Abstract
Blood-brain barrier (BBB) membrane proteins play crucial roles in the proper functioning of the BBB as well as in disease progression. Previously we developed a novel approach for identifying membrane proteins expressed at the BBB, which we referred to as Multiplex Expression Cloning (MEC). In the current study, the proteome coverage of the MEC approach was expanded to allow the identification of a total of 30 BBB membrane proteins that are diverse in function and abundance. To unveil those membrane proteins that are enriched at the BBB and hence partially responsible for some of its unique characteristics, the transcript abundance levels for all 30 BBB membrane proteins were compared to those found in microvessels derived from lung, liver, heart and kidney. Such quantitative PCR profiling (qPCR) of RNA samples from laser capture microdissected microvessels revealed that the transcripts for 5 membrane proteins, namely Lutheran glycoprotein, carbonic anhydrase IV, uncoupling protein 2 (UCP2), podocalyxin and solute carrier family 38, member 5 (SLC38A5) were BBB selective in that expression was elevated in brain microvessels when compared to all of the vascular beds tested. Many other membrane protein transcripts, while not as BBB-restricted, showed selective expression within subsets of tissues indicating other potential parallels and contrasts between vascular beds in the body. The identification of BBB membrane proteins could help better understand the molecular mechanisms responsible for BBB function and those with selective expression may have utility for BBB-targeted therapies.
Keywords: Blood-brain barrier, Multiplex expression cloning, membrane proteomics, laser capture microdissection, quantitative PCR
INTRODUCTION
Membrane proteins play a significant role in defining several functional aspects of the blood-brain barrier (BBB), ranging from its characteristic tight junctions that prevent paracellular transport of molecules to the highly specialized transport systems that regulate the exchange of nutrients and signaling agents across the BBB. Since the BBB only comprises 1/1000 the volume of the brain (Pardridge 2007), proteomic studies aimed at membrane proteomics of the whole brain, such as the ones described by Nielsen et al. (Nielsen et al. 2005) and Wang et al. (Wang et al. 2006), although quite comprehensive, typically underrepresent the BBB membrane proteome. To date, there are very few studies that have directly attempted moderate to high throughput analysis of the BBB proteome. Haqqani et al. (Haqqani et al. 2007, Haqqani et al. 2005) combined laser capture microdissection (LCM) isolation of brain microvessels with quantitative mass spectrometry (MS) to directly characterize the expression levels of 57 and 160 BBB proteins that were differentially expressed in blood vessels subjected to cerebral ischemia. More recently, the identification of 881 different proteins expressed at the mouse brain microvasculature was enabled by MS analysis of lysates obtained from brain microvessels selectively isolated by the use of LCM (Lu et al. 2008). In addition, absolute quantification of 34 known membrane resident transporters at the BBB has been performed using MS combined with isotope-labeled peptide standards (Kamiie et al. 2008). While these studies have broken ground in terms of BBB proteomics, they were not focused primarily on membrane proteins or as in the latter example, focused only on known BBB transporters. Moreover, use of the aforementioned MS-based methods tends to underrepresent membrane proteins as a result of their relative insolubility and / or low abundance levels in vivo (Grant & Wu 2007).
To complement these existing studies and MS-based proteomic techniques, we recently described a novel proteomics approach that is capable of sampling both highly hydrophobic and low abundance membrane proteins (Agarwal & Shusta 2009). Briefly, the approach referred to as Multiplex Expression Cloning (MEC) involves the expression of a BBB cDNA library in a mammalian host cell line where subsequently, with the aid of a BBB membrane protein specific polyclonal antiserum (BMSPA), BBB-resident membrane proteins can be identified. In this study, we have expanded the coverage of the MEC procedure by several technical adaptations leading to the identification of many more BBB membrane proteins. Although important, the identification of these proteins merely serves as a first step towards their characterization. In particular, the expression profile of these proteins in tissues and vascular beds other than the brain could be highly useful in understanding their significance at the BBB during health and disease. Thus, the transcript expression levels of BBB membrane proteins identified using the MEC approach were measured across several vascular beds by combining MEC with LCM and quantitative PCR. In this way, it was discovered that five membrane protein encoding transcripts, Lutheran glycoprotein, carbonic anhydrase IV, uncoupling protein 2 (UCP2), podocalyxin and solute carrier family 38, member 5 (SLC38A5), were selectively expressed at the BBB.
MATERIALS AND METHODS
Cell culture
The HEK293 T/17 (human embryonic kidney cell line) cell line was obtained from the American Type Culture Collection (Manassas, VA) and is a clone of the parent HEK293 cell line that has been stably transfected with the SV40 large T-antigen and hence supports efficient episomal replication of SV40-origin containing plasmids. HEK293 T/17 cells were cultured in a humidified incubator at 37°C with 5% carbon dioxide using DMEM media (Gibco, Invitrogen, Carlsbad, CA) buffered with 2.0 g/l sodium bicarbonate and 7.2 g/l HEPES to pH 7.2 and supplemented with 4.5 g/l dextrose and 10% fetal bovine serum (Gibco, Invitrogen).
Multiplex expression cloning
The generation of the BBB cDNA library and the BBB membrane protein-specific polyclonal antiserum (BMSPA) has been described previously (Agarwal & Shusta 2009, Shusta et al. 2002b, Pardridge et al. 1990). Enrichment of the BBB cDNA library using the MEC approach was carried out as described previously (Agarwal & Shusta 2009), except for the following modifications (Figure 1). The clone recovery process after FACS isolation was greatly improved by switching the host cell line from HEK293 to HEK293 T/17. Due to constitutive expression of the SV40 large T-antigen, the HEK293 T/17 cells allow the SV40 origin containing plasmids from the BBB cDNA library to replicate to greater numbers thus facilitating their recovery from cells. Additionally, the transfected cells were permeablized by the use of 0.5% saponin in PBS prior to immunolabeling to allow the BMSPA to access intracellular components of the cell membrane (Figure 1, path 2). The preparation and sequencing of the cDNA pools and individual cDNA clones was conducted as described previously (Agarwal & Shusta 2009). The BMSPA was incubated with an untransfected HEK293 T/17 cell monolayer at 4°C for 1 hour to remove any background antigenicity prior to immunolabeling.
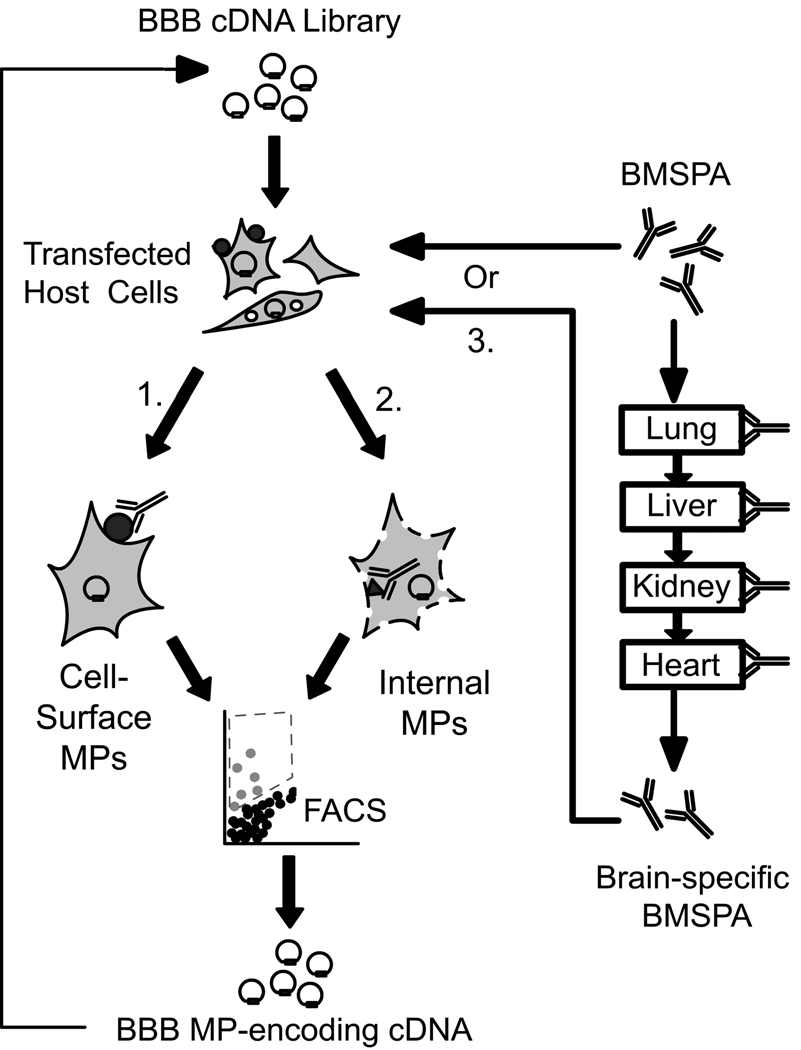
Figure 1.
