Abstract
The calcitonin receptor-like receptor (CLR) acts as a receptor for the calcitonin gene-related peptide (CGRP) but in order to recognize CGRP, it must form a complex with an accessory protein, receptor activity modifying protein 1 (RAMP1). Identifying the protein/protein and protein/ligand interfaces in this unusual complex would aid drug design. The role of the extreme N-terminus of CLR (Glu23-Ala60) was examined by an alanine scan and the results were interpreted with the help of a molecular model. The potency of CGRP at stimulating cAMP production was reduced at Leu41Ala, Gln45Ala, Cys48Ala and Tyr49Ala; furthermore, CGRP-induced receptor internalization at all of these receptors was also impaired. Ile32Ala, Gly35Ala and Thr37Ala all increased CGRP potency. CGRP specific binding was abolished at Leu41Ala, Ala44Leu, Cys48Ala and Tyr49Ala. There was significant impairment of cell surface expression of Gln45Ala, Cys48Ala and Tyr49Ala. Cys48 takes part in a highly conserved disulfide bond and is probably needed for correct folding of CLR. The model suggests that Gln45 and Tyr49 mediate their effects by interacting with RAMP1 whereas Leu41 and Ala44 are likely to be involved in binding CGRP. Ile32, Gly35 and Thr37 form a separate cluster of residues which modulate CGRP binding. The results from this study may be applicable to other family B GPCRs which can associate with RAMPs.
Abbreviations: CGRP, calcitonin gene-related peptide; CLR, calcitonin receptor-like receptor; ECD, extracellular domain; GPCR, G-protein coupled receptor; HA, hemagglutinin; pEC50, –log(EC50); PBS, phosphate buffered saline; RAMP, receptor activity modifying protein; WT, wild-type
Keywords: CLR, CGRP, RAMP1, Family B GPCRs, Molecular modelling, Alanine scan
1. Introduction
Calcitonin gene-related peptide (CGRP) is an abundant 37 amino acid peptide found throughout the sensory nervous system. It is an extremely potent vasodilator and it is an important mediator of neurogenic inflammation. The CGRP receptor is unusual as its subunits include a family B G-protein coupled receptor (GPCR) called the calcitonin receptor-like receptor (CLR) and a receptor activity modifying protein (RAMP1) as well as an additional peripheral membrane protein, receptor component protein [8]. Interaction of CLR with the homologous proteins RAMP2 and RAMP3 gives receptors for the peptide adrenomedullin (although the RAMP3-containing complex retains reasonable affinity for CGRP) [19]. Recent work suggests that the CGRP receptor is formed by the association of a RAMP1 monomer with a dimer of CLR [12]. Understanding this receptor on a mechanistic level presents considerable challenges. However this is not only important in understanding the intrinsic properties of the CGRP receptor, but may provide insight on how a number of other family B GPCRs interact with RAMPs, most notably the calcitonin receptor which generates amylin receptors [4,21].
A general model is now emerging of ligand binding to family B GPCRs in which the C-termini of the ligands lie alongside the extreme N-termini of the corresponding receptors [22,25]. It is tempting to assume that CGRP adopts a similar mode of binding to its receptor. However, the requirement for RAMP1 complicates matters. The extreme N-terminus of CLR is an important site for both CGRP [2] and RAMP association [14], so it is plausible that it could interact directly with CGRP and RAMP1. However, the functional role of individual amino acids in this region has not been examined.
To further investigate the role of the extreme N-terminus of CLR, we report the results of an alanine scan on residues 23–60 of this protein, representing the first 38 residues of the receptor after removal of the signal peptide. Two clusters of residues are shown to be important for CGRP action. At the end of the epitope, modeling suggests Gln45 and Tyr49 are chiefly involved in interacting with RAMP1, whereas Leu41 and Ala45 facilitate CGRP binding. In the middle of the epitope, Ile32, Gly35 and Thr37 also influence CGRP binding. It is possible that this model can be extended to amylin and calcitonin receptors.
2. Methods
2.1. Materials
Human αCGRP was from Calbiochem (Beeston, Nottingham, UK). 125I-iodo8histidyl-human αCGRP was from PerkinElmer Life and Analytical Sciences (Waltham, MA). [8-3H] Adenosine 3′, 5′, cyclic phosphate, NH4 salt was from Amersham Biosciences (Chalfont, UK). Chemicals were from Sigma–Aldrich UK.
2.2. Expression constructs and mutagenesis
Human CLR with an N-terminal hemagglutinin (HA) epitope tag and human RAMP1 with an N-terminal myc epitope tag were mutated using the Quik Change II site-directed mutagenesis kit (Stratagene, Cambridge, UK), as described previously [6].
2.3. Cell culture and transfection
Cos-7 cells were cultured and transfected with polyethyleneimine as described previously [28].
