Abstract
Notwithstanding the well established association of HLA-DRB1 shared epitope alleles, interest remains in identifying additional Major Histocompatibility Complex (MHC) region variants associated with rheumatoid arthritis (RA). We used a panel of 1,201 haplotype tagging single nucleotide polymorphisms (SNPs) designed for African Americans to find genetic variants associated with RA in a 3.8 Mb region encompassing the MHC. Conditioning on seven covariates including HLA-DRB1 risk alleles and population structure, we identified a SNP in HLA-DOA (rs9276977) significantly associated with RA; minor allele frequency (MAF) 0.27 in cases versus 0.21 in controls: OR(±95% CI) = 2.86 (1.61, 5.31). Genotyping of rs9276977 in an independent sample of African-American RA patients and controls did not replicate the association (MAF 0.28 in cases versus 0.27 in controls). This study points to the potential association of a SNP in the HLA-DOA gene with RA in African-Americans, but also underscores the importance of replication of findings in larger patient cohorts.
Keywords: genetic association, MHC, HLA-DOA, African American, HLA-DRB1
Introduction
Genetic and environmental factors are known to influence the complex etiology of rheumatoid arthritis (RA). The most significantly associated genetic contribution is with HLA-DRB1 alleles containing the shared epitope1; however, the pathogenesis of RA has a polygenic basis. Recent studies have associated polymorphisms in the class II Major Histocompatibility Complex (MHC) region with RA among persons of European descent, independent of HLA-DRB1 shared epitope presence.2–3 In the understudied African American population, our laboratory has established that the association of HLA-DRB1 alleles in a genetically admixed African American population was likely due to the introgression of alleles of European descent.4 In addition, we have used the haplotype block structure of a 3.8 Mb region of the MHC to design a population specific panel of haplotype-tagging SNPs for future association studies.5
The purpose of the current study is to utilise known patterns of linkage disequilibrium and a panel of haplotype-tagging single nucleotide polymorphisms (htSNPs) across the MHC to aid in detecting single marker associations with RA in an African American population. Building on the previous work, we report genotyping 1,384 htSNPs from a 3.8Mb region surrounding the MHC in 276 African American RA patients using Illumina GoldenGate BeadXpress technology. Control data, SNP selection, and recruitment details for patients used for the current study are fully described in Hughes et al.4 and Kelley et al.5 In the current analysis, we conditioned on estimates of population structure and the number of HLA-DRB1 risk alleles to identify SNP variants, using the panel of htSNPs. We are able to significantly associate with RA susceptibility a SNP, rs9276977, in the HLA-DOA gene.
Results and discussion
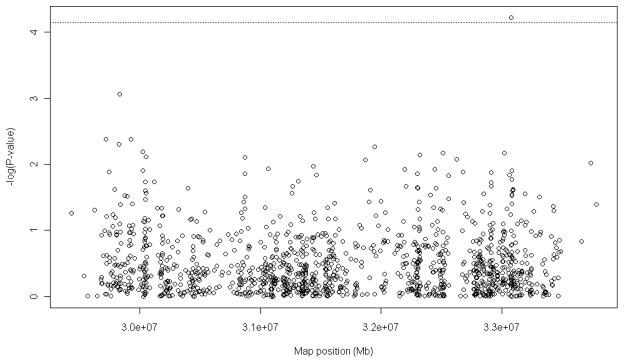
Of the 1,384 markers suggested as htSNPs in Kelley et al.5, 1,201 were analysed in this study, which included those SNPs with minor allele frequency (MAF) > 0.05, < 20% missing values, and control genotypes in Hardy-Weinberg Equilibrium (p < 0.05). We have demonstrated that one SNP, rs9276977, of the 1201 SNPs analysed for RA association was significant at the Bonferroni corrected alpha level of 7.12 × 10−5 after the potentially confounding effects of the seven covariates, HLA-DRB1 risk alleles (0, 1, 2), gender, age, current smoking status (Yes/No), admixture (proportion of European ancestry), anti-CCP antibody status (Pos/Neg), IgG serum RF factor (Pos/Neg), and the SNP genotype (0, 1, 2) had been accounted for (Figure 1). Each of the 720,600 SNP by SNP and 1201 SNP by smoking status interactions were modeled using logistic regression including the respective main effects and the same covariates used for the single marker tests, but no significant interaction effects were detected. The log odds of having RA, given a unit increase in the rs9276977 minor allele, was 1.05 corresponding to an odds ratio (± 95% confidence limits) of 2.86 (1.61, 5.31). The high odds ratio underscores the limited power to detect association of SNPs with moderate effect in this sample of 94 controls and 276 cases. At the lower 95% confidence limit the statistical power to detect an association of rs9276977 is 0.1, and increases to 0.96 at the mean estimate for a sample of 370 individuals at the given Bonferroni corrected alpha level. The MAF of rs9276977 in the controls was 0.21 and in cases was 0.27. The MAF for controls was reasonable for this African American study population considering population genetic estimates from the HapMap project. The frequency of rs9276977 in controls was intermediate by comparison to MAF for the Yoruba population (0.23) and the European population (0.17). rs9276977 is in a transcribed but untranslated region of the HLA-DOA gene, which has been a focus of interest for its potential role in autoimmunity.8–9 It is possible that other nearby causal loci may be linked to this SNP.
