE65 displayed a dramatically reduced ability (4–9 fold) to transactivate the nuclear signaling (Fig. 2, A and B), despite comparable levels of expression of p60FE65 and p97FE65 (Fig. 2B). However, p60FE65 did maintain some transactivation potential. While results varied between experiments, normalized luciferase activities were significantly higher in cells expressing p60FE65/APP-Gal4 compared to the p60FE65/Gal4 control (Fig. 2). This result is only partially consistent with the observation by Telese et al., who showed that the first 9 amino acids of the WW domain were not essential for transcriptional activity (Telese et al. 2005). The different results may be explained by the fact that these authors also incorporated extensive additional deletions at the C-terminus of FE65, including deletions of both PID1 and PID2 domains, which have been shown to be able to modulate the transactivation potential (Cao and Sudhof 2004; Telese et al. 2005; Yang et al. 2006).
Fig. 2. p60FE65 weakly transactivates APP-Gal4 nuclear signaling.
COS (A) and HEK293 (B) cells were co-transfected with Gal4 or APP-Gal4, p97FE65 or p60FE65 [KOFE65(M327L)], luciferase (a reporter gene), and β-galactosidase control, followed by a luciferase activity assay. Relative levels of luciferase activities were normalized to β–galactosidase activities in order to control for transfection efficiencies. Western blotting with antibody FE518 revealed similar levels of p97FE65 and p60FE65 [KOFE65(M327L)] expressed in the transfected cells (B, insert).
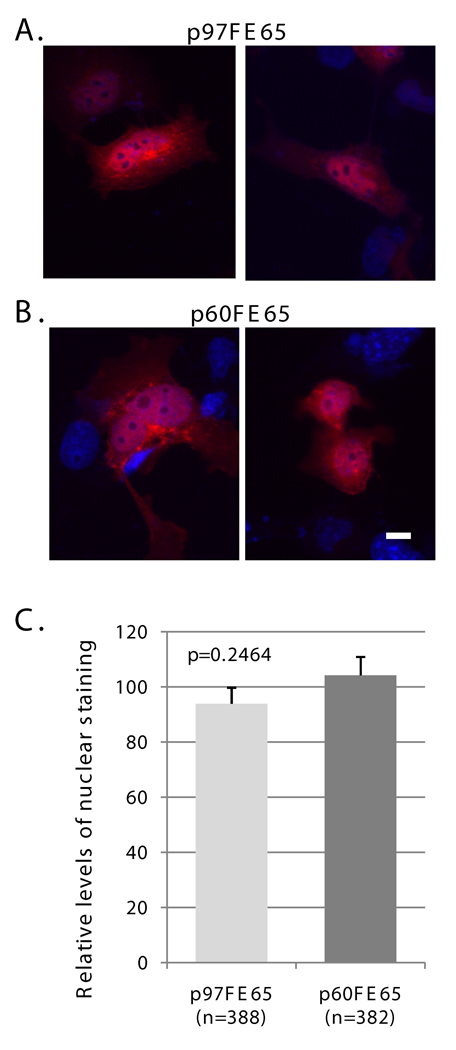
Previous evidence showed that a truncated WW domain inhibited nuclear translocation of FE65 (Minopoli et al. 2001), which could consequently impair transactivation of the reporter genes. We, therefore, compared the subcellular localizations of transfected p97FE65 and p60FE65 in COS cells by fluorescence quantitation of stained nuclei (Fig. 3). Immunostaining with the FE65-specific antibody FE518 (Hu et al. 2002) (Fig. 1D) revealed that p60FE65, expressed from pcDNA3.1-KOFE65(M327L), also localized to the nuclei of COS cells as its longer counterpart (Fig. 3, A and B). Fluorescence quantitation of stained nuclei in a large number of transfected cells revealed comparable levels of the two isoforms in the nucleus (P = 0.2464; Fig. 3C). The results suggest that the reduced nuclear signaling function found in p60FE65 is not due to the inability of this protein form to enter the nucleus, at least in cell culture models.