2.4. Radioligand binding
Confluent cells from five 160-cm2 Petri dishes (pre-coated with poly-d-lysine), were washed with phosphate buffered saline (PBS), followed by the addition of 15 ml of ice-cold sterile double distilled water to induce cell lysis. Following 5 min incubation on ice, the ruptured cells were washed with ice-cold PBS before being scraped from the plates and pelleted in a bench-top centrifuge (13,000 × g for 30 min). The membrane pellet was resuspended in 1 ml binding buffer (25 mM HEPES pH 7.4, 2.5 mM CaCl2, 1 mM MgCl2, 50 mg/L bacitracin) and forced through a 23G needle. 0.1 ml aliquots were snap-frozen in liquid nitrogen and stored at −70 °C. Membranes were slowly thawed on ice before diluting to a concentration that gave total radioligand binding of <10% total counts added. In a reaction volume of 200 μl, 75 pM (∼60,000 cpm) 125I-CGRP with or without 1 μM unlabelled CGRP and membranes were combined, all diluted in binding buffer. Assays were carried out for 1 h in MultiScreen 96-well filtration plates (glass fibre filters, 0.65 μm pore size, Millipore, Bedford, MA) pre-soaked in 1% non-fat milk/PBS. After the incubation, membrane-associated radioligand was harvested by transferring the assay mixture to the filtration plate housed in a vacuum manifold. The wells of the filtration plate were washed three times with 0.2 ml PBS before harvesting the filter discs. Filter-bound radioactivity was measured in a gamma counter (RiaStar 5405 counter; PerkinElmer Life and Analytical Sciences, Waltham, MA). Non-specific binding was ∼1% of total counts added.
2.5. Assay of cAMP production
48-Well plates were transiently transfected with WT receptor (HA CLR/myc RAMP1) alongside a mutant receptor in every experiment, to take account of day-to-day differences in transfection and coupling efficiencies. Stimulation with agonists and assay of cAMP was by a radioreceptor assay as described elsewhere [28].
2.6. Analysis of cell surface expression of mutants by ELISA and agonist dependent internalization
24-Well plates were transiently transfected with WT receptor a mutant receptor in every experiment. A negative control of myc RAMP1/empty pcDNA3.1(−) was used. The ELISA was carried out as described previously to measure both cell surface and total expression of CLR [28]. Cell surface expression data was normalized to make mean WT expression 100% and the mean myc RAMP1/empty pcDNA3.1(−) vector as 0%. Receptor internalization was measured after 1 h treatment with 100 nM human αCGRP at 37 °C by ELISA as described previously [7].
2.7. Molecular modeling
A comparative structure of the extracellular domain (ECD) of CLR, from residues 23–134 was generated using Modeller9v3 [9] based on the gastric inhibitory peptide receptor- and parathyroid hormone type 1 receptor-ECDs [22,25]. This gave 500 models which were ranked by Modeller9v3 energy objective function. The stereochemical quality of the top 20 structures was assessed by PROCHECKv3.5.4 [17] to select the best model. The program LOOPY [5] was used to build 2000 initial loop conformations for loops 2 and 3 and 4000 conformations for loop 1. LOOPY was used to attach the loops to the protein by a random tweak method and then to perform a fast energy minimization on torsional space to allow the program SCAP to predict and build the protein side-chains. Initial loop conformations were ranked by DFIRE 2.0 [29] statistical energy function. The top conformers underwent a physical-based scoring method [24] using AMBER99 in the presence of Generalised Born solvation model [26] implemented in the TINKER program package (http://dasher.wustl.edu/tinker/). Minimized fragments were subsequently ranked by total energy potential by the program ANALYZE in TINKER. A conformation from the top 10 structures was selected based on the architecture of the loops in other family B GPCR ECDs and the total energy potential score of the loop conformer. The H++ web server (http://biophysics.cs.vt.edu/H++/) was used to calculate the protonation states of titratable sites in CLR ECD (external dielectric constant = 80, internal dielectric constant = 6, salinity = 0.15 M, pH = 7.2). Gromacs v4.0 [13] was used for the refinement of the CLR ECD.
The CLR ECD was docked against itself with GRAMMv1.03 [1] using the generic low resolution settings to produce 1000 complexes. These were ranked based on residue level pair potentials scoring, using the 3D-Dock suite [20]. The dimer interface was refined using the MULTIDOCK program from the 3D-Dock suite. RAMP1 ECD was docked onto the CLR ECD complex using the strategy described above. The resulting trimer was refined with Gromacs v4.0, utilizing the OPLS-AA/L force field parameters to perform a deepest descent energy minimization in the presence of an explicit SPC/E water model with neutralizing Na+ ions.
2.8. Data analysis
For the radioligand binding assays, specific binding of mutants was compared to WT by Tukey's test following a one-way ANOVA. For cAMP measurements, Prism 4 (Graphpad Software Inc., San Diego, CA) was used to generate sigmoidal concentration–response curves. A two-tailed unpaired t-test was used to compare WT and mutant pEC50 values, basal and maximum cAMP production. For ELISA-based analysis of total receptor expression, cell surface expression and internalization, a Mann–Whitney test was used to compare WT with mutants.
3. Results
3.1. Stimulation of cAMP production
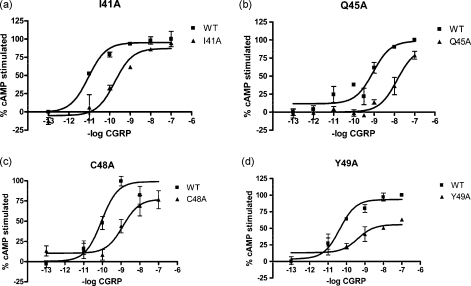
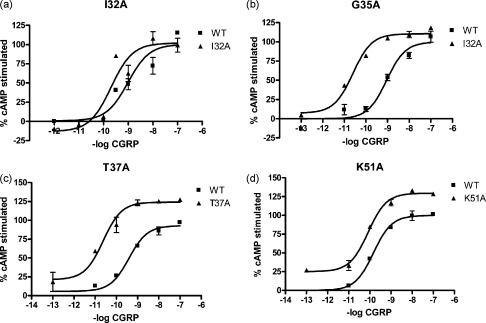
Each mutant was challenged with human αCGRP and cAMP production was measured. Ile41Ala, Gln45Ala, Cys48Ala and Tyr49Ala all had a reduced potency to CGRP (Fig. 1, Table 1). The mutant Ala44Leu showed an 8-fold decrease in potency, although this did not reach statistical significance. Ile32Ala, Gly35Ala and Thr37Ala showed an approximate 10-fold increase in potency (Fig. 2, Table 1). There were no significant potency changes at any other mutant. Glu39Ala showed an increased maximum response (80.9 ± 8.0% increase above WT) and Lys51Ala showed increased constitutive activity (30.2 ± 7.0% increase in basal cAMP production compared to WT) (Fig. 2). Neither of these parameters were significantly changed for any other mutant (Supplementary Table 1).