Figure 1.
−log P values of additive htSNP effect on case (+RA) - control status plotted by map position. Generalized linear models (binomial link function) were used to model case (N = 276)/control (N = 94) status with seven predictors for each of the 1,201 SNP markers using the statistical software package [R].6 The patients and controls studied and the procedures for genotyping were fully described in Hughes et al.4 and Kelley et al.5 The predictors included the seven covariates, HLA-DRB1 risk alleles (0, 1, 2), gender, age, current smoking status (Yes/No), admixture (proportion of European ancestry), anti-CCP antibody status (Pos/Neg), and IgG serum RF factor (Pos/Neg), and the SNP with minor alleles coded 0, 1, or 2. The horizontal line is the adjusted type 1 error rate based on 702 effectively independent markers. The vertical line is the map position of the HLA-DRB1 locus. A permutation test based procedure (N = 100,000 permutations) was used to calculate an empirical p-value for rs9276977 of 6 × 10−5. The genotypic scores but not the responses were permuted in order to preserve the linkage among the potentially correlated covariates and disease status for each individual in the study. This permutation procedure has equivalent statistical properties as permuting the residuals of the regression of the response on the covariates.7
The SNP rs9276977 was genotyped in 371 additional cases and 131 controls from individuals of the CLEAR2 cohort and an additional 377 African-American controls. A Fisher’s exact test was used to demonstrate no significant association between MAF in cases (0.28) versus control (0.27) in the replication study. The obvious discrepancy between the replication study and the original cohort is the MAF estimates in the control population. Control MAF for rs9276977 may have been undersampled in the original cohort or oversampled in the replication cohort. Because the MAF frequency in the replication cohort far exceeds the range expected in an admixed African American population based on the HapMap data, we consider the latter to be more likely. In the future, more samples will need to be analyzed for an accurate assessment of the MAF in controls in the African American population.
In the current analysis we used the number of HLA-DRB1 risk alleles and estimates of population stratification as covariates, which was similar to the approach of Vignal et al.3 We also used assessments of anti-cyclic citrullinated peptide (anti-CCP) antibody status as a covariate, which has specificity for RA in African Americans similar to that in persons of European ancestry.10 Anti-CCP antibody status consistently had a large effect in explaining variation between RA cases and controls in the current analysis. Ding et al.2 found SNPs varied by case-control status only in the anti-CCP antibody positive group; therefore, we modeled the explanatory power of the genetic effect after controlling for the variation explained by anti-CCP status and other covariates.
DNA sequence data indicate HLA-DOA has limited amino acid sequence variation in patients with RA8; however, these variants were synonymous changes and were not shown to vary between patients or controls. Nevertheless, HLA-DOA has been proposed to have functional implications in autoimmunity since it can inhibit the activity of HLA-DM genes in vitro possibly to help regulate the antigen loading and presentation in B cells.9 One hypothesis is that this inhibition may cause increased antigen loading thereby increasing the pool of MHC presenting cell surface antigens, which could promote T-cell activity.9 Neither Ding et al.2 nor Vignal et al.3 reported a significant association between rs9276977 and RA. Therefore it appears that rs9276977 may have a population specific effect on RA. African Americans are a recently admixed population with unique patterns of linkage disequilibrium, haplotype block structure, and recombination rates. Thus, it may not be unusual that this population specific haplotype tagging SNP panel revealed an association with RA that has not been reported from predominantly Caucasian populations.