Schematic of multiplex expression cloning approaches. Host HEK293 T/17 cells are transfected with the BBB cDNA library and then probed with either the unsubtracted BMSPA or the ‘brain-specific’ BMSPA to identify extracellular (1. Unpermeablized cells; antibody cannot access interior cellular compartments) and intracellular (2. Saponin permeablized cells allowing intracellular access to the antibody) BBB membrane protein epitopes. The ‘brain-specific’ BMSPA (3.) was generated by serial subtraction against acetone powders of lung, liver, kidney and heart as shown to the right and used with saponin permeabilized cells (path 2.). The target cells are isolated via fluorescence activated cell sorting (FACS) and the plasmid they harbor is recovered. The process is repeated until the pool is sufficiently enriched for cDNA that encode BBB membrane proteins (usually 4 rounds).
Generation of tissue acetone powders, BMSPA subtraction and Western blotting
Fresh bovine lung, liver, kidney and heart tissue were obtained from a local meat processor. Tissues were cut into small pieces which were then homogenized using a Polytron tissue homogenizer (PowerGen 125, Fisher Scientific, Pittsburgh, PA). Ten volumes of ice cold acetone were added to the homogenized tissue and the precipitate was collected by filtering through a Whatman filter paper (Fisher Scientific). The semi-dry precipitate was blended again with the same volume of ice cold acetone used in the previous step and filtered again. The precipitate formed a cake which was pulverized with a mortar and pestle, spread out on a surface and left to dry overnight at room temperature. The dried powder was further pulverized the next day and stored at −20°C. The BMSPA was subtracted by the acetone powders by incubating it at a dilution of 1:150 in 10 mL PBSG (40 % goat serum in 0.01 M PBS, pH 7.4) with 0.5 g of acetone powder of the appropriate tissue for 1 hour at 37°C in a shaker. For subtraction by several tissues, this process was serially repeated for each individual tissue powder. Finally the subtracted BMSPA was filter sterilized and stored at 4°C for later use. Western blotting under reducing conditions was performed as described previously (Shusta et al. 2002b).
Protein expression profiling using tissue-subtracted BMSPA antisera
HEK293 T/17 cells were transfected with individual membrane encoding cDNA clones using Lipofectamine 2000 per the manufacturer’s protocol (Invitrogen). The transfection mix was replaced with fresh media 24 hours after the transfection step. At 60 hours post-transfection, the culture medium was aspirated and the transfected monolayers were washed once with PBSF (0.01 M PBS, pH 7.4 supplemented with 10% fetal bovine serum). The cells were then blocked for 30 minutes with PBSG at 4°C, washed once in PBSF and incubated for 1 hour at 4°C in HEK293 T/17-subtracted BMSPA or tissue-subtracted BMSPA, as appropriate, diluted 1:500 in PBSG. The cells were then washed twice in PBSF, incubated for 30 minutes in HEK293 T/17-depleted phycoerythrin (PE) conjugated anti-rabbit IgG (Sigma Aldrich) diluted 1:200 in PBSG at 4°C and then washed twice with PBSF. The cells were subsequently detached from the plates via gentle pipetting and subjected to flow cytometric analyses. Only live cells capable of excluding the nuclear stain, propidium iodide, were analyzed. A BD FACSCalibur instrument equipped with a 488 nm laser for excitation and a 670 LP filter for detection of Propidium Iodide and a 585/42 nm filter for detection of the PE signal was used for flow cytometry analyses. The mean fluorescence intensity of the immunolabeled cells was measured.
Preparation of tissue sections
In order to preserve the integrity of mRNA, tissue specimens from bovine brain, lung, liver, kidney and heart were freshly isolated and immediately frozen by embedding in a cryomold using Tissue-Tek O.C.T Compound (Sakura Finetek, Zoeterwoude, The Netherlands), which was subsequently floated in a bath of isopentane (Sigma Aldrich, St. Louis, MO) cooled by liquid nitrogen. The tissue sections were handled using RNase free techniques. Seven micron thick tissue sections were generated using a Microm HM505E (Microm GmbH, Walldorf, Germany) and placed onto MMi cell cut slides (Molecular machines & industries inc., Rockledge, FL) in an RNase free environment. The slides were stored at −70°C until LCM was performed.
Laser capture microdissection of blood vessels and RNA recovery
Tissue sections were removed from −70°C storage and thawed at room temperature for 1 minute. All subsequent labeling steps were conducted in RNase free conditions using RNase free reagents at room temperature. The tissue section was fixed in 100% ethanol for 1 minute, air dried for 3 minutes, incubated in 50 µg/ml of fluorescein-labeled Griffonia simplicifolia agglutinin-I-B4 (Vector Laboratories, Burlingame, CA) for 10 minutes, washed three times in RNase free water, and dried for 5 minutes in a dessicator. The sample was then dehydrated by two 30 second incubations in 95% ethanol followed by another two in 100% ethanol and finally a 3 minute incubation in 100% isopropyl alcohol. The tissue section was air dried for 2 minutes and then placed in a dessicator for 10 minutes to remove residual moisture. The lectin labeled blood vessels were cut out from the sections and captured on an adhesive cap using the MMI Cell Cut instrument (Molecular Machines & Industries Inc.). The isolation was restricted to microvessels (~10 microns or less in size) while large vessels as well as other agglomerates such as the glomeruli in the kidney were avoided to prevent enrichment of other cell types such as epithelial cells. Each cap could accommodate 200–300 blood vessels and a minimum of 3 caps were obtained for each tissue in order to have a sufficient amount of RNA. Total RNA was extracted from the microdissected blood vessels with the RNeasy micro kit (Qiagen, Valencia, CA) using the manufacturer’s protocol for purification of total RNA from microdissected cryosections. Carrier RNA was not used to avoid issues related to RNA amplification. RNA was also extracted from whole tissue sections by simply dissolving the section in the lysis buffer and following the same protocol used for the microdissected samples. Additionally, the RNA samples were subjected to a DNase I digestion during the extraction protocol to remove any contaminating genomic DNA. The recovered RNA was subsequently amplified using the Quantitect whole transcriptome kit (Qiagen) as per the manufacturer’s recommendation for a high yield reaction (8 hours of amplification time). This kit utilizes the multiple displacement amplification technology and yields amplified cDNA. After amplification, the cDNA yields were about 0.8–1 µg/µl based on optical density readings at 260 nm.
Quantitative PCR
Quantitative PCR was carried out using a Bio-Rad iCycler thermocycler (Bio-Rad, Hercules, CA) and the Quantitect SYBR Green PCR kit (Qiagen) following the manufacturer’s protocol. 1–2 ng of cDNA (1:250 dilution of amplified samples) was used as the template per reaction. Gene-specific primers were designed using PerlPrimer v1.1.14 (http://perlprimer.sourceforge.net/) and the oligo design and analysis tools on the website of Integrated DNA technologies (http://www.idtdna.com). The primer sequences are listed in Supplemental Table 1. Whenever possible, the primers were designed to span intron-exon boundaries. Gene expression levels were measured by calculating the delta-delta cycle threshold values (ΔΔCt), which represent the difference in gene expression levels in terms of PCR cycles. Ct represents the PCR cycle at which the fluorescence from the PCR product reaches a defined threshold value, which was selected at a point where the amplification process was exponential in nature. ΔCt represents normalization of the gene expression to a control gene such as β-actin. Thus ΔCt = Ct,β-actin − Ct,gene. ΔΔCt represents the normalized difference in gene expression levels between the sample and a control (ΔΔCt = ΔCt, sample − ΔCt,control). Two genes were initially tested as the reference gene, namely β-actin and the 60S acidic large ribosomal protein (RPLP0) and it was found that the ΔΔCt values obtained within and across tissues were similar for both these genes. Hence all of the remaining ΔΔCt values were calculated using just the β-actin gene as the reference. Approximate fold differences in Table 2 are reported as 2ΔΔCt.
Table 2.