Fig. 1.
CGRP-stimulated cAMP responses of the mutated receptors where there is a decrease in potency. (a) I41A, (b) Q45A, (c) C48A and (d) Y49A. Cos-7 cells were transfected with WT/RAMP1 or mutant/RAMP1 and assayed for CGRP-stimulated cAMP production. WT type receptors; open squares. Mutant receptors; as indicated. Data are representative of three to five similar experiments.
Table 1.
Ability of mutant receptors to stimulate cAMP production.
| Mutant | N | pEC50 WT | pEC50 mutant | Mutant | N | pEC50 WT | pEC50 mutant |
|---|---|---|---|---|---|---|---|
| E23A | 4 | 8.98 ± 0.32 | 9.18 ± 0.37 | M42A | 6 | 9.81 ± 0.26 | 9.86 ± 0.10 |
| L24A | 4 | 9.09 ± 0.35 | 9.39 ± 0.42 | T43A | 3 | 9.77 ± 0.41 | 9.74 ± 0.37 |
| E25A | 6 | 10.08 ± 0.40 | 9.72 ± 0.39 | A44L | 5 | 9.32 ± 0.20 | 8.41 ± 0.45 |
| E26A | 5 | 9.33 ± 0.57 | 9.33 ± 0.21 | Q45A | 3 | 9.11 ± 0.08 | 8.10 ± 0.18** |
| S27A | 4 | 9.26 ± 0.14 | 9.51 ± 0.34 | Y46A | 4 | 9.17 ± 0.19 | 9.12 ± 0.44 |
| P28A | 4 | 9.11 ± 0.33 | 9.90 ± 0.37 | E47A | 5 | 9.57 ± 0.21 | 9.40 ± 0.32 |
| E29A | 4 | 9.41 ± 0.14 | 9.26 ± 0.20 | C48A | 7 | 10.09 ± 0.23 | 8.77 ± 0.38** |
| D30A | 4 | 9.66 ± 0.29 | 9.68 ± 0.41 | Y49A | 6 | 9.71 ± 0.19 | 8.76 ± 0.18** |
| S31A | 7 | 9.11 ± 0.31 | 9.00 ± 0.37 | Q50A | 5 | 9.49 ± 0.10 | 9.61 ± 0.49 |
| I32A | 4 | 8.95 ± 0.14 | 9.63 ± 0.13* | K51A | 4 | 9.45 ± 0.37 | 9.66 ± 0.25 |
| Q33A | 3 | 9.28 ± 0.22 | 9.04 ± 0.03 | I52A | 6 | 9.79 ± 0.24 | 9.86 ± 0.29 |
| L34A | 7 | 9.55 ± 0.30 | 9.51 ± 0.23 | M53A | 5 | 9.72 ± 0.30 | 10.20 ± 0.24 |
| G35A | 6 | 9.07 ± 0.12 | 10.09 ± 0.24** | Q54A | 4 | 9.38 ± 0.15 | 9.60 ± 0.33 |
| V36A | 4 | 9.27 ± 0.18 | 9.90 ± 0.30 | D55A | 5 | 9.33 ± 0.30 | 9.76 ± 0.13 |
| T37A | 5 | 9.15 ± 0.17 | 10.08 ± 0.23* | P56A | 3 | 9.74 ± 0.14 | 9.21 ± 0.18 |
| R38A | 5 | 9.17 ± 0.15 | 9.32 ± 0.35 | I57A | 3 | 9.23 ± 0.27 | 9.27 ± 0.33 |
| N39A | 4 | 9.24 ± 0.34 | 9.55 ± 0.42 | Q58A | 6 | 10.12 ± 0.36 | 10.28 ± 0.37 |
| K40A | 4 | 9.61 ± 0.15 | 10.01 ± 0.25 | Q59A | 5 | 9.68 ± 0.26 | 9.70 ± 0.32 |
| I41A | 7 | 10.02 ± 0.34 | 9.22 ± 0.096* | A60L | 6 | 9.65 ± 0.44 | 9.56 ± 0.43 |
Values are means ± s.e.m. *, **, p > 0.05 and 0.01 compared to WT, Student's, t-test.
Fig. 2.
CGRP-stimulated cAMP response of the mutated receptors where there is either an increase in potency or constitutive activity. (a) I32A, (b) G35A, (c) T37A and (d) K51A. Cos-7 cells were transfected with WT/RAMP1 or mutant/RAMP1 and assayed for CGRP-stimulated cAMP production. WT type receptors; open squares. Mutant receptors; as indicated. Data are representative of three to five similar experiments.