Fig. 3. p60FE65 is able to translocate to the nucleus.
A–B: COS cells were transfected with pcDNA3.1-p97FE65 (A) or pcDNA3.1-KOFE65(M327L) (B) and stained with antibody FE518 (red). Nuclei were visualized with DAPI (blue). C: Relative levels of nuclear p97FE65 (A) and p60FE65 (B) were determined by fluorescence quantitation of stained nuclei in p97FE65-positive cells (n =388), and p60FE65 [KOFE65(M327L)]-positive cells (n=382), respectively. Data were presented as mean ± SE, and analyzed by two-tailed t-test. The results showed that there was no significant difference in FE65 levels in the nuclei of the two groups of cells. Size bar equals 10 microns.
FE65 localization in the brain
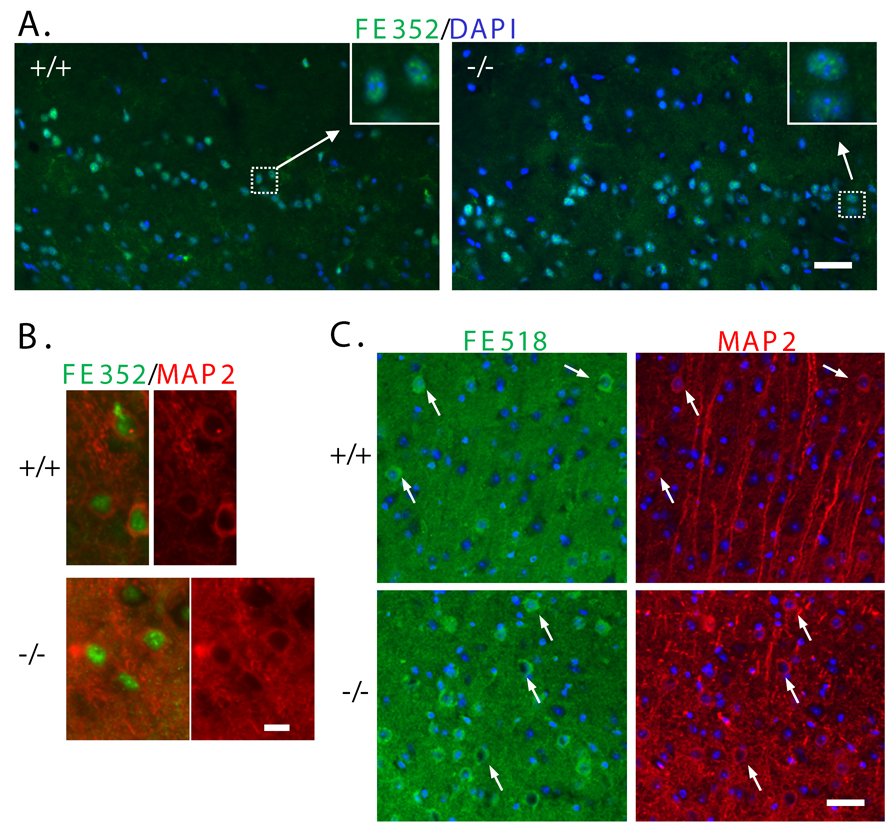
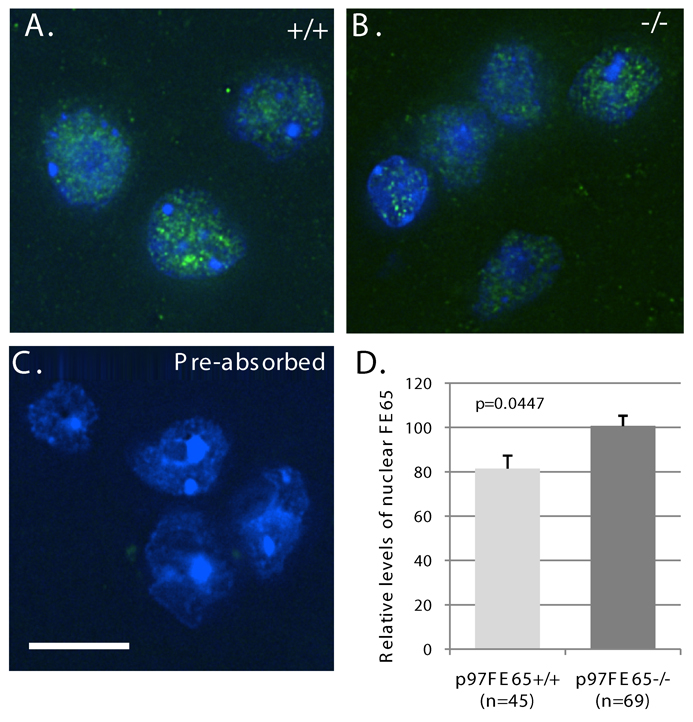
We then examined the nuclear localization of endogenous p60FE65 and p97FE65 in brain tissues. p97FE65 wild-type (+/+) and null (−/−) mouse brains were analyzed by immunohistochemistry with antibody FE352 that recognizes both p60FE65 and p97FE65 (Wang et al. 2004) (Fig. 1D). FE65-specific immunoreactivity was found in the nuclei of cerebral cortex in both p97FE65 knockout (expressing only p60FE65) and FE65 wild-type (expressing both p97FE65 and p60FE65) mice (Fig. 4A). The results indicate that a FE65-mediated nuclear signaling pathway may exist in the central nervous system. Many of the FE352-positive cells were neurons, as they were also stained positive with the neuronal marker MAP2 (Fig. 4B). The nuclear localization of FE65 was further confirmed in Z-series images collected by DeltaVision (Applied Precision, Issaquah, WA) deconvolution microscopy (Fig. 5A–B). The staining intensity was significantly diminished when sections were incubated with antibody FE352 that had been pre-absorbed with excess epitope peptides (a representative image was shown in Fig. 5C), indicating the specificity of the antibody. We noted that some of the nuclei were diffusely positive for FE65 and some were more punctuate (Fig. 5A–B), which may reflect the hetero-functional status of the brain cells at the moment of the sample collected. The green dots outside the nuclei may indicate those FE65 located in the cytoplasm. We determined relative levels of nuclear FE65 