Gene expression profiles of BBB versus other microvasculatures.
| Gene | Lung | Liver | Kidney | Heart |
|---|---|---|---|---|
| Lutherana | 4.4 ± 1.0 (21) | 4.0 ± 1.0 (16) | 3.7 ± 1.1 (13) | 1.9 ± 1.0 (4) |
| CAIVa | 6.7 ± 1.7 (104) | 4.7 ± 1.4 (26) | 4.9 ± 1.6 (30) | 8.4 ± 1.2 (338) |
| CD46 | 4.5 ± 1.9 (23) | 2.2 ± 1.1 (5) | N.D. | 2.5 ± 1.2 (6) |
| RACK1 | N.D. | −1.5 ± 1.3 (−3) | N.D. | −2.2 ± 1.8 (−5) |
| ITM2B | 3.5 ± 1.5 (11) | N.D. | 1.6 ± 1.2 (3) | N.D. |
| MHCI | N.D. | N.D. | 1.7 ± 1.6 (3) | N.D. |
| UCP2 | 2.0 ± 1.8 (4) | 4.7 ± 1.4 (26) | 7.3 ± 1.8 (158) | 5.4 ± 1.5 (42) |
| LAMP2 | 3.5 ± 1.4 (11) | −1.7 ± 1.2 (−3) | 1.8 ± 1.3 (3) | N.D. |
| FIH1-like | 2.4 ± 1.6 (5) | N.D. | N.D. | −4.1 ± 1.1 (−17) |
| RPLP1 | 2.8 ± 1.3 (7) | −2.8 ± 1.2 (−7) | N.D. | −3.3 ± 1.4 (−10) |
| ARHGAP21 | 4.8 ± 1.6 (28) | 2.6 ± 1.2 (6) | N.D. | N.D. |
| SCP2 | 3.5 ± 1.0 (11) | −6.6 ± 1.1 (−97) | −1.3 ± 1.2 (−2) | −4.9 ± 1.0 (−30) |
| LOC613429 | 4.0 ± 1.1 (16) | N.D. | 2.3 ± 1.1 (5) | N.D. |
| PODXLa | 3.4 ± 1.0 (11) | 9.7 ± 1.0 (832) | 7.7 ± 1.1 (208) | 7.5 ± 1.1 (181) |
| OCIAD1 | 5.7 ± 1.0 (52) | N.D. | N.D. | N.D. |
| SPARC | 2.5 ± 1.2 (6) | 3.5 ± 1.2 (11) | 1.9 ± 1.1 (4) | 1.3 ± 1.3 (2) |
| SCAMP1 | 5.0 ± 1.4 (32) | 2.2 ± 1.1 (5) | N.D. | 3.3 ± 1.1 (10) |
| SVIL | 2.3 ± 1.1 (5) | N.D. | N.D. | −2.9 ± 1.2 (−7) |
| ND4 | 4.7 ± 1.2 (26) | −1.6 ± 1.2 (−3) | 1.4 ± 1.1 (3) | −1.7 ± 1.2 (−3) |
| CTNNAL1 | 3.0 ± 1.5 (8) | 1.6 ± 1.1 (3) | N.D. | N.D. |
| P85α | 3.9 ± 1.4 (15) | −1.2 ± 1.2 (−2) | N.D. | −3.0 ± 1.2 (−6) |
| SDF1 | −1.3 ± 1.0 (−2) | −1.7 ± 1.0 (−3) | N.D. | N.D. |
| NDFIP1b | N.D. | N.D. | N.D. | N.D. |
| CHID1 | 4.0 ± 1.2 (16) | N.D. | N.D. | −1.5 ± 1.0 (−3) |
| SLC38A5a | 18.5 ± 0.4 (4×105) | 7.4 ± 0.5 (169) | 15.4 ± 0.4 (4×104) | 17.9 ± 1.1 (4×105) |
| LMNA | N.D. | 4.0 ± 1.0 (16) | 1.9 ± 1.1 (4) | N.D. |
| DSTN | 2.5 ± 1.2 (6) | 1.6 ± 1.1 (3) | N.D. | N.D. |
| WAVE2b | N.D. | N.D. | N.D. | N.D. |
| RAB11FIP5 | 7.2 ± 1.8 (147) | 4.5 ± 1.4 (23) | 2.8 ± 1.4 (7) | N.D. |
| RPL10 | 0.9 ± 0.2 (2) | −3.3 ± 0.2 (−10) | N.D. | −1.6 ± 0.7 (−3) |
| vWFa | 1.8 ± 1.2 (3) | 5.4 ± 1.5 (42) | 3.3 ± 1.2 (10) | 2.7 ± 1.0 (6) |
| PECAM1 | N.D. | 3.3 ± 1.1 (10) | 1.2 ± 1.0 (2) | N.D. |
| GLUT1a | 10.2 ± 1.0 (1176) | 7.3 ± 1.2 (158) | 6.2 ± 1.0 (73) | 7.6 ± 1.0 (194) |
The mean ΔΔCt values are given along with standard deviation for four measurements spanning two independent PCR experiments each. A positive value represents enrichment of the gene at the BBB when compared to microvessels from the tissue indicated and vice versa. Approximate fold differences are indicated in parentheses. N.D. = No difference in transcript abundance as determined by a 2-tailed student’s t-test (p < 0.05).
Membrane proteins that have enriched transcript abundance at the BBB compared to peripheral vascular beds.
Membrane proteins that have comparable transcript abundance levels at all vascular beds examined.
Note: In order to compare transcript abundance in other vascular beds, simply subtract the ΔΔCt values given in the table. For example, the average ΔΔCt value for Lutheran glycoprotein transcript in lung compared to heart microvasculature = 4.4 – 1.9 = 2.5 or approximately 6-fold, implying that the transcript abundance of Lutheran glycoprotein in lung microvessels is 6-fold higher than in heart microvessels.
RESULTS
Multiplex Expression Cloning (MEC) of BBB Membrane Proteins
As described previously, the MEC approach involves the enrichment of a bovine BBB cDNA library for those clones that encode membrane proteins by using a BBB membrane protein-specific polyclonal antiserum (BMSPA) (Figure 1) (Agarwal & Shusta 2009). Briefly, the cDNA library is transfected into the HEK293 T/17 mammalian host cell line for membrane protein expression. HEK293 T/17 cells were chosen as they are unlikely to naturally express BBB-selective membrane proteins and hence can be used to expression clone these proteins from a cDNA library. Subsequently, the transfected HEK293 cells are probed with the BMSPA that was raised against membrane preparations of bovine brain microvessels and hence specifically recognizes a panel of BBB membrane proteins (Figure 2) (Agarwal & Shusta 2009, Pardridge et al. 1990). Cells expressing BBB membrane proteins on the cell surface are thus tagged by the BMSPA and isolated via fluorescence activated cell sorting (FACS) (Figure 1, path 1). Subsequently the plasmid cDNA harbored by the BMSPA-labeled cells is recovered, pooled together and amplified in an E.coli host. This completes one round of enrichment for those cDNA clones that encode for BBB membrane proteins. Once the plasmid pool has been sufficiently enriched for membrane protein-encoding clones after several rounds of MEC, the clones can then be individually analyzed and their sequences determined.