3.2. Receptor expression and radioligand binding
A change in potency could be either because the mutants no longer bound CGRP with high affinity, or because the receptors were not at the cell surface. Accordingly, cell surface expression of receptors was measured (Table 2). Large reductions were seen for Gln45Ala, Cys48Ala and Tyr49Ala with small reductions for Thr43Ala and Ala44Leu. Efficient expression of CLR requires its association with RAMP1 for transport to the cell surface [19]. Total CLR production as measured for these mutants by a whole-cell ELISA was only significantly reduced for Ala44Leu (81.7 ± 6.5% of WT) and Tyr49Ala (77.6 ± 3.8% of WT). This suggests that all these mutants are synthesized with reasonable efficiency so the reduction in cell surface expression is because of defective trafficking or insertion into the membrane. Whilst nine mutants showed a significant increase in cell surface expression, only for Arg38Ala and Tyr46Ala was this increase greater than 50% of the WT; these were also the only two mutants to show a greater than 50% increase in total cell expression (158.8 ± 7.6% and 196.6 ± 30.0% of WT respectively, Table 3).
Table 2.
Cell surface expression and binding properties of mutant receptors.
| Mutant | % Expression | % Binding | Mutant | % Expression | % Binding |
|---|---|---|---|---|---|
| E23A | 108.7 ± 5.1 | 81.7 ± 4.0 | M42A | 83.2 ± 6.3 | 97.2 ± 12.0 |
| L24A | 111.1 ± 9.2 | 101.6 ± 3.5 | T43A | 83.6 ± 5.3* | 87.6 ± 12.3 |
| E25A | 143.0 ± 16.4* | 90.6 ± 12.5 | A44L | 82.7 ± 6.0* | 4.3 ± 1.6* |
| E26A | 123.7 ± 8.6 | 71.9 ± 10 | Q45A | 66.6 ± 3.3*** | 61.4 ± 3.7 |
| S27A | 117.1 ± 7.5 | 116.4 ± 22.2 | Y46A | 167.0 ± 8.6*** | 71.4 ± 16.7 |
| P28A | 95.9 ± 9.9 | 90.2 ± 14 | E47A | 81.7 ± 4.7 ** | 90 ± 10.7 |
| E29A | 108.6 ± 9.7 | 90.5 ± 0.8 | C48A | 45.4 ± 3.9*** | 2.7 ± 1.2* |
| D30A | 123.4 ± 6.3** | 74.1 ± 5 | Y49A | 33.8 ± 5.6*** | 5.5 ± 3.1* |
| S31A | 111.4 ± 9.7 | 98.6 ± 2.4 | Q50A | 98.5 ± 9.4 | 93.5 ± 2.5 |
| I32A | 114.3 ± 9.9 | 114.7 ± 13.7 | K51A | 82.0 ± 8.4 | 85.9 ± 6.7 |
| Q33A | 141.0 ± 8.3*** | 59.2 ± 14.2 | I52A | 123.9 ± 5.8* | 86.2 ± 1.9 |
| L34A | 129.4 ± 14.6 | 103 ± 14.2 | M53A | 109.6 ± 5.1 | 95.8 ± 6.7 |
| G35A | 147.6 ± 9.2** | 103 ± 8.4 | Q54A | 106.5 ± 9.3 | 103.1 ± 15.8 |
| V36A | 116.1 ± 9.3* | 87.8 ± 6 | D55A | 100.5 ± 7.9 | 86.7 ± 10.4 |
| T37A | 108.5 ± 8.6 | 88.3 ± 8.1 | P56A | 131.1 ± 7.0** | 83.3 ± 8.8 |
| R38A | 161.7 ± 9.0*** | 89.0 ± 8.6 | I57A | 105.4 ± 9.9 | 102.1 ± 4.3 |
| N39A | 147.6 ± 10.0** | 94.9 ± 13.5 | Q58A | 88.0 ± 10.6 | 78.8 ± 7.9 |
| K40A | 83.9 ± 4.0 | 91.1 ± 9.0 | Q59A | 102.9 ± 7.9 | 82.1 ± 6.6 |
| I41A | 133.6 ± 8.7 | 2.0 ± 2.7 | A60L | 79.4 ± 7.1 | 104.9 ± 6 |
Expression shows cell surface expression of HA-tagged receptors, 3–6 experiments; WT expression = 100%. *, **, ***, p > 0.05, 0.01 and 0.001 compared to WT, Mann–Whitney. Binding shows % specific binding of 125I-CGRP; WT receptor binding = 100%, n = 3. Values are means ± s.e.m. *, **, ***, p > 0.05, 0.01 and 0.001 compared to WT, Student's t-test.
Table 3.
Total expression of mutated receptors.
| Mutant | N | Total expression (% of WT) | Mutant | N | Total expression (% of WT) |
|---|---|---|---|---|---|
| E23A | 9 | 91.4 ± 3.4 | I41A | 9 | 100.1 ± 6.7 |
| E25A | 9 | 88.7 ± 1.7 | T43A | 15 | 116.0 ± 9.4 |
| E47A | 9 | 99.5 ± 2.0 | A44L | 9 | 81.7 ± 6.5* |
| D30A | 12 | 141.5 ± 11.0* | Q45A | 14 | 109.1 ± 13.8 |
| I32A | 9 | 138.1 ± 7.6*** | Y46A | 15 | 196.6 ± 30.0*** |
| Q33A | 9 | 135.0 ± 11.9 | C48A | 9 | 91.5 ± 3.08 |
| G35A | 12 | 126.7 ± 5.3** | Y49A | 9 | 77.6 ± 3.8* |
| T37A | 9 | 112.9 ± 9.3 | I52A | 9 | 176.1 ± 13.3* |
| R38A | 12 | 158.8 ± 7.6*** | P56A | 9 | 111.6 ± 14.9 |
| N39A | 12 | 130.6 ± 7.1** |
Total cellular expression of mutants that showed a significant change in cell surface expression were measured by a whole-cell ELISA. Values are expressed as % WT expression and are means ± s.e.m. **p > 0.05 and 0.01 compared to WT, Mann–Whitney test.