in the deconvolved Z-sections by fluorescence quantitation. Levels of FE65 immunoreactivity in cortical nuclei of p97FE65 null mouse brains were significantly higher than those in p97FE65 wild-type (P = 0.0447; Fig. 5D). The increase in nuclear FE65 may reflect the up-regulated expression of p60FE65 found in p97FE65 null mouse brains (Wang et al. 2004). Consistent with the results from cell cultures (Fig. 3), the results indicate that endogenous p60FE65 is also able to enter the nucleus in brain cells.
Fig. 4. Immunohistochemical analysis of FE65 localization in p97FE65 wild-type (+/+) and null (−/−) cerebral cortex.
A: Frozen sagittal brain sections were stained with the FE65-specific antibody FE352 (green). Nuclei were counterstained with DAPI (blue). B: Co-staining with antibodies FE352 (green) and neuronal marker MAP2 (red). C: Frozen sagittal sections were co-stained with the FE65-specific antibody FE518 (green) and the neuronal marker MAP2 (red). Nuclei were visualized with DAPI (blue). The MAP2+ neurons show cytoplasmic immunoreactivity for FE65. Arrows are included for reference. Size bars in A and C equal 50 microns. Size bar in B equals 10 microns.
Fig. 5. FE65 nuclear localization in cerebral cortices was confirmed via DeltaVision deconvolution microscopy.
A–B. Frozen brain sections from p97FE65 wild type (+/+) (A) and p97FE65 null (−/−) (B) mice were stained with the FE65-specific antibody FE352. Representative images were shown. Note the co-localization of FE65 (green) with the DNA-binding dye DAPI (blue). C. Nuclear immunoreactivity was not evident when brain sections of p97FE65 null and p97FE65 wild type (not shown) mice were stained with FE352 that had been pre-absorbed with 200x molar excess of an epitope peptide. The results show that the antibody is specific for the immunogenic peptide. Blue is DNA stained with DAPI; brighter blue areas are nucleoli. D. Relative levels of nuclear FE65 staining in randomly selected cells in p97FE65 wild type (+/+) and p97FE65 null (−/−) mouse cortexes were determined by fluorescence quantitation. All images and image analysis represent deconvolved Z-sections. Data were presented as mean ± SE, and analyzed by two-tailed t-test. Size bar is 10 microns.