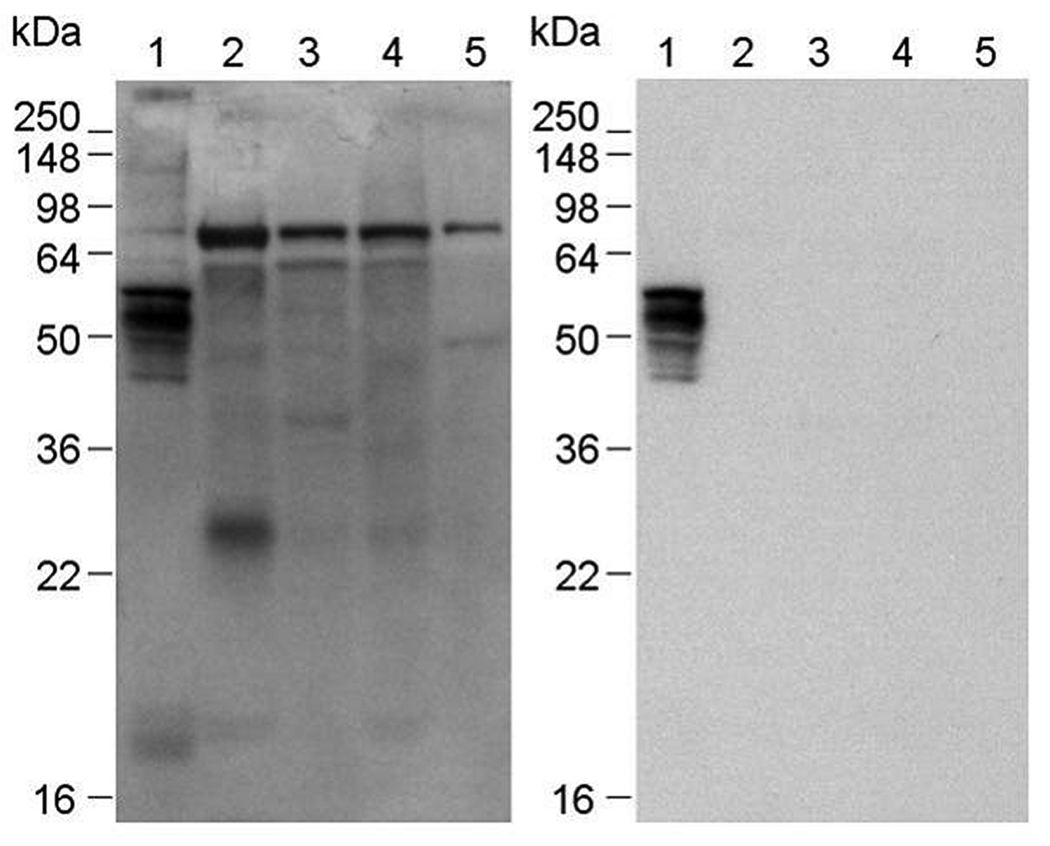
Figure 2.
Antigens recognized by BMSPA. Western blot of bovine tissue lysates separated by reducing SDS-PAGE and stained with the unsubtracted BMSPA (left panel) or with the ‘brain-specific’ subtracted version of BMSPA (right panel). Lane 1: Brain microvessels, lane 2: total lung, lane 3: total liver, lane 4: total kidney and lane 5: total heart lysates. (Note: The blot exposure times for the panels were selected so that the intensity of the bands in lane 1 of each panel was comparable)
In the current study we extended the MEC approach in three significant ways. First, we sampled about 5–10 million transfected cells in each round to oversample the diversity of the BBB cDNA library and give a higher probability for identification of rare membrane protein-encoding cDNA clones. Next, to allow the sampling of intracellularly-localized membrane-associated proteins or intracellular epitopes of integral membrane proteins that may be recognized by the BMSPA, the HEK293 cells were permeablized with 0.5% saponin solution prior to immunolabeling with the BMSPA (Figure 1, path 2.). A total of 304 cDNA clones obtained after the fourth round were individually analyzed by flow cytometry, yielding 27 cDNA clones whose membrane protein products were specifically recognized by the BMSPA. Sequencing of the 27 cDNA clones revealed 15 nonredundant sequences of BBB membrane proteins (Table 1). Finally, in addition to identifying membrane proteins that are generally expressed at the BBB using paths 1 (Agarwal & Shusta 2009) and 2 (this study) (Figure 1), another key goal of the current study was to adapt the MEC approach for identification of those membrane proteins that possess a certain level of specificity to the BBB. Accordingly, the BMSPA was serially “subtracted” by adsorption against acetone powders of bovine lung, liver, kidney and heart to remove antigenicity against those proteins that are commonly expressed both at the BBB and these tissues (Figure 1, path 3). In principle, the subtraction renders the BMSPA brain-specific with respect to these organs. The acetone powder adsorption approach was validated by the Western blot of tissue lysates in Figure 2, which reveals that while the unsubtracted BMSPA recognizes several proteins in the tissue lysates of lung, liver, kidney and heart, the ‘brain-specific’ BMSPA demonstrates selectivity to several proteins present at the BBB. Four rounds of MEC utilizing the ‘brain-specific’ BMSPA (Figure 1, path 2) were conducted and 186 clones obtained after the final enrichment round were individually analyzed. Of these, 40 clones were found to encode for BBB membrane proteins representing 7 nonredundant clones. Of these 7 clones, Lutheran glycoprotein, RPLP1 and PODXL had already been identified by the unsubtracted BMSPA, whereas ARHGAP21, RPL10, LOC613429 and OCIAD1 were newly identified (Table 1). Table 1 also lists the nature of the membrane-association of these proteins, transcript abundance levels in vivo as determined in a previous BBB genomic study (Enerson & Drewes 2006) and a list of previous reports in the literature that have identified these proteins to be expressed at the BBB. Most notably, previous MS analyses have only identified 2 of the 30 BBB membrane proteins (Haqqani et al. 2007, Haqqani et al. 2005, Lu et al. 2008) listed in Table 1, thus highlighting the complementary capability of the MEC approach to effectively sample those membrane proteins that are not particularly accessible to mass spectrometry.
Table 1.
BBB Membrane proteins identified using the MEC approach.
| Symbol | Protein Name | Genbank a | Location b | Transcript Abundancec |
BBBd |
|---|---|---|---|---|---|
| CD46 | Membrane cofactor protein | NM_183080.1 | TM | N.D. | (Shusta et al. 2002c) |
| CAIV | Carbonic anhydrase IV | NM_173897 | GPI | 4 | (Enerson & Drewes 2006, Ghandour et al. 1992) |
| RACK1 | Receptor for activated C-kinase | BC102286 | MPA | 2 | (Enerson & Drewes 2006) |
| LAMP2 | Lysosomal associated membrane protein 2 | BC102819 | TM | 9 | (Enerson & Drewes 2006) |
| LU e | Lutheran glycoprotein (Auberger B antigen included) | NM_174741 | TM | 4 | (Shusta et al. 2002b, Enerson & Drewes 2006) |
| SPARC | Secreted protein acidic cysteine rich / osteonectin | BT030548 | ECM | 295 | (Haqqani et al. 2007, Enerson & Drewes 2006, Pen et al. 2007) |
| ITM2B | Integral membrane protein 2B | XM_587730 | TM | 29 | (Enerson & Drewes 2006, Calabria & Shusta 2008) |
| MHCI | Major histocompatibility complex class I, A | AY960156 | TM | 4 | (Pardridge 2007, Enerson & Drewes 2006, Shusta 2005) |
| SCAMP1 | Secretory carrier membrane protein 1 | NM_001076054 | TM | N.D. | |
| UCP2 | Mitochondrial uncoupling protein 2 | NM_001033611 | TM | 4 | (Enerson & Drewes 2006) |
| SVIL | Supervillin | BC113265 | MPA | N.D. | |
| RPLP1 e | Ribosomal acidic protein large P1 | BC102695 | MA | 116 | (Enerson & Drewes 2006) |
| ND4 | Mitochondrial NADH dehydrogenase subunit 4 | YP_209214 | TM | 108 | (Enerson & Drewes 2006) |
| CTNNAL1 | α-catenin related protein | XM_872743 | MPA | 3 | (Enerson & Drewes 2006) |
| P85α | Phosphatidylinositol 3-kinase regulatory subunit 1 | NM_174575 | MPA | N.D. | |
| SDF1 | Stromal cell derived factor 1 | NM_001113174.1 | MPA / MA | N.D. | (McCandless et al. 2008) |
| NDFIP1 f | Nedd4 Family interacting protein 1 | NW_001504192.1 | TM | N.D. | |
| CHID1 | Chitinase Domain Containing 1 | NM_001015515.1 | MPA | N.D. | |
| SLC38A5 | Solute carrier family 38, member 5 | NM_001015580.1 | TM | 8 | (Enerson & Drewes 2006, Lyck et al. 2009) |
| LMNAs f | Lamin A/C | NW_001494724.1 | MA | 10 | (Lu et al. 2008, Enerson & Drewes 2006) |
| DSTN | Destrin (Actin Depolymerizing Factor) | NM_001015586.1 | MPA | 20 | (Enerson & Drewes 2006) |
| WAVE2 | WAS protein family, member 2 | NW_001494698.2 | MPA | 2 | (Enerson & Drewes 2006) |
| PODXL e | Podocalyxin like protein | XM_868371.3 | TM | 12 | (Enerson & Drewes 2006, Shusta et al. 2002a, Testa et al. 2009) |
| RIP11 f | RAB11 family interacting protein 5 (class I) | XM_616110.3 | MA | N.D. | |
| FIH1-like | Similar to factor inhibiting hypoxia-inducible factor subunit α | NW_001494359.2 | ? | N.D. | |
| SCP2 | Sterol Carrier Protein 2 | NM_001033990.2 | MA | 20 | (Enerson & Drewes 2006) |
| ARHGAP21 f,e | Rho GTPase activating protein 21 | XM_581232.3 | MPA / MA | 28 | (Enerson & Drewes 2006) |
| RPL10 f,e | Ribosomal protein L10 | NM_174760.2 | MA | 8 | (Enerson & Drewes 2006) |
| LOC613429 e | Hypothetical protein LOC613429 | BC103023.1 | TM | N.D. | |
| OCIAD1 e | OCIA Domain containing 1 | NM_001015648.1 | TM | N.D. |
Accession numbers from a BLAST search against the nucleotide collection database (nr/nt) for Bos taurus derived sequences – underlined entries refer to the Bos taurus genomic sequence and represent novel mRNA transcripts, that were named based on homology to the corresponding protein from other mammals;
TM-transmembrane, MPA-Membrane protein associated, MA-Membrane associated, ECM-Extracellular matrix, GPI-glycosylphosphatidylinositol anchored, ?-Not yet known;
Transcript abundance reported as tags per 100,000, N.D. = Not detected [from (Enerson & Drewes 2006), GSE3696 record [GEO; NCBI]];
References indicating BBB localization;
Proteins identified to be BBB-enriched by MEC using ‘brain-specific’ BMSPA;
Proteins identified by MEC that require cell membrane permeabilization by saponin for detection (intracellular epitope). The first 15 entries in this table were identified by our initial study using path 1 (Agarwal & Shusta 2009), many of which were again identified in the MEC processes performed in this study, the next 15 clones were newly identified in this study using path 2 (using either the unsubtracted or subtracted BMSPA).
