Abstract
Glucocorticoid (GC) insensitivity represents a profound challenge in managing patients with asthma. The mutual inhibition of transcriptional activity between GC receptor (GR) and other regulators is one of the mechanisms contributing to GC resistance in asthma. We recently reported that interferon regulatory factor (IRF)-1 is a novel transcription factor that promotes GC insensitivity in human airway smooth muscle (ASM) cells by interfering with GR signaling (Tliba et al., Am J Respir Cell Mol Biol 2008;38:463–472). Here, we sought to determine whether the inhibition of GR function by IRF-1 involves its interaction with the transcriptional co-regulator GR-interacting protein 1 (GRIP-1), a known GR transcriptional co-activator. We here found that siRNA-mediated GRIP-1 depletion attenuated IRF-1–dependent transcription of the luciferase reporter construct and the mRNA expression of an IRF-1–dependent gene, CD38. In parallel experiments, GRIP-1 silencing significantly reduced GR-mediated transactivation activities. Co-immunoprecipitation and GST pull-down assays showed that GRIP-1, through its repression domain, physically interacts with IRF-1 identifying GRIP-1 as a bona fide transcriptional co-activator for IRF-1. Interestingly, the previously reported inhibition of GR-mediated transactivation activities by either TNF-α and IFN-γ treatment or IRF-1 overexpression was fully reversed by increasing cellular levels of GRIP-1. Together, these data suggest that the cellular accumulation of IRF-1 may represent a potential molecular mechanism mediating altered cellular response to GC through the depletion of GRIP-1 from the GR transcriptional regulatory complexes.
Keywords: glucocorticoid, cytokine, airway smooth muscle, IRF-1, GRIP-1
CLINICAL RELEVANCE.
In steroid-resistant asthma, inflammatory factors lead to an excess of transcription factors that antagonize steroid signaling via the competition with glucocorticoid receptor (GR)-associated co-activators. We here showed that competition between interferon regulatory factor-1 and GR for GR interacting protein-1 is a novel molecular mechanism promoting steroid dysfunction in inflammatory diseases.
Glucocorticoids (GCs) remain the cornerstone treatment for chronic inflammatory diseases such as asthma (1). Five to ten percent of patients with asthma, however, develop steroid insensitivity. Therefore, an unmet need requires the exploration of alternative therapeutic targets to be used in conjunction with or separately from GCs (2). A strong correlation between steroid insensitivity and inflammatory diseases, such as asthma, nasal polyps, and inflammatory bowel disease (3–5), prompted investigators to examine whether inflammatory mediators modulate the cellular responses to steroids. Using peripheral blood mononuclear cells (PBMCs), investigators showed that GC effects were dramatically reduced in the presence of cytokines. Kam and colleagues were the first to show that IL-2 and IL-4 reduced the inhibitory effect of methylprednisolone on mitogen-induced T cell proliferation (6). Other cytokines such as IL-1β, IL-6, IFN-γ, IL-2, IL-7, IL-13, IL-15, or IL-8 also attenuate dexamethasone effects in PBMCs (7–10), proliferating T cells, monocytes, and neutrophils. Thus, cytokines may promote the development of steroid insensitivity seen in patients with asthma by reducing cell/tissue sensitivity to GCs.
Despite considerable efforts in immune cells, the alteration of steroid responsiveness in structural cell types of the target tissues remains poorly defined. We have been studying the modulation of steroid responsiveness in airway smooth muscle (ASM), which is increasingly recognized as an important player in the pathogenesis of asthma by driving airway inflammation (11) and may therefore be a target for inhaled GCs (12). Accordingly, we and others showed that GCs were effective in abrogating the expression of a number of proinflammatory cytokines, chemokines, and adhesion molecules in ASM cells when exposed to a “single” proinflammatory stimulus (13). Yet, we recently found that steroid insensitivity can develop in ASM exposed to a “mixture” of proasthmatic cytokines. Treatment of ASM cells with the specific combination of IFNs with TNF-α impairs the ability of GCs to inhibit the expression of calcium regulatory protein CD38, the chemokines RANTES and fractalkine, and cell surface proteins such as intercellular adhesion molecule-1 and Toll-like receptor-2 (8).
The mechanisms underlying cytokine-induced steroid insensitivity in ASM cells have not been completely defined, but we recently showed that a “short-term” treatment of ASM cells with IFNs and TNF-α partially inhibits steroid transactivation through the accumulation of interferon regulatory factor (IRF)-1 (14), an early response gene involved in diverse transcriptional regulatory processes (15). Interestingly, polymorphism in IRF-1 has been associated with childhood atopic asthma (16). Because expression of IRF-1 is increased after viral infections (17) and because IRF-1 suppresses steroid effectiveness in ASM cells (14), we proposed that IRF-1 may mediate the reduced steroid responsiveness seen in patients with asthma experiencing viral infections (18).