Inconsistent with previously published results using different FE65 antibodies (Kesavapany et al. 2002; Bao et al. 2007), nuclear FE65, as detected by antibody FE352, was not readily apparent in most neurons of the hippocampus. Little FE65 reactivity was evident within the neurons of the CA1–3 and dentate gyrus regions. In contrast, cytoplasmic staining was consistently detected in smaller cells (probably glia) of the stratum radiatum and stratum oriens (data not shown).
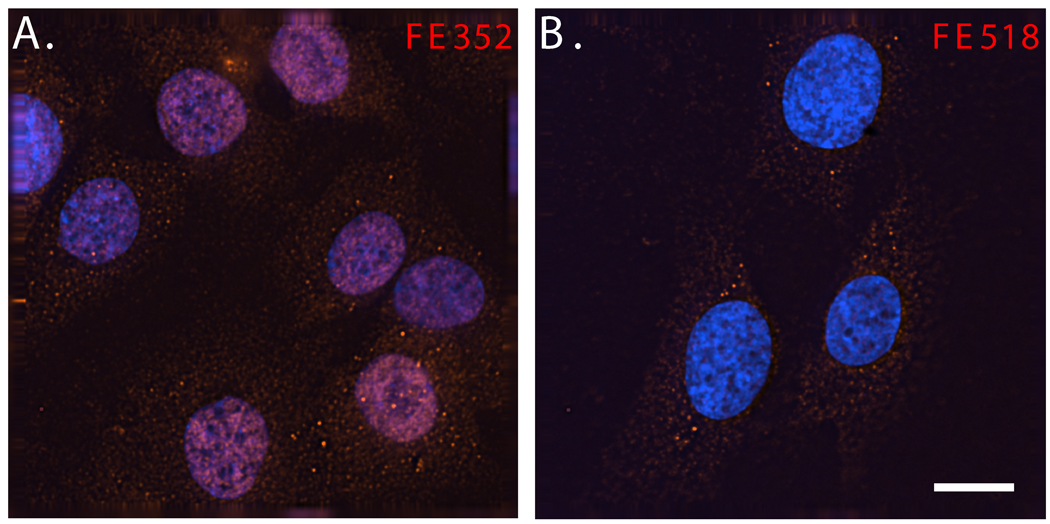
Interestingly, FE518, another FE65-specific antibody that also recognizes both p60FE65 and p97FE65, gave different immunohistochemical results. We observed FE518 immunoreactivity within the cytoplasm of neurons of the cortex (Fig. 4C), and other brain regions including the hippocampus (data not shown), and of cultured neuroblastoma cells, SK-N-SH (Fig. 6). Despite different staining patterns between the two FE65-specific antibodies, again, comparable subcellular localizations of FE65 were observed in both p97FE65 wild-type and null mice. The distinct FE352 and FE518 staining results might reflect the existence of different conformational states of FE65 within the nuclei and cytoplasm of neurons, which are preferably recognized by different FE65 antibodies. The lack of FE352 nuclear staining within the hippocampi of laboratory mice, at least those not stimulated with learning/memory-relevant paradigms, suggests that the FE65 nuclear signaling pathway might be tightly regulated in this brain region.
Fig. 6. FE65-specific antibodies FE352 and FE518 reveal endogenous FE65 localizations of SK-N-SH neuroblastoma cells via DeltaVision deconvolution microscopy.
SK-N-SH cells were stained with antibody FE352 (A) or FE518 (B). Nuclei were visualized with DAPI (blue). Size bar equals 10 microns.