Expression Profiling of BBB Membrane Proteins Across Other Tissues
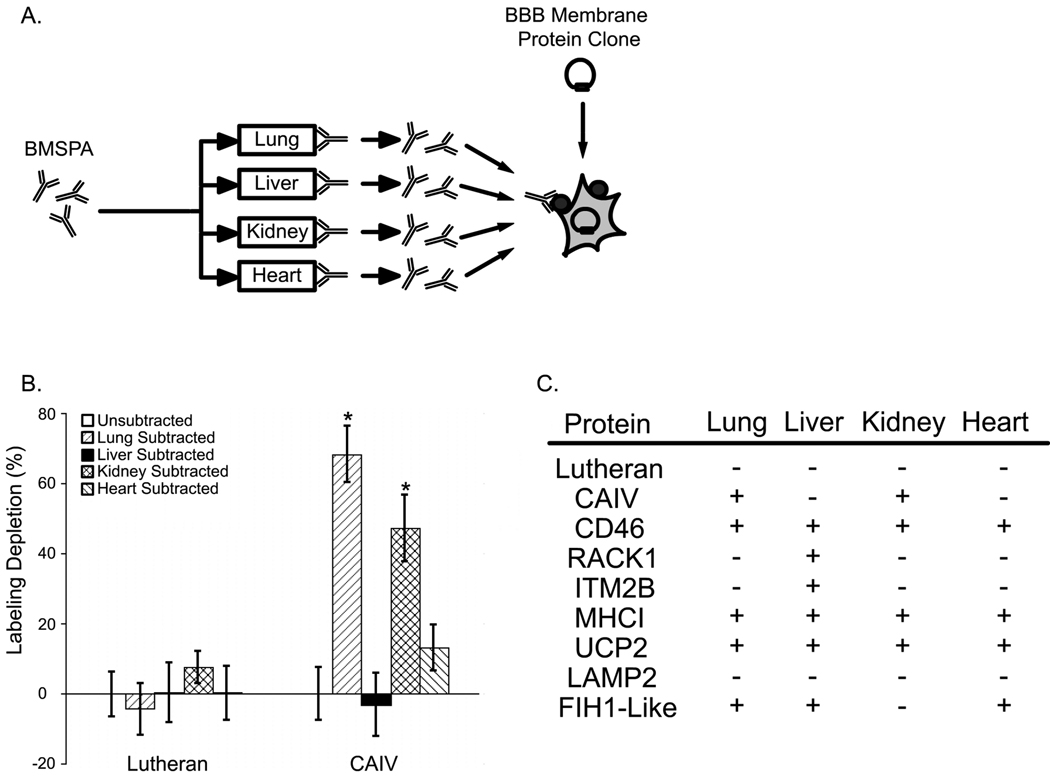
Next, to better understand the tissue distribution of these BBB membrane proteins, two different approaches were employed. First, the comparative protein expression profiles of individual BBB membrane proteins across whole tissues were evaluated by using single-organ subtracted BMSPA antisera as depicted in Figure 3A. If a BBB membrane protein that is expressed by the transfected cells is also present on another tissue, for example lung, it will be recognized by the unsubtracted BMSPA but will display a reduction in labeling when probed with the lung-subtracted version of BMSPA. Thus, by probing cells transfected with the cDNA of an individual BBB membrane protein with lung, liver, kidney or heart-subtracted BMSPA, one can generate an approximate expression profile for that protein. The bar graph in Figure 3B depicts the results of this tissue-based protein expression profiling approach for two representative genes, namely Lutheran glycoprotein and carbonic anhydrase IV (CAIV). As shown, there is a significant depletion in the antigenicity of the lung-subtracted and kidney-subtracted BMSPA for CAIV, indicating that CAIV is also expressed at lung and kidney in addition to the BBB. In contrast, Lutheran glycoprotein appears to be quite BBB-specific as no organ subtraction was capable of depleting the antigenic signal. In this way, qualitative tissue expression profiles were generated for 9 BBB membrane proteins as noted in Figure 3C. Notably, this analysis revealed that CD46, MHCI and UCP2 were widely expressed across all tissues considered, whereas the expression of the Lutheran glycoprotein was more restricted to the brain. RACK1 and ITM2B were expressed in the brain and the liver, whereas LAMP2 had an elevated expression at the lung in addition to brain. FIH1-like protein was expressed in all organs tested except kidney.
Figure 3.
Protein expression profiling across tissues. A. The BMSPA was subtracted by adsorption with acetone powders of lung, liver, kidney or heart to generate single tissue-subtracted versions of BMSPA, which were subsequently used to probe HEK293/T17 cells transfected with individual BBB membrane protein clones. B. Percent depletion in the cell surface immunolabeling intensity for HEK293/T17 cells transfected with individual cDNA clones. Cells were probed with single tissue-subtracted versions of BMSPA and immunolabeling intensity was measured using flow cytometry. Error bars denote standard deviation for 2 independent samples. C. Protein expression profiles across tissues based on percent depletion. A ‘+’ sign indicates that the protein is “expressed” in the tissue indicated (vascular, avascular or both) in addition to the BBB since there is a statistically significant depletion in the level of subtracted BMSPA labeling as determined by a two-tailed student’s t-test (100% in Figure 3B, p < 0.05, also indicated with a *), whereas a ‘−‘ sign indicates negligible depletion suggesting that the protein is “not expressed” in the tissue or expression is at too low of a level to yield a detectable depletion in antiserum binding (0% in Figure 3B).