3.3. Radioligand binding
For four mutants, Ile41Ala, Ala44Leu, Cys48Ala and Tyr49Ala, high affinity CGRP binding was abolished (Table 2), consistent with the reduced potency and/or expression shown by these receptors. It should be noted that radioligand binding assays only detect high affinity binding (Kd < 10 nM), whereas a functional assay will continue to respond to much weaker associations; hence the apparent discrepancy that Ile41Ala and Cys48Ala give a cAMP response in the absence of detectable binding of radioligand There was a reduction in the binding seen with Gln45Ala, although this did not reach significance.
3.4. Receptor internalization
Following an hour pre-treatment with 100 nM CGRP, the cell surface expression of the WT receptor was typically reduced by about 60% (Table 4). Fourteen mutants showed changes in receptor internalization. Large effects were seen with Ile41Ala, Ala44Leu, Gln45Ala, Cys48Ala and Tyr49Ala, where it was either greatly impaired or totally abolished. These residues show impaired cAMP responsiveness, confirming their importance.
Table 4.
Internalization of receptors.
| Mutant | N | % Internalization |
Mutant | N | % Internalization |
||
|---|---|---|---|---|---|---|---|
| WT | Mutant | WT | Mutant | ||||
| E23A | 14 | 60.87 ± 5.94 | 61.15 ± 5.45 | M42A | 15 | 52.15 ± 6.68 | 60.32 ± 5.58 |
| L24A | 14 | 57.47 ± 6.38 | 63.81 ± 3.77 | T43A | 9 | 66.03 ± 5.30 | 69.75 ± 3.56** |
| E25A | 12 | 57.65 ± 3.92 | 57.42 ± 4.45 | A44L | 12 | 71.16 ± 2.30 | 51.67 ± 3.59 |
| E26A | 12 | 71.10 ± 5.76 | 71.66 ± 3.79 | Q45A | 9 | 54.84 ± 2.69 | 27.45 ± 7.51** |
| S27A | 12 | 58.40 ± 6.98 | 53.36 ± 7.22 | Y46A | 9 | 89.19 ± 2.05 | 56.83 ± 3.73*** |
| P28A | 14 | 62.65 ± 5.48 | 58.27 ± 2.85 | E47A | 9 | 68.72 ± 2.89 | 61.44 ± 2.85 |
| E29A | 21 | 63.40 ± 5.64 | 60.34 ± 2.86 | C48A | 15 | 57.04 ± 3.36 | −14.65 ± 4.2*** |
| D30A | 9 | 65.40 ± 2.68 | 57.55 ± 5.80* | Y49A | 18 | 49.80 ± 4.81 | 0.00 ± 9.34*** |
| S31A | 18 | 59.86 ± 5.98 | 64.01 ± 5.71 | Q50A | 15 | 68.58 ± 3.42 | 65.79 ± 3.06 |
| I32A | 15 | 64.96 ± 5.86 | 46.42 ± 4.43* | K51A | 9 | 73.14 ± 2.87 | 76.48 ± 3.57 |
| Q33A | 15 | 63.92 ± 4.20 | 52.42 ± 4.14* | I52A | 14 | 75.73 ± 4.89 | 51.41 ± 4.06*** |
| L34A | 11 | 54.84 ± 4.78 | 48.11 ± 4.84 | M53A | 9 | 68.28 ± 4.54 | 65.22 ± 3.41 |
| G35A | 20 | 62.51 ± 5.34 | 61.18 ± 3.24 | Q54A | 18 | 65.97 ± 6.18 | 63.03 ± 4.29 |
| V36A | 16 | 55.77 ± 6.11 | 44.86 ± 8.79 | D55A | 12 | 72.80 ± 1.96 | 65.32 ± 2.74 |
| T37A | 12 | 61.89 ± 5.12 | 54.01 ± 5.50 | P56A | 15 | 72.16 ± 2.78 | 56.22 ± 2.71*** |
| R38A | 15 | 54.54 ± 5.99 | 33.79 ± 5.02* | I57A | 15 | 68.28 ± 6.03 | 59.61 ± 7.04 |
| N39A | 13 | 54.44 ± 3.13 | 62.20 ± 5.60 | Q58A | 9 | 69.32 ± 4.83 | 75.50 ± 3.29 |
| K40A | 9 | 67.16 ± 5.22 | 64.61 ± 4.02 | Q59A | 9 | 49.42 ± 3.45 | 59.90 ± 3.33 |
| I41A | 9 | 50.06 ± 6.03 | 16.92 ± 9.94*** | A60L | 9 | 55.80 ± 3.23 | 57.09 ± 4.14 |
The internalization of the WT receptor (as assessed by cell surface ELISA) was compared with that of each mutant in paired experiments. Values are % internalization, normalized against the cell surface expression prior to addition of CGRP and are means ± s.e.m, n = 9–18 derived from 3 to 6 independent experiments. *, **, ***, p > 0.05, 0.01 and 0.001, Mann–Whitney.