Here we were able to build on previous work in African Americans by incorporating population specific information on admixture and haplotype block structure.4–5 Two covariates of particular interest are population admixture and the number of HLA-DRB1 risk alleles. A concern for association studies in admixed populations is that allele frequency variation between source populations may cause spurious associations with genotype and disease status.11 We were able to explicitly model global admixture, estimated by Hughes et al.4 as the proportion of the genome of European ancestry, which allowed us to avoid potential genetic confounding at the genome-wide level. In addition we were able to control for the association of HLA-DRB1 risk alleles with RA that has been well-documented in virtually all racial/ethnic groups.
Population based genetic association studies provide a powerful approach for mapping common variants that are related to variation in disease status. However, successfully detecting genetic association is limited by the well known statistical issue of “large p/small n”. In particular the theoretically valid Bonferroni correction of type 1 error for multiple independent testing of genetic markers can vastly reduce the power to detect association. One recently proposed solution is to reduce the number of markers from the number actually tested by accounting for correlations of marker genotypes.12 We applied the approach of Gao et al.12 in dividing the markers into haplotype blocks. We used regions between recombination hotspots previously defined by Kelley et al.5 to calculate the effective number of independent markers (Table 1). We were able to use this information to set the alpha level at 7.12 × 10−5 based on 702 effective markers compared to 4.12 × 10−5 from the 1,201 markers.
Table 1.
The effective number of independent markers across eight regions spanning the MHC. Principal components analysis, using the R function prcomp, of the pairwise genotype correlation matrix within each of eight regions delineated in Kelley et al.5 demonstrated that the effective number of independent markers was 702. The effective number of independent markers (i) was selected by summing the sorted (descending order) eigenvalues from 1:(i) until the proportion of the sum of all eigenvalues equaled 0.995; i.e., the number of principal components needed to explain 99.5 % of the total variation as illustrated in Gao et al. 2008.
| Distance between recombination hotspots(Map distance in Mb) | Total markers | Independent markers | Proportion |
|---|---|---|---|
| 1 (x <=29.9) | 69 | 43 | 0.62 |
| 2 (29.9 < x <= 30.3) | 127 | 66 | 0.52 |
| 3 (30.3 < x <=30.98) | 146 | 94 | 0.64 |
| 4 (30.98 < x <= 31.25) | 123 | 78 | 0.63 |
| 5 (31.25 < x <= 32.24) | 269 | 140 | 0.52 |
| 6 (32.24 < x <= 32.51) | 115 | 71 | 0.62 |
| 7 (32.51< x <= 32.9) | 129 | 68 | 0.53 |
| 8 (x > 32.9) | 223 | 142 | 0.64 |
| Total | 1201 | 702 | 0.58 |
In summary, we found that a SNP, located in the HLA-DOA gene is associated with RA after conditioning on the number of HLA-DRB1 risk alleles, the potential confounding effects of proportion European ancestry in an African American population, and other covariates including anti-CCP antibody status. The present study adds another genetic locus, HLA-DOA, to a growing list of genetic loci in the MHC, independent of the HLA-DRB1 locus, that may be significantly associated with RA. This result sets the stage for future studies of genetic association with RA in larger African American populations.
Acknowledgments
The CLEAR Investigators are as follows: Graciela S. Alarcon, MD, MPH and George Howard, DrPH from the University of Alabama at Birmingham, Birmingham, AL; Doyt L. Conn, MD from Emory University, Atlanta, GA; Beth L. Jonas, MD and Leigh F. Callahan, PhD from the University of North Carolina, Chapel Hill, NC; Edwin A. Smith, MD from the Medical University of South Carolina, Charleston, SC; Richard D. Brasington, Jr., MD from Washington University, St. Louis, MO; and Larry W. Moreland, MD, University of Pittsburgh.
We gratefully acknowledge the following physicians who enrolled patients: Jacob Aelion, MD, Jackson, TN; Charles Bell, Birmingham, AL; Sohrab Fallahi, MD, Montgomery, AL; Richard Jones, PhD, MD, Tuscaloosa, AL; Maura Kennedy, MD, Birmingham, AL; Adahli Estrada Massey, MD, Auburn, AL; John Morgan, MD, Birmingham, AL; Donna Paul, MD, Montgomery, AL; Runas Powers, MD, Alexander City, AL; William Shergy, MD, Huntsville, AL; Cornelius Thomas, MD, Birmingham, AL; Ben Wang, MD, Memphis, TN.