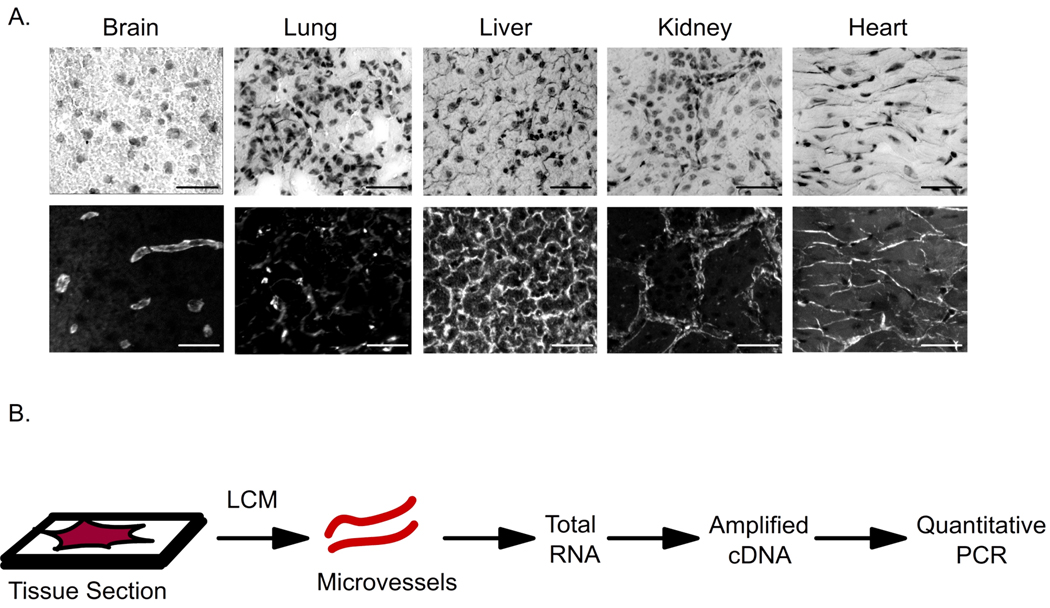
Although this approach provides qualitative expression profiles for membrane proteins across whole tissues (as a result of the adsorption of antisera with whole tissue acetone powders), it does not reveal any information about the expression levels of these proteins within the microvasculature of these tissues. Moreover, statistically significant differences in antibody labeling upon depletion could only be distinguished for those BBB membrane proteins that display a high level of basal labeling with the unsubtracted antiserum as a result of the associated errors of the experiment. Hence this approach was limited to 9 of the 30 BBB membrane proteins identified in Table 1. Accordingly, we used a second, more quantitative approach and measured the relative abundance of the BBB membrane protein transcripts in the microvasculature of the peripheral tissues. As shown in Figure 4A, the microvessels in bovine tissue sections of brain, lung, liver, kidney and heart were fluorescently labeled using the endothelial binding lectin, Griffonia simplicifolia agglutinin-I-B4. Isolation of individual microvessels was then performed using laser capture microdissection (LCM) (Figure 4B). Total RNA was recovered from the microdissected blood microvessels and amplified in order to facilitate the measurement of transcript abundance levels via quantitative PCR analyses (Figure 4B). The LCM process led to selective isolation of blood vessels as indicated by substantial enrichment in endothelial markers von Willibrand factor (vWF) and PECAM1 in the microdissected samples compared to total tissue isolates (supplemental Figure 1B). To ensure that the RNA amplification step did not introduce bias, transcript abundance levels of amplified and unamplified RNA samples were compared. There was no difference (p>0.1) in the values obtained for amplified and unamplified samples either for endothelial markers, vWF and PECAM1 (supplemental Figure 1A). Importantly, while a BBB marker, glucose transporter (GLUT1), was enriched in the BBB RNA isolates, transcripts from potential contaminating cells like neuron specific enolase (NSE, neurons) and glial fibrillary acidic protein (GFAP, astrocytes) are both decreased in the microvessel RNA isolates indicating the selective enrichment of BBB-derived transcripts (supplemental Figure 1A).
Figure 4.
Microvessel processing for LCM and qPCR. A. Phase contrast image of hematoxylin-stained tissue sections (top) with the corresponding fluorescence image displaying fluorescent labeling of endothelial cells using a fluorescein-linked Griffonia simplicifolia agglutinin-I-B4 lectin (bottom). B. The fluorescently labeled microvessels were subsequently excised using laser capture microdissection (LCM) as shown and the total RNA was extracted from these samples, amplified and subsequently used for transcript abundance profiling. (Scale bar = 20 µm)
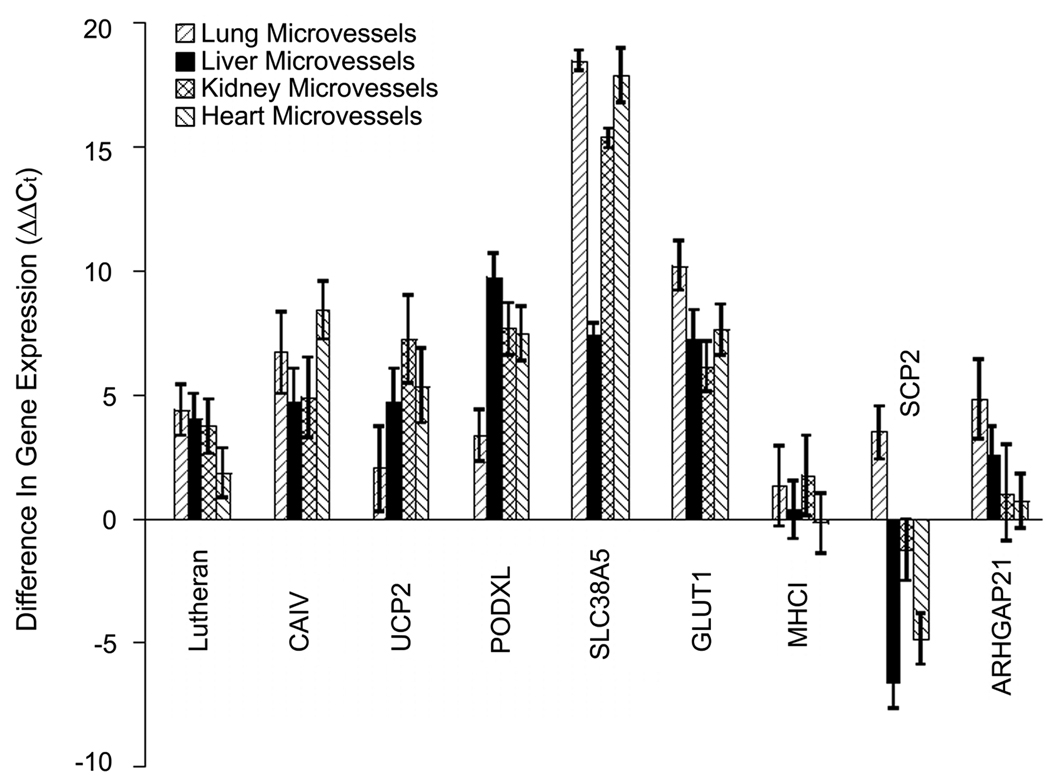
Using these LCM-derived isolates, the relative transcript abundance values for all 30 membrane proteins were measured and BBB expression compared to lung, liver, kidney and heart microvascular expression (Table 2). While transcripts for all membrane proteins were detected at the BBB as expected given the nature of the MEC approach that relies on membrane protein expression at the BBB, differential transcript expression profiles for these proteins across tissues were common. For example, SCP2 is more highly enriched in blood vessels of liver and heart compared with the BBB (Figure 5). Other proteins, such as MHCI display similar expression levels across each vascular bed whereas transcripts encoding proteins such as ARHGAP21 are BBB enriched only when compared to certain vascular beds in the body like lung and liver. Notably, of the BBB membrane proteins listed in Table 1, Lutheran glycoprotein, CAIV, UCP2, PODXL and SLC38A5 all display elevated expression levels at the BBB as compared to all other vascular beds tested indicating a high level of BBB exclusivity for these membrane proteins (compare to BBB-resident GLUT-1 in Figure 5). Indicative of the vascular origin within the brain, each of these five transcripts was also enriched in the BBB isolates compared with total brain whereas non-vascular transcripts were decreased (compare to BBB-resident GLUT1 and contrast to non-resident transcripts NSE and GFAP in Supplemental Figure 1A).
Figure 5.
Expression profiles for selected genes from Table 2 comparing transcript abundance at the BBB when compared to microvessels in the tissue indicated. Positive ΔΔCt values represent enrichment of the gene at the BBB when compared to microvessels in the tissue indicated. Error bars denote standard deviation for duplicate PCR reactions of two independent PCR runs.
DISCUSSION
In the current study we have employed several variations of MEC to clone BBB membrane proteins. The process required protein presence in isolated BBB membranes used to raise the BMSPA, transcript presence in cDNA libraries derived from freshly isolated microvessels, and transcript presence in LCM-derived RNA samples. Thus, several redundant yet distinct approaches indicated the presence of these membrane proteins at the BBB. Indeed, several of the identified membrane proteins have been localized to the BBB by targeted experiments or generalized genomic studies. This study now confirms the presence of such proteins at the BBB while also providing new information regarding BBB membrane proteins that have not been previously ascribed to the BBB (Table 1).