DISCUSSION
We have described a novel 60 kDa protein, p60FE65, which is expressed in the brains of wild type mice and whose expression is up-regulated in p97FE65 null mice (Wang et al. 2004). This protein is detected in Western blots by several FE65-specific antibodies that recognize epitopes located in the middle or the C-terminal region of FE65. However, an antibody that is specific for a more N-terminal epitope does not recognize the protein. We propose that p60FE65 is translated from an alternative downstream methionine on the p97FE65 transcript. In this paper, we show evidence that M261 is the translation initiation site of p60FE65. The isoform lacks the N-terminal residues 1–260 in p97FE65, including the N-terminus of the WW domain along with one of two signature tryptophan residues (W260), which may participate in the stabilization of the folding characteristic of this domains (Meiyappan et al. 2007). As a result, p60FE65 likely fails to efficiently bind proteins that interact with the WW domain in p97FE65, and to execute those biological functions that require the N-terminal sequences.
A transcriptional function has been proposed for the FE65-centered protein network (Cao and Sudhof 2001 and 2004; Perkinton et al. 2004; Hass and Yankner 2005; Telese et al. 2005; Yang et al. 2006; Kim et al. 2007). FE65 is capable of transactivating Gal4-mediated transcription, both when directly fused to the Gal4 DNA binding domain (Yang et al. 2006) and when bridged to Gal4 via its interaction with an APP C-terminal moiety of APP-Gal4 fusion proteins (Cao and Sudhof, 2001). Thus, FE65 is believed to play a central role in a transcription-activation complex. While APP has been considered as another central player in this transcription complex, recent evidence challenges this assertion. Results from our lab show that both co-expressions with APP and knockdown of APP with siRNA has little effect on the magnitude of reporter gene expression driven by FE65-Gal4 (Yang et al. 2006). Thus, in the APP-Gal4-mediated assays, we believe that APP simply acts as a linker that allows FE65 to drive Gal4-dependent reporter expression.
Because the WW domain has been shown to be important for the role of FE65 in the activation of transcription (Cao and Sudhof 2001 and 2004), we hypothesized that p60FE65 might be deficient in nuclear signaling. Indeed, in this study, we show that p60FE65 has an attenuated ability to activate transcription in the Gal4 reporter assay—only 11–25% of activity is preserved, when compared to p97FE65 (Fig. 2). The observations suggest that intact FE65 nuclear signaling may be important for maintaining a normal learning/memory process, because p97FE65 isoform-specific KO mice exhibit marked learning/memory deficits (Wang et al. 2004). In these KO mice, up-regulated expression of p60FE65 was found; however, the up-regulation was apparently unable to rescue the phenotype of memory deficiency. Although we are unclear what other functions are also lost or gained in this short form of FE65, the reduced nuclear signaling activity may be, at least, partially relevant to the abnormal memory processing.
Although the WW domain was thought to be important for the ability of FE65 to enter the nucleus (Minopoli et al. 2001), our evidence shows that p60FE65 is present in the nucleus at levels comparable to those of p97FE65-transfected cells. These results indicate that p60FE65 contains sufficient structural information for moving into the nucleus; alternatively, the amino acid residues 1–260 in p97FE65 are dispensable for this particular function. In addition, our results also suggest that the attenuated ability of p60FE65 in gene transactivation is resulted from its intrinsic inability to trigger those nuclear events required for the activity.
The conclusion is further supported by the immunohistochemical results on brain sections from p97FE65 wild type and null mice. Antibody FE352 revealed nuclear FE65 staining in neurons of both genotypes. Quantitative analysis showed that higher levels of nuclear FE65 immunoreactivity were found in p97FE65 null mouse brains than those in p97FE65 wild type brains, which is consistent with the evidence of the up-regulated expression of p60FE65 in the null mice. In contrast to antibody FE352, another FE65-specific antibody, FE518, showed predominantly cytoplasmic staining in neurons of brain sections of both genotypes, and in non-transfected SK-N-SH cells (Fig. 6). Other laboratories have also reported either nuclear or cytoplasmic immunoreactivity for endogenous FE65 in neurons of the brain using antibodies not described in this paper (Kesavapany et al. 2002; Bao et al. 2007). The discrepancies may be explained by the availability of the different FE65 epitopes in different cellular compartments, likely due to the conformation changes of FE65, and/or the occupancy by the FE65-binding proteins.