3.5. Molecular modeling
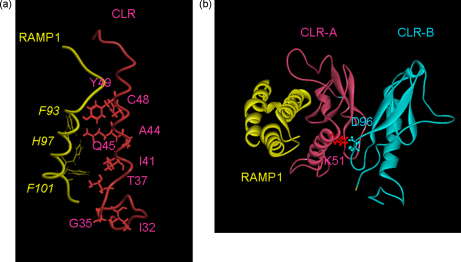
To assist with the interpretation of the mutagenesis, a speculative model has been produced of the CLR/RAMP1 complex (Fig. 3). It is proposed that the CGRP receptor ECD is an asymmetrical trimer, consistent with recent data [12].
Fig. 3.
Model of the CLR: RAMP1 complex. (a) RAMP1 and CLR ECD-A interface residues in italics are on RAMP1, those in normal type are on CLR. (b) Side view of the predicted orientations of the subunits of CGRP receptor ECD; the proposed interaction of K51 with D96 is also shown for one of the CLR pairs.
An initial homology model of the CLR ECD was built based on the gastric inhibitory peptide receptor- and parathyroid hormone type 1 receptor-ECDs [22,25], as described in the methods. The CLR ECD contains three loop regions, loop1 (Asp55-Arg67), loop2 (Asn76-Gly81) and loop3 (Pro89-Ser98). These were modeled with the program LOOPY [5]. A conformation from the top 10 structures was selected based on the architecture of the loops in other family B GPCR ECDs and the total energy potential score of the loop conformer, as described in the methods.
The CLR ECD was docked against itself to produce 1000 complexes. The top 10 complexes were visually inspected and assessed based on molecule orientation. The C-terminus region of both the CLR ECDs (CLR ECD-A and CLR ECD-B, respectively) are predicted to face the same direction, as would be expected in vivo, as this region in both molecules would be expected to be facing the lipid bilayer. A complex that satisfied these requirements was selected. The dimer interface was refined using the MULTIDOCK program from the 3D-Dock suite as described in the methods.
The model suggests that the two CLR ECDs dock together to produce a roughly symmetrical complex. The interface between them is located in loop3 between Pro89 and Ser98. The corresponding loop in the mouse corticotrophin receptor ECD is predicted to be relatively stable as evidenced by very broad cross-peaks in the NMR spectrum [10]. Consequently, this may be a plausible docking region of the two molecules, which could be further stabilized by neighboring beta strands contributed by both CLR molecules. CLR has three putative N-glycosylation sites; Asn66, Asn118 and Asn123; glycosylation of either Asn66 or Asn118 seems essential for normal pharmacological functioning [11]. On the model, all three sites on the CLR ECD-B are freely accessible for post-translational modifications.
4. Discussion
The results of this investigation suggest a small number of residues in the extreme N-terminus of CLR are important for receptor function, in particular, Ile41, Ala44, Gln45, Cys48 and Tyr49. Mutation of these residues either disrupts CGRP-stimulated cAMP production, CGRP binding or CLR expression at the cell surface. There is also some evidence for a second cluster of important residues consisting of Ile32, Gly35 and Thr37.
Cys48 is predicted to take part in one of the highly conserved disulfide bonds that characterize the ECDs of family B GPCRs and its mutation to alanine in other receptors also causes a loss of signaling [18]. The other residues, apart from Tyr49 are not widely conserved and so must have receptor-specific roles.
The extreme N-terminus of the CLR is predicted to adopt an alpha helical conformation between Asn39 and Gln54. Ile41, Gln45 and Tyr49 are predicted to lie on the same face of the extreme N-terminus helix. Gln45A and Tyr49A appear to be expressed normally within the cell but are not trafficked to the cell surface. As cell surface expression of CLR requires association with RAMPs, it is possible that the mutations block this process. Our model suggests that the CLR N-terminal helix of CLR-A is predicted to dock against helix 3 of RAMP1. It has previously been suggested that Phe93, His97 and Phe101 of RAMP1 participate in a CLR binding interface [16,28]. The model predicts that a hydrogen bond occurs between the side chain amide of Gln45 of CLR-A and the epsilon nitrogen of His97 of RAMP1; the residues are appropriately orientated to allow this interaction. Tyr49 of CLR-A and Phe93 of RAMP1 may pack together.
Ile41 appears to be too far away from RAMP1 to make a significant hydrophobic interaction, consistent with its normal cell surface expression. However, Ile41 impairs cAMP accumulation and specific binding suggesting that it is involved directly in CGRP binding. Therefore, the RAMP1/CLR-A interface may also provide a platform for CGRP docking. This is further supported by Ala44, which is predicted to face away from RAMP1, as Ala44Leu selectively decreases CGRP responsiveness with little effect on the expression of the CGRP receptor. It is possible that Gln45 and Tyr49 may also participate in CGRP binding, in addition to their roles in interacting with RAMP1. In the presence of 50% trifluoroethanol (TFE) CGRP has an 80% α helical content [27]; our model suggests that there is sufficient space to accommodate a linear peptide (corresponding to an extended form of CGRP) in the groove between RAMP1 and the α helix of the A monomer of CLR.
Ile32, Gly35 and Thr37 form a cluster of residues on the extreme N-terminus of CLR that increase in CGRP potency. At this part of the N-terminus, the accuracy of family B alignments decreases. Accordingly, it is difficult to use the model to make any useful comments as to the mechanism for this effect, beyond noting that they cluster at the base of the known helix. However, it suggests that there are extended contact points for CGRP along the entire region. Deletion of the first 18 residues of this section (as far as Lys40) gives a receptor that cannot respond to CGRP [15] and an alanine scan of the same region found that the mutants Leu24Ala and Leu34Ala decreased CGRP potency, but not binding [2]. Whilst there is agreement with the current study that this part of the N-terminus has a role in CGRP binding, the residues identified are different. There may be cell-line specific factors at work; as the authors observed a decrease in binding but not in signaling, their receptors may have been very efficiently coupled to Gs.