We gratefully acknowledge staff and coordinators at the following sites: University of Alabama at Birmingham: Stephanie Ledbetter, MS; Zenoria Causey, MS; Selena Luckett, RN, CRNC; Laticia Woodruff, RN, MSN; Candice Miller; Emory University: Joyce Carlone, RN, FNP-BC; Karla Caylor, BSN, RN; Sharon Henderson, RN; University of North Carolina: Diane Bresch, BSN; Medical University of South Carolina: Trisha Sturgill.
We gratefully acknowledge Nicholas Pajewski, Kelly Vaughan, and Miguel Padilla for discussions of the data analysis.
The CLEAR Registry is a national resource, with clinical data, DNA, and other biological samples available. For details, see the following website: http://www.dom.uab.edu/rheum/CLEAR%20home.htm.
This work was supported by NIH N01 AI40068, N01 AR02247, P01 AR40894, R01 AR51394, and NIH GCRC/CTSA grants M01 RR00032, U54 RR025777, R01 GM069430, T32 HL072757 (University of Alabama at Birmingham) and M01 RR 000046 (University of North Carolina).
References
- 1.Gregersen PK, Silver J, Winchester RJ. The shared epitope hypothesis: An approach to understanding the molecular genetics of susceptibility to rheumatoid arthritis. Arthritis Rheum. 1987;30:1205–1213. doi: 10.1002/art.1780301102. [DOI] [PubMed] [Google Scholar]
- 2.Ding B, Padyukov L, Lundstrom E, Seielstad M, Plenge RM, Oksenberg JR, et al. Different patterns of associations with anti-citrullinated protein antibody-positive and anti-citrullinated protein antibody-negative rheumatoid arthritis in the extended major histocompatibility complex region. Arthritis Rheum. 2009;60:30–38. doi: 10.1002/art.24135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vignal C, Bansal AT, Balding DJ, Binks MH, Dickson MC, Montgomery DS, et al. Genetic association of the major histocompatibility complex with rheumatoid arthritis implicates two non-DRB1 loci. Arthritis Rheum. 2009;60:53–62. doi: 10.1002/art.24138. [DOI] [PubMed] [Google Scholar]
- 4.Hughes L, Morrison D, Kelley JM, Padilla MA, Vaughn LK, Westfall AO, et al. The HLA-DRB1 shared epitope is associated with susceptibility to rheumatoid arthritis in African Americans through European genetic admixture. Arthritis Rheum. 2008;58:349–358. doi: 10.1002/art.23166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kelley JM, Hughes LB, Feng R, Liu N, Padilla MA, Vaughan LK, et al. Evaluating linkage disequilibrium and recombination provides a haplotype-tagging SNP panel of the major histocompatibility complex in African Americans. Genes Immun. 2008;9:271–273. doi: 10.1038/gene.2008.6. [DOI] [PubMed] [Google Scholar]
- 6.R Development Core Team. R: A Language and Environment for Statistical Computing. 2008 http://www.R-project.org.
- 7.O’Gorman TW. The performance of randomization tests that use permutations of independent variables. Commun Stat Simulat. 2005;34:895–908. [Google Scholar]
- 8.van Lith M, van Ham M, Neefjes J. Novel polymorphisms in HLA-DOA and HLA-DOB in B-cell malignancies. Immunogenetics. 2008;54:591–595. doi: 10.1007/s00251-002-0500-6. [DOI] [PubMed] [Google Scholar]
- 9.Denzin LK, Fallas JL, Prendes M, Yi W. Right place, right time, right peptide: DO keeps DM focused. Immunol Rev. 2005;207:279–292. doi: 10.1111/j.0105-2896.2005.00302.x. [DOI] [PubMed] [Google Scholar]
- 10.Mikuls TR, Holers VM, Parrish L, Kuhn KA, Conn DL, Gilkeson G, et al. Anti-cyclic citrullinated peptide antibody and rheumatoid factor isotypes in African Americans with early rheumatoid arthritis. Arthritis Rheum. 2006;54:3057–3059. doi: 10.1002/art.22200. [DOI] [PubMed] [Google Scholar]
- 11.Balding DJ. A tutorial on statistical methods for population association studies. Nat Rev Gen. 2006;7:781–791. doi: 10.1038/nrg1916. [DOI] [PubMed] [Google Scholar]
- 12.Gao X, Starmer J, Martin ER. A multiple testing correction method for genetic association studies using correlated single nucleotide polymorphisms. Genet Epidemiol. 2008;32:361–369. doi: 10.1002/gepi.20310. [DOI] [PubMed] [Google Scholar]