In conclusion, based on our Gal4 assay results in two different cell lines, we argue that nuclear p60FE65 is unable to function as efficiently as nuclear p97FE65 in neurons, in vivo, and feel that we have the beginnings of an argument for how lost p97FE65 expression might contribute to the impaired learning/memory evident in p97FE65 null mice. Importantly, levels of p97FE65 (but not p60FE65) were reduced in brain during aging (Wang et al. 2004). It will be interesting to determine whether the reduced expression, presumably resulting in attenuated FE65 nuclear signaling, might partially contribute to declining learning and memory during normal aging. While we were unable to detect nuclear FE65 within hippocampal neurons using the antibodies described in this study, others have done so using different FE65 antibodies (Bao et al. 2007). Thus, it is possible that the FE65 nuclear function is highly regulated in this brain region. Specific stimuli, such as activation of synaptic activity in response to a learning/memory paradigm, might induce the necessary translocation and/or conformational change that makes FE65 available for executing a function in the nucleus as well as visible in this compartment via immunohistochemistry. Future work to further validate this hypothesis should also include the identification of functional targets regulated by the FE65 nuclear signaling pathway, and the examination of their effects on learning/memory related functions.
Supplementary Material
1. cDNA sequences of the isolated transcript from the p97FE65 isoform specific knockout mice
ACKNOWLEDGEMENTS
This study was supported, in part, by R01 AG19711 and T32 AG00057 from the National Institute on Aging.
List of abbreviations
- DAT
Dementias of the Alzheimer type
- APP
Beta-amyloid precursor protein
- AICD
APP intracellular domain
- PID
Phosphotyrosine interaction domain
- Map2
Microtubule associated protein 2
- Gal4DB
DNA binding domain of transcription factor Gal4
REFERENCES
- 1.Bao J, Cao C, Zhang X, Jiang F, Nicosia SV, Bai W. Suppression of beta-amyloid precursor protein signaling into the nucleus by estrogens mediated through complex formation between the estrogen receptor and Fe65. Mol Cell Biol. 2007;27:1321–1333. doi: 10.1128/MCB.01280-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cao X, Sudhof TC. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 3.Cao X, Sudhof TC. Dissection of amyloid-beta precursor protein-dependent transcriptional transactivation. J Biol Chem. 2004;279:24601–24611. doi: 10.1074/jbc.M402248200. [DOI] [PubMed] [Google Scholar]
- 4.Ermekova KS, Zambrano N, Linn H, Minopoli G, Gertler F, Russo T, Sudol M. The WW domain of neural protein FE65 interacts with proline-rich motifs in Mena, the mammalian homolog of Drosophila enabled. J Biol Chem. 1997;272:32869–32877. doi: 10.1074/jbc.272.52.32869. [DOI] [PubMed] [Google Scholar]
- 5.Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Sitaraman S, Carlson E, Sagi SA, Chevallier N, Jin K, Greenberg DA, Bredesen DE. Reversal of Alzheimer's-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acad Sci U S A. 2006;103:7130–7135. doi: 10.1073/pnas.0509695103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guenette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. Embo J. 2006;25:420–431. doi: 10.1038/sj.emboj.7600926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hardy J. Amyloid, the presenilins and Alzheimer's disease. Trends Neurosci. 1997;20:154–159. doi: 10.1016/s0166-2236(96)01030-2. [DOI] [PubMed] [Google Scholar]