Lys51, which increased basal activity, faces away from RAMP1, towards Asp96 on the second CLR of the dimer (Fig. 3). It is possible that the mutation Lys51A disrupts the CLR dimer interface leading to an alteration in the conformation of the N-terminus and so to enhanced constitutive activity.
For a number of mutants, there was an increase in cell surface expression. This generally correlates with an increase in total expression. At present, it is difficult to suggest any detailed mechanism for this.
It has been proposed that family B GPCRs share a common fold in their ECDs [23]. Consequently, the RAMP interface identified here may also be shared amongst other family B GPCRs that have been found to complex with the RAMP family [3]. It is clear that there are at differences as to how different RAMPs interact with CLR or other GPCRs. For example, residues 23–40 of CLR can be replaced by the corresponding sequence of the porcine calcitonin receptor and still form a functional CGRP receptor; the same replacement is not enough to preserve the RAMP2 interaction and adrenomedullin binding [15]. It is plausible that the overall architecture of the GPCR–RAMP complex may be conserved for these receptors, but the influence of different RAMPs may alter the position of the extreme N-terminus to reveal or mask certain binding epitopes.
In conclusion, this study has revealed a cluster of residues likely to lie on a helix at the extreme N-terminus of CLR that are important for RAMP association and the direct binding of CGRP.
Acknowledgements
We would like to thank Dr. Dan Rathbone (Aston University) and Dr. John Simms (Monash University) for helpful discussions. This work was funded by a studentship awarded to James Barwell by the British Heart Foundation (FS/05/054) and a BBSRC CASE award in collaboration with GlaxoSmithKline to Philip Miller.
Footnotes
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.peptides.2009.10.021
Contributor Information
James Barwell, Email: Barwellj@aston.ac.uk.
Philip S. Miller, Email: philipsaxon@gmail.com.
Dan Donnelly, Email: D.Donnelly@leeds.ac.uk.
David R. Poyner, Email: D.R.Poyner@aston.ac.uk.
Appendix A. Supplementary data
Display e-component
References
- 1.Aiyar N., Daines R.A., Disa J., Chambers P.A., Sauermelch C.F., Quiniou M. Pharmacology of SB-273779, a nonpeptide calcitonin gene-related peptide 1 receptor antagonist. J Pharmacol Exp Ther. 2001;296:768–775. [PubMed] [Google Scholar]
- 2.Banerjee S., Evanson J., Harris E., Lowe S.L., Thomasson K.A., Porter J.E. Identification of specific calcitonin-like receptor residues important for calcitonin gene-related peptide high affinity binding. BMC Pharmacol. 2006;6:9. doi: 10.1186/1471-2210-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Christopoulos A., Christopoulos G., Morfis M., Udawela M., Laburthe M., Couvineau A. Novel receptor partners and function of receptor activity-modifying proteins. J Biol Chem. 2003;278:3293–3297. doi: 10.1074/jbc.C200629200. [DOI] [PubMed] [Google Scholar]
- 4.Christopoulos G., Perry K.J., Morfis M., Tilakaratne N., Gao Y., Fraser N.J. Multiple amylin receptors arise from receptor activity-modifying protein interaction with the calcitonin receptor gene product. Mol Pharmacol. 1999;56:235–242. doi: 10.1124/mol.56.1.235. [DOI] [PubMed] [Google Scholar]
- 5.Soto C.S., Fasnacht M., Zhu J., Forrest L., Honig B. Loop modeling: sampling, filtering, and scoring. Proteins: Struct Funct Bioinform. 2008;70:834–843. doi: 10.1002/prot.21612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conner A.C., Hay D.L., Simms J., Howitt S.G., Schindler M., Smith D.M. A key role for transmembrane prolines in calcitonin receptor-like receptor agonist binding and signalling: implications for family B G-protein-coupled receptors. Mol Pharmacol. 2005;67:20–31. [PubMed] [Google Scholar]
- 7.Conner M., Hicks M.R., Dafforn T., Knowles T.J., Ludwig C., Staddon S. Functional and biophysical analysis of the C-terminus of the CGRP-receptor; a family B GPCR. Biochemistry. 2008;47:8434–8444. doi: 10.1021/bi8004126. [DOI] [PubMed] [Google Scholar]
- 8.Evans B.N., Rosenblatt M.I., Mnayer L.O., Oliver K.R., Dickerson I.M. CGRP-RCP, a novel protein required for signal transduction at calcitonin gene-related peptide and adrenomedullin receptors. J Biol Chem. 2000;275:31438–31443. doi: 10.1074/jbc.M005604200. [DOI] [PubMed] [Google Scholar]
- 9.Fiser A., Sali A., Charles W., Carter JaRM.S. Methods in enzymology. Academic Press; 2003. Modeller: generation and refinement of homology-based protein structure models. pp. 461–91. [DOI] [PubMed] [Google Scholar]
- 10.Grace C.R., Perrin M.H., Gulyas J., Digruccio M.R., Cantle J.P., Rivier J.E. Structure of the N-terminal domain of a type B1 G protein-coupled receptor in complex with a peptide ligand. Proc Natl Acad Sci USA. 2007;104:4858–4863. doi: 10.1073/pnas.0700682104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gujer R., Aldecoa A., Buhlmann N., Leuthauser K., Muff R., Fischer J.A. Mutations of the asparagine117 residue of a receptor activity-modifying protein 1-dependent human calcitonin gene-related peptide receptor result in selective loss of function. Biochemistry. 2001;40:5392–5398. doi: 10.1021/bi002497v. [DOI] [PubMed] [Google Scholar]