A particular focus of this study was an attempt to identify membrane proteins with highly selective expression at the BBB to help elucidate the unique functional attributes of the BBB membrane proteome. First, using whole tissue antibody subtraction in the MEC process to help identify BBB-selective membrane proteins, Lutheran glycoprotein and podocalyxin were amongst the proteins identified. Further analysis using single-organ subtracted antiserum confirmed that Lutheran glycoprotein was highly specific to the BBB. While this approach of whole tissue antiserum subtraction may yield clones having BBB-selective expression like Lutheran glycoprotein, it will miss potentially BBB vascular-specific clones because while they may be expressed in a certain tissue (and hence subtracted), they may not be of vascular origin. For example, CD46 has a significant expression level in all the tissues tested (Figure 3B). However, the widespread expression of CD46 is not only vascular in origin (Perez de la Lastra et al. 1999) but also includes localization at the epithelium of lung and kidney (McQuaid & Cosby 2002). Therefore, to better compare expression of the membrane proteins on a vascular basis, we generated a quantitative transcript profile across 5 vascular beds, and vascular-enriched BBB membrane proteins were identified. In particular those membrane proteins that are specifically expressed at the BBB when compared to other vascular beds are of interest since they may contribute to the unique properties of the BBB. Twenty-eight of thirty proteins were differentially expressed at the BBB versus at least one of the peripheral vascular beds indicating a unique “fingerprint” for membrane protein expression at the BBB.
Of special interest are the five BBB membrane proteins that have transcripts with highly specific BBB expression compared with all vascular beds tested. Namely, Lutheran glycoprotein, carbonic anhydrase IV, podocalyxin, uncoupling protein 2 (UCP2) and the amino acid transporter SLC38A5 are all preferentially expressed at the BBB with comparative elevation in transcript levels from 4–105 fold. The Lutheran glycoprotein is involved in the attachment of endothelial cells to laminin present in the basement membrane (Udani et al. 1998). Previous studies have confirmed the BBB-enrichment of the Lutheran glycoprotein compared with liver and kidney tissues (Shusta et al. 2002b) and it is upregulated in human brain tumor vessels (Boado et al. 2000). Carbonic anhydrase IV is a GPI-anchored protein that has been reported to be BBB-resident and localized to the luminal endothelial surface suggesting an important role in regulating the carbon dioxide-bicarbonate balance in the brain (Ghandour et al. 1992). PODXL is expressed in vascular endothelial cells throughout the body including in the brain (Testa et al. 2009, Horvat et al. 1986), glomerular podocytes (Kerjaschki et al. 1984), platelets and megakaryocytes (Miettinen et al. 1999). Interestingly, the podocalyxin protein acts both as an anti-adhesin in its role in keeping open the glomerular slits in podocyte foot processes in the kidney (Dekan et al. 1991), but has also been shown to act as an adhesin as a ligand for L-selectin and can support lymphocyte adhesion and rolling (Sassetti et al. 1998). Moreover, podocalyxin, when overexpressed ectopically in MDCK epithelial cells can disrupt tight junctions and relocalize tight junction proteins (Takeda et al. 2000). Our current results indicate the BBB-enrichment of PODXL, a finding also suggested in our previous suppression subtractive hybridization (SSH) genomics study (Shusta et al. 2002a). PODXL has also been shown to be overexpressed in proliferating endothelial cells found around glioblastomas indicating potential clinical importance for this membrane protein (Hayatsu et al. 2008). While its function at the BBB is unclear, the preferential expression of podocalyxin at the BBB suggests an important role. UCP2 is a mitochondrial uncoupling protein expressed in the brain that has been largely studied in terms of neuronal function (Horvath et al. 1999, Andrews et al. 2005). Interestingly, it has been shown that UCP2 helps maintain a low level of reactive oxygen species (ROS) thereby preventing oxidative stress to cells (Andrews et al. 2005), and as a result has been shown to serve as a neuroprotector in traumatic brain injury and ischemia (reviewed in (Paradis et al. 2003)). Thus, the cloning of UCP2 at the BBB and its moderate selectivity compared with peripheral vascular beds suggests a potentially intriguing role for the BBB in brain stress response. Finally, the most BBB-selective membrane protein identified was the amino acid transporter SLC38A5, a carrier of glutamine, asparagine, and histidine, among other amino acids (Nakanishi et al. 2001). A 2.2 kb transcript of SLC38A5 is abundant in human brain and lung, whereas a larger 2.6 kb transcript is expressed in liver and in kidney (Nakanishi et al. 2001). In the brain, SLC38A5 has been shown to be present in astrocyte processes (Cubelos et al. 2005), and very recently the transcript was shown to be one of the most highly expressed in freshly isolated and purified brain endothelial cells (Lyck et al. 2009). Our data confirms the expression of this membrane protein at the BBB and indicates the extremely restrictive expression when compared to the peripheral vasculature indicating a potential BBB biomarker. In conclusion, the BBB serves as a critical interface between the brain and the rest of the body. As a result, BBB membrane proteins play important roles in facilitating the exchange of material between the blood and brain, and the data presented in this study begin to unearth the membrane protein heterogeneity between the BBB and peripheral vascular beds.
Supplementary Material
ACKNOWLEDGEMENTS
The authors wish to acknowledge Kurt Vogel and the Meat Science and Muscle Biology Laboratory for supplying bovine tissue, Tom Pier and Dr. Shelly Cook of the Department of Pathology and Laboratory Medicine for help with the LCM, Ally Pen and Dr. Danica Stanimirovic at the University of Ottawa (Ottawa, Canada) for LCM advice and the staff at the UW Paul P. Carbone Comprehensive Cancer Center Flow Cytometry Facility for their assistance with FACS. This project was funded by National Institutes of Health Grant NS052649.
List of abbreviations
- BBB
Blood-brain barrier
- BMSPA
BBB membrane protein-specific polyclonal antiserum
- FACS
Fluorescence activated cell sorting
- MEC
Multiplex expression cloning
- SV40
Simian Virus 40
- LCM
Laser capture microdissection
REFERENCES
- Agarwal N, Shusta EV. Multiplex expression cloning of blood-brain barrier membrane proteins. Proteomics. 2009;9:1099–1108. doi: 10.1002/pmic.200800368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews ZB, Diano S, Horvath TL. Mitochondrial uncoupling proteins in the CNS: in support of function and survival. Nature reviews.Neuroscience. 2005;6:829–840. doi: 10.1038/nrn1767. [DOI] [PubMed] [Google Scholar]
- Boado RJ, Li JY, Pardridge WM. Selective Lutheran glycoprotein gene expression at the blood-brain barrier in normal brain and in human brain tumors. Journal of cerebral blood flow and metabolism. 2000;20:1096–1102. doi: 10.1097/00004647-200007000-00009. [DOI] [PubMed] [Google Scholar]
- Calabria AR, Shusta EV. A genomic comparison of in vivo and in vitro brain microvascular endothelial cells. Journal of cerebral blood flow and metabolism. 2008;28:135–148. doi: 10.1038/sj.jcbfm.9600518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubelos B, Gonzalez-Gonzalez IM, Gimenez C, Zafra F. Amino acid transporter SNAT5 localizes to glial cells in the rat brain. Glia. 2005;49:230–244. doi: 10.1002/glia.20106. [DOI] [PubMed] [Google Scholar]
- Dekan G, Gabel C, Farquhar MG. Sulfate contributes to the negative charge of podocalyxin, the major sialoglycoprotein of the glomerular filtration slits. Proc Natl Acad Sci U S A. 1991;88:5398–5402. doi: 10.1073/pnas.88.12.5398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enerson BE, Drewes LR. The rat blood-brain barrier transcriptome. Journal of cerebral blood flow and metabolism. 2006;26:959–973. doi: 10.1038/sj.jcbfm.9600249. [DOI] [PubMed] [Google Scholar]