- 8.Hass MR, Yankner BA. A {gamma}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J Biol Chem. 2005;280:36895–36904. doi: 10.1074/jbc.M502861200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Muller U, De Strooper B. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep. 2006;7:739–745. doi: 10.1038/sj.embor.7400704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu Q, Hearn MG, Jin LW, Bressler SL, Martin GM. Alternatively spliced isoforms of FE65 serve as neuron-specific and non-neuronal markers. J Neurosci Res. 1999;58:632–640. doi: 10.1002/(sici)1097-4547(19991201)58:5<632::aid-jnr4>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 11.Hu Q, Cool BH, Wang B, Hearn MG, Martin GM. A candidate molecular mechanism for the association of an intronic polymorphism of FE65 with resistance to very late onset dementia of the Alzheimer type. Hum Mol Genet. 2002;11:465–475. doi: 10.1093/hmg/11.4.465. [DOI] [PubMed] [Google Scholar]
- 12.Hu Q, Wang L, Yang Z, Cool BH, Zitnik G, Martin GM. Endoproteolytic cleavage of FE65 converts the adaptor protein to a potent suppressor of the sAPPalpha pathway in primates. J Biol Chem. 2005;280:12548–12558. doi: 10.1074/jbc.M411855200. [DOI] [PubMed] [Google Scholar]
- 13.Hu Q, Kukull WA, Bressler SL, Gray MD, Cam JA, Larson EB, Martin GM, Deeb SS. The human FE65 gene: genomic structure and an intronic biallelic polymorphism associated with sporadic dementia of the Alzheimer type. Hum Genet. 1998;103:295–303. doi: 10.1007/s004390050820. [DOI] [PubMed] [Google Scholar]
- 14.Ikin AF, Sabo SL, Lanier LM, Buxbaum JD. A macromolecular complex involving the amyloid precursor protein (APP) and the cytosolic adapter FE65 is a negative regulator of axon branching. Mol Cell Neurosci. 2007;35:57–63. doi: 10.1016/j.mcn.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kesavapany S, Banner SJ, Lau KF, Shaw CE, Miller CC, Cooper JD, McLoughlin DM. Expression of the Fe65 adapter protein in adult and developing mouse brain. Neuroscience. 2002;115:951–960. doi: 10.1016/s0306-4522(02)00422-0. [DOI] [PubMed] [Google Scholar]
- 16.Kim SY, Kim MY, Mo JS, Park HS. Notch1 intracellular domain suppresses APP intracellular domain-Tip60-Fe65 complex mediated signaling through physical interaction. Biochim Biophys Acta. 2007;1773:736–746. doi: 10.1016/j.bbamcr.2007.02.001. [DOI] [PubMed] [Google Scholar]
- 17.Lambert JC, Mann D, Goumidi L, Harris J, Pasquier F, Frigard B, Cottel D, Lendon C, Iwatsubo T, Amouyel P, Chartier-Harlin MC. A FE65 polymorphism associated with risk of developing sporadic late-onset alzheimer's disease but not with Abeta loading in brains. Neurosci Lett. 2000;293:29–32. doi: 10.1016/s0304-3940(00)01477-4. [DOI] [PubMed] [Google Scholar]
- 18.Meiyappan M, Birrane G, Ladias JA. Structural Basis for Polyproline Recognition by the FE65 WW Domain. J Mol Biol. 2007 doi: 10.1016/j.jmb.2007.06.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minopoli G, de Candia P, Bonetti A, Faraonio R, Zambrano N, Russo T. The beta-amyloid precursor protein functions as a cytosolic anchoring site that prevents Fe65 nuclear translocation. J Biol Chem. 2001;276:6545–6550. doi: 10.1074/jbc.M007340200. [DOI] [PubMed] [Google Scholar]
- 20.Neve RL. Alzheimer's disease sends the wrong signals--a perspective. Amyloid. 2008;15:1–4. doi: 10.1080/13506120701814608. [DOI] [PubMed] [Google Scholar]
- 21.Perkinton MS, Standen CL, Lau KF, Kesavapany S, Byers HL, Ward M, McLoughlin DM, Miller CC. The c-Abl tyrosine kinase phosphorylates the Fe65 adaptor protein to stimulate Fe65/amyloid precursor protein nuclear signaling. J Biol Chem. 2004;279:22084–22091. doi: 10.1074/jbc.M311479200. [DOI] [PubMed] [Google Scholar]