- 12.Heroux M., Hogue M., Lemieux S., Bouvier M. Functional calcitonin gene-related peptide receptors are formed by the asymmetric assembly of a calcitonin receptor-like receptor homo-oligomer and a monomer of receptor activity-modifying protein-1. J Biol Chem. 2007;282:31610–31620. doi: 10.1074/jbc.M701790200. [DOI] [PubMed] [Google Scholar]
- 13.Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS, 4 algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 14.Ittner L.M., Koller D., Muff R., Fischer J.A., Born W. The N-terminal extracellular domain 23–60 of the calcitonin receptor-like receptor in chimeras with the parathyroid hormone receptor mediates association with receptor activity-modifying protein 1. Biochemistry. 2005;44:5749–5754. doi: 10.1021/bi048111o. [DOI] [PubMed] [Google Scholar]
- 15.Koller D., Born W., Leuthauser K., Fluhmann B., McKinney R.A., Fischer J.A. The extreme N-terminus of the calcitonin-like receptor contributes to the selective interaction with adrenomedullin or calcitonin gene-related peptide. FEBS Lett. 2002;531:464–468. doi: 10.1016/s0014-5793(02)03585-8. [DOI] [PubMed] [Google Scholar]
- 16.Kusano S., Kukimoto-Niino M., Akasaka R., Toyama M., Terada T., Shirouzu M. Crystal structure of the human receptor activity-modifying protein 1 extracellular domain. Protein Sci. 2008;17:1907–1914. doi: 10.1110/ps.036012.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laskowski R.A. PDBsum: summaries and analyses of PDB structures. Nucleic Acids Res. 2001;29:221–222. doi: 10.1093/nar/29.1.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lisenbee C.S., Dong M., Miller L.J. Paired cysteine mutagenesis to establish the pattern of disulfide bonds in the functional intact secretin receptor. J Biol Chem. 2005;280:12330–12338. doi: 10.1074/jbc.M414016200. [DOI] [PubMed] [Google Scholar]
- 19.McLatchie L.M., Fraser N.J., Main M.J., Wise A., Brown J., Thompson N. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 20.Moont G., Gabb H.A., Sternberg M.J. Use of pair potentials across protein interfaces in screening predicted docked complexes. Proteins. 1999;35:364–373. [PubMed] [Google Scholar]
- 21.Muff R., Buhlmann N., Fischer J.A., Born W. An amylin receptor is revealed following co-transfection of a calcitonin receptor with receptor activity modifying proteins-1 or -3. Endocrinology. 1999;140:2924–2927. doi: 10.1210/endo.140.6.6930. [DOI] [PubMed] [Google Scholar]
- 22.Parthier C., Kleinschmidt M., Neumann P., Rudolph R., Manhart S., Schlenzig D. Crystal structure of the incretin-bound extracellular domain of a G protein-coupled receptor. Proc Natl Acad Sci USA. 2007;104:13942–13947. doi: 10.1073/pnas.0706404104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parthier C., Reedtz-Runge S., Rudolph R., Stubbs M.T. Passing the baton in class B GPCRs: peptide hormone activation via helix induction? Trends Biochem Sci. 2009;34:303–310. doi: 10.1016/j.tibs.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.de Bakker P.I.W., Depristo M.A., Burke D.F., Blundell T.L. Ab initio construction of polypeptide fragments: accuracy of loop decoy discrimination by an all-atom statistical potential and the AMBER force field with the Generalized Born solvation model. Proteins: Struct Funct Genet. 2003;51:21–40. doi: 10.1002/prot.10235. [DOI] [PubMed] [Google Scholar]
- 25.Pioszak A.A., Xu H.E. Molecular recognition of parathyroid hormone by its G protein-coupled receptor. Proc Natl Acad Sci USA. 2008;105:5034–5039. doi: 10.1073/pnas.0801027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qiu D., Shenkin P.S., Hollinger F.P., Still W.C. The GB/SA continuum model for solvation. A fast analytical method for the calculation of approximate born radii. J Phys Chem A. 1997;101:3005–3014. [Google Scholar]
- 27.Robinson S.D., Aitken J.F., Bailey R.J., Poyner D.R., Hay D.L. Novel peptide antagonists of adrenomedullin and calcitonin gene-related peptide receptors; identification, pharmacological characterization and interactions with position 74 in RAMP1/3. J Pharmacol Exp Ther. 2009 doi: 10.1124/jpet.109.156448. [DOI] [PubMed] [Google Scholar]
- 28.Simms J., Hay D.L., Bailey R.J., Konycheva G., Bailey G., Wheatley M. Structure-function analysis of RAMP1 by alanine mutagenesis. Biochemistry. 2009;48:198–205. doi: 10.1021/bi801869n. [DOI] [PubMed] [Google Scholar]
- 29.Yuedong Yang Y.Z. Ab initio folding of terminal segments with secondary structures reveals the fine difference between two closely related all-atom statistical energy functions. Protein Sci. 2008;17:1212–1219. doi: 10.1110/ps.033480.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.