- Ghandour MS, Langley OK, Zhu XL, Waheed A, Sly WS. Carbonic anhydrase IV on brain capillary endothelial cells: a marker associated with the blood-brain barrier. Proceedings of the National Academy of Sciences of the United States of America. 1992;89:6823–6827. doi: 10.1073/pnas.89.15.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant KJ, Wu CC. Advances in neuromembrane proteomics: efforts towards a comprehensive analysis of membrane proteins in the brain. Brief Funct.Genomic Proteomic. 2007;6:59–69. doi: 10.1093/bfgp/elm001. [DOI] [PubMed] [Google Scholar]
- Haqqani AS, Kelly J, Baumann E, Haseloff RF, Blasig IE, Stanimirovic DB. Protein markers of ischemic insult in brain endothelial cells identified using 2D gel electrophoresis and ICAT-based quantitative proteomics. Journal of proteome research. 2007;6:226–239. doi: 10.1021/pr0603811. [DOI] [PubMed] [Google Scholar]
- Haqqani AS, Nesic M, Preston E, Baumann E, Kelly J, Stanimirovic D. Characterization of vascular protein expression patterns in cerebral ischemia/reperfusion using laser capture microdissection and ICAT-nanoLC-MS/MS. FASEB. 2005;19:1809–1821. doi: 10.1096/fj.05-3793com. [DOI] [PubMed] [Google Scholar]
- Hayatsu N, Kaneko MK, Mishima K, Nishikawa R, Matsutani M, Price JE, Kato Y. Podocalyxin expression in malignant astrocytic tumors. Biochemical and biophysical research communications. 2008;374:394–398. doi: 10.1016/j.bbrc.2008.07.049. [DOI] [PubMed] [Google Scholar]
- Horvat R, Hovorka A, Dekan G, Poczewski H, Kerjaschki D. Endothelial cell membranes contain podocalyxin--the major sialoprotein of visceral glomerular epithelial cells. The Journal of cell biology. 1986;102:484–491. doi: 10.1083/jcb.102.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath TL, Warden CH, Hajos M, Lombardi A, Goglia F, Diano S. Brain uncoupling protein 2: uncoupled neuronal mitochondria predict thermal synapses in homeostatic centers. The Journal of neuroscience. 1999;19:10417–10427. doi: 10.1523/JNEUROSCI.19-23-10417.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamiie J, Ohtsuki S, Iwase R, et al. Quantitative atlas of membrane transporter proteins: development and application of a highly sensitive simultaneous LC/MS/MS method combined with novel in-silico peptide selection criteria. Pharm Res. 2008;25:1469–1483. doi: 10.1007/s11095-008-9532-4. [DOI] [PubMed] [Google Scholar]
- Kerjaschki D, Sharkey DJ, Farquhar MG. Identification and characterization of podocalyxin--the major sialoprotein of the renal glomerular epithelial cell. J Cell Biol. 1984;98:1591–1596. doi: 10.1083/jcb.98.4.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Murugesan N, Macdonald JA, Wu SL, Pachter JS, Hancock WS. Analysis of mouse brain microvascular endothelium using immuno-laser capture microdissection coupled to a hybrid linear ion trap with Fourier transform-mass spectrometry proteomics platform. Electrophoresis. 2008;29:2689–2695. doi: 10.1002/elps.200700936. [DOI] [PubMed] [Google Scholar]
- Lyck R, Ruderisch N, Moll AG, Steiner O, Cohen CD, Engelhardt B, Makrides V, Verrey F. Culture-induced changes in blood-brain barrier transcriptome: implications for amino-acid transporters in vivo. J Cereb Blood Flow Metab. 2009 doi: 10.1038/jcbfm.2009.72. DOI jcbfm200972 [pii] 10.1038/jcbfm.2009.72. [DOI] [PubMed] [Google Scholar]
- McCandless EE, Piccio L, Woerner BM, Schmidt RE, Rubin JB, Cross AH, Klein RS. Pathological expression of CXCL12 at the blood-brain barrier correlates with severity of multiple sclerosis. The American journal of pathology. 2008;172:799–808. doi: 10.2353/ajpath.2008.070918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuaid S, Cosby SL. An immunohistochemical study of the distribution of the measles virus receptors, CD46 and SLAM, in normal human tissues and subacute sclerosing panencephalitis. Laboratory investigation. 2002;82:403–409. doi: 10.1038/labinvest.3780434. [DOI] [PubMed] [Google Scholar]
- Miettinen A, Solin ML, Reivinen J, Juvonen E, Vaisanen R, Holthofer H. Podocalyxin in rat platelets and megakaryocytes. The American journal of pathology. 1999;154:813–822. doi: 10.1016/S0002-9440(10)65328-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi T, Sugawara M, Huang W, Martindale RG, Leibach FH, Ganapathy ME, Prasad PD, Ganapathy V. Structure, function, and tissue expression pattern of human SN2, a subtype of the amino acid transport system N. Biochemical and biophysical research communications. 2001;281:1343–1348. doi: 10.1006/bbrc.2001.4504. [DOI] [PubMed] [Google Scholar]
- Nielsen PA, Olsen JV, Podtelejnikov AV, Andersen JR, Mann M, Wisniewski JR. Proteomic mapping of brain plasma membrane proteins. Molecular & cellular proteomics : MCP. 2005;4:402–408. doi: 10.1074/mcp.T500002-MCP200. [DOI] [PubMed] [Google Scholar]
- Paradis E, Clavel S, Bouillaud F, Ricquier D, Richard D. Uncoupling protein 2: a novel player in neuroprotection. Trends in molecular medicine. 2003;9:522–525. doi: 10.1016/j.molmed.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Pardridge WM. Blood-brain barrier genomics. Stroke. 2007;38:686–690. doi: 10.1161/01.STR.0000247887.61831.74. [DOI] [PubMed] [Google Scholar]
- Pardridge WM, Yang J, Buciak JL, Boado RJ. Differential Expression of 53-and 45-kDa Brain Capillary-Specific Proteins by Brain Capillary Endothelium and Choroid Plexus in Vivo and by Brain Capillary Endothelium in Tissue Culture. Molecular and cellular neurosciences. 1990:20–28. doi: 10.1016/1044-7431(90)90038-6. [DOI] [PubMed] [Google Scholar]
- Pen A, Moreno MJ, Martin J, Stanimirovic DB. Molecular markers of extracellular matrix remodeling in glioblastoma vessels: microarray study of laser-captured glioblastoma vessels. Glia. 2007;55:559–572. doi: 10.1002/glia.20481. [DOI] [PubMed] [Google Scholar]
- Perez de la Lastra JM, Hanna SM, Morgan BP. Distribution of membrane cofactor protein (MCP/CD46) on pig tissues. Relevance To xenotransplantation. Immunology. 1999;98:144–151. doi: 10.1046/j.1365-2567.1999.00830.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassetti C, Tangemann K, Singer MS, Kershaw DB, Rosen SD. Identification of podocalyxin-like protein as a high endothelial venule ligand for L-selectin: parallels to CD34. J Exp Med. 1998;187:1965–1975. doi: 10.1084/jem.187.12.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusta EV. Blood-brain barrier genomics, proteomics, and new transporter discovery. NeuroRx. 2005;2:151–161. doi: 10.1602/neurorx.2.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shusta EV, Boado RJ, Mathern GW, Pardridge WM. Vascular genomics of the human brain. Journal of cerebral blood flow and metabolism. 2002a;22:245–252. doi: 10.1097/00004647-200203000-00001. [DOI] [PubMed] [Google Scholar]
- Shusta EV, Boado RJ, Pardridge WM. Vascular proteomics and subtractive antibody expression cloning. Molecular & cellular proteomics. 2002b;1:75–82. doi: 10.1074/mcp.t100008-mcp200. [DOI] [PubMed] [Google Scholar]
- Shusta EV, Zhu C, Boado RJ, Pardridge WM. Subtractive expression cloning reveals high expression of CD46 at the blood-brain barrier. Journal of neuropathology and experimental neurology. 2002c;61:597–604. doi: 10.1093/jnen/61.7.597. [DOI] [PubMed] [Google Scholar]
- Takeda T, Go WY, Orlando RA, Farquhar MG. Expression of podocalyxin inhibits cell-cell adhesion and modifies junctional properties in Madin-Darby canine kidney cells. Mol Biol Cell. 2000;11:3219–3232. doi: 10.1091/mbc.11.9.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Testa JE, Chrastina A, Li Y, Oh P, Schnitzer JE. Ubiquitous yet distinct expression of podocalyxin on vascular surfaces in normal and tumor tissues in the rat. J Vasc Res. 2009;46:311–324. doi: 10.1159/000189792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udani M, Zen Q, Cottman M, Leonard N, Jefferson S, Daymont C, Truskey G, Telen MJ. Basal cell adhesion molecule/lutheran protein. The receptor critical for sickle cell adhesion to laminin. The Journal of clinical investigation. 1998;101:2550–2558. doi: 10.1172/JCI1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qian WJ, Chin MH, et al. Characterization of the mouse brain proteome using global proteomic analysis complemented with cysteinyl-peptide enrichment. Journal of proteome research. 2006;5:361–369. doi: 10.1021/pr0503681. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.