- 22.Pietrzik CU, Yoon IS, Jaeger S, Busse T, Weggen S, Koo EH. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci. 2004;24:4259–4265. doi: 10.1523/JNEUROSCI.5451-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price DL, Sisodia SS. Mutant genes in familial Alzheimer's disease and transgenic models. Annu Rev Neurosci. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- 24.Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The Alzheimer amyloid precursor protein (APP) and FE65, an APP-binding protein, regulate cell movement. J Cell Biol. 2001;153:1403–1414. doi: 10.1083/jcb.153.7.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sabo SL, Ikin AF, Buxbaum JD, Greengard P. The amyloid precursor protein and its regulatory protein, FE65, in growth cones and synapses in vitro and in vivo. J Neurosci. 2003;23:5407–5415. doi: 10.1523/JNEUROSCI.23-13-05407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sabo SL, Lanier LM, Ikin AF, Khorkova O, Sahasrabudhe S, Greengard P, Buxbaum JD. Regulation of beta-amyloid secretion by FE65, an amyloid protein precursor-binding protein. J Biol Chem. 1999;274:7952–7957. doi: 10.1074/jbc.274.12.7952. [DOI] [PubMed] [Google Scholar]
- 27.Saganich MJ, Schroeder BE, Galvan V, Bredesen DE, Koo EH, Heinemann SF. Deficits in synaptic transmission and learning in amyloid precursor protein (APP) transgenic mice require C-terminal cleavage of APP. J Neurosci. 2006;26:13428–13436. doi: 10.1523/JNEUROSCI.4180-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schroeter EH, Kisslinger JA, Kopan R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature. 1998;393:382–386. doi: 10.1038/30756. [DOI] [PubMed] [Google Scholar]
- 29.Struhl G, Adachi A. Nuclear access and action of notch in vivo. Cell. 1998;93:649–660. doi: 10.1016/s0092-8674(00)81193-9. [DOI] [PubMed] [Google Scholar]
- 30.Telese F, Bruni P, Donizetti A, Gianni D, D'Ambrosio C, Scaloni A, Zambrano N, Rosenfeld MG, Russo T. Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 2005;6:77–82. doi: 10.1038/sj.embor.7400309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trommsdorff M, Borg JP, Margolis B, Herz J. Interaction of cytosolic adaptor proteins with neuronal apolipoprotein E receptors and the amyloid precursor protein. J Biol Chem. 1998;273:33556–33560. doi: 10.1074/jbc.273.50.33556. [DOI] [PubMed] [Google Scholar]
- 32.Turner PR, O'Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70:1–32. doi: 10.1016/s0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 33.Wang B, Hu Q, Hearn MG, Shimizu K, Ware CB, Liggitt DH, Jin LW, Cool BH, Storm DR, Martin GM. Isoform-specific knockout of FE65 leads to impaired learning and memory. J Neurosci Res. 2004;75:12–24. doi: 10.1002/jnr.10834. [DOI] [PubMed] [Google Scholar]
- 34.Yang Z, Cool BH, Martin GM, Hu Q. A dominant role for FE65 (APBB1) in nuclear signaling. J Biol Chem. 2006;281:4207–4214. doi: 10.1074/jbc.M508445200. [DOI] [PubMed] [Google Scholar]
- 35.Zambrano N, Minopoli G, de Candia P, Russo T. The Fe65 adaptor protein interacts through its PID1 domain with the transcription factor CP2/LSF/LBP1. J Biol Chem. 1998;273:20128–20133. doi: 10.1074/jbc.273.32.20128. [DOI] [PubMed] [Google Scholar]
- 36.Zambrano N, Bruni P, Minopoli G, Mosca R, Molino D, Russo C, Schettini G, Sudol M, Russo T. The beta-amyloid precursor protein APP is tyrosine-phosphorylated in cells expressing a constitutively active form of the Abl protoncogene. J Biol Chem. 2001;276:19787–19792. doi: 10.1074/jbc.M100792200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. cDNA sequences of the isolated transcript from the p97FE65 isoform specific knockout mice