The precise transcriptional mechanism by which IRF-1 interferes with GC signaling remains to be determined. Most anti-inflammatory effects of steroids are conferred by the GC receptor (GR), a ligand-dependent transcriptional regulator that suppresses the expression of inflammatory genes (3). Steroid receptor coactivator (SRC)/p160 family members (SRC1, SRC2/transcriptional intermediary factor 2 [TIF2]/GR interacting protein-1 [GRIP-1], and SRC3/receptor-associated co-activator 3 [RAC3]/p300/CREB binding protein [CBP]-co-integrator protein [pCIP]/amplifie in breast 1 [AIB1]) serve as co-activators for all nuclear receptors including GR. As such, this family of proteins interacts with ligand-bound GR to recruit histone acetyltransferases (CBP/p300) and coactivator-associated arginine methyltransferase 1 (CARM1) that unpack the condensed chromatin, thereby facilitating the access of transcription factors to target genes. Unlike other p160 s, GRIP-1 also possesses a unique GR co-repressor activity facilitating GC-mediated repression of activator protein (AP)-1 and NF-κB activities (19, 20). Although originally identified as a nuclear receptor co-factor, GRIP-1 was later shown to engage in physical and functional interactions with an IRF family member (IRF-3) and serves as an IRF-3 co-activator in macrophages (21). However, the role of GRIP-1 in ASM cells has not been assessed. Here we characterized whether GRIP-1 is a component of the IRF-1 transcriptional regulatory complexes and whether such interactions affect the response of ASM cells to GCs.
MATERIALS AND METHODS
ASM Cell Culture and Characterization
Primary human ASM cells were isolated from the trachealis muscle of lung transplant donors and purified as described (22) in accordance with procedures approved by the University of Pennsylvania Committee on Studies Involving Human Beings.
Transfection of ASM Cells
ASM cells were transfected using Basic Nucleofector Kit for Primary Smooth Muscle Cells according to manufacturer's instructions using Amaxa Nucleofector II device (program U-25; Amaxa Biosystems, Cologne, Germany) (14). Small interfering RNA experiments were performed using siRNA Test Kit for Cell Lines and Adherent Primary Cells according to manufacturer's instructions (Amaxa Biosystems) (14). ASM cells were transfected (14) using 2 μg of IRF-1 (kindly provided by Dr. Yokosawa, Hokkaido University, Japan [23]), GRIP-1 (21), and/or 2 μg of GC-responsive element (GRE)-dependent luciferase reporter (Clontech Laboratories Inc., Mountain View, CA), and/or 2 μg of IRF-1–dependent luciferase reporter (Panomics, Inc., Fremont, CA), and/or 100 nM of the combination of three different Silencer Pre-designed siRNAs to GRIP-1 (CACAGGAAUGAUUGGUAA, GGAGAUUGAUAGAGCCUUA, GACAAGGGUUGAAUAUGA; Santa Cruz Biotechnology, Santa Cruz, CA), and 1 μg of β-galactosidase vector (to normalize transfection efficiency) (Promega, Madison, WI). Controls included pcDNA3 empty vector (Stratagene, La Jolla, CA) and/or scrambled siRNA (Santa Cruz Biotechnology). The activities of luciferase and β-galactosidase were evaluated using luciferase and β-galactosidase detection kits (Promega), respectively, according to the manufacturer's instructions (14). The reporter luciferase activities were normalized to β-galactosidase activity and expressed as relative luminescence unit (RLU). Data were then expressed as percentage of controls (means ± SEM).
Immunoblotting and Immunoprecipitations
Immunoblot analysis for GRIP-1, IRF-1, and GR was performed as described (14). To ensure equal loading, the membranes were stripped and reprobed with anti–β-actin antibody (Santa Cruz Biotechnology). Immunoprecipitations using the IRF-1, GRIP-1 (Santa Cruz Biotechnology), and GR (Affinity BioReagents, Golden, CO) antibodies were performed as described (14). To quantify GRα/GRIP-1 association in co-immunoprecipitation experiments, a semiquantitative measure was performed. To this goal, the densitometry of three Western blot replicates was assessed using on a Gel-Pro Analyzer (Media Cybernetics, Silver Spring, MD). The background was subtracted from the respective band intensity to obtain the net intensity of GRIP-1 and GRα. The net bound GRIP1 was normalized by its respective net GRα to correct for different loading. The relative change in normalized bound GRIP-1 in fluticasone propionate (FP)-treated cells compared with untreated cells was calculated as follows: {[GRIP-1 band intensity − GRIP-1 background]/[GRα band intensity − GRα background]} in FP-treated cells divided by {[GRIP-1 band intensity − GRIP-1 background]/[GRα band intensity − GRα background]} in untreated cells.
In Vitro Binding
Glutathione S-Transferase (GST)-tagged IRF-1 was expressed in Escherichia coli, purified, and tested for its ability to interact with in vitro transcribed/translated GRIP-1 derivatives as described (21). The gels were stained with Coomassie blue (not shown) and autoradiographed.
RT-PCR Analysis
Total RNAs were extracted from human ASM cells using RNeasy Mini Kit (Qiagen, Valencia, CA) as previously described (14), reverse-transcribed, and subjected to PCR with CD38, MPK-1, and GAPDH primers as reported earlier (14, 24, 25). GRIP-1 primers were purchased from Santa Cruz Biotechnology. Linear amplification range was determined in preliminary experiments; therefore, only one representative gel within the linear range is shown. To quantify the data, the intensity of the area density of each PCR band was assessed on a Gel-Pro Analyzer (Media Cybernetics), and expressed as a ratio of area density of MKP-1 or CD38 to GAPDH. All experiments were performed in three different cell lines.
Materials and Reagents
Tissue culture reagents were obtained from Invitrogen (Carlsbad, CA). Human recombinant (r) TNF-α and rIFN-γ were provided by Roche Diagnostics (Indianapolis, IN). FP was purchased from Sigma (St. Louis, MO).
Statistical Analysis
Data points from individual assays represent the mean values of triplicate measurements. Significant differences among groups were assessed with ANOVA (Bonferroni-Dunn test) or by t test analysis, with values of P < 0.05 sufficient to reject the null hypothesis for all analyses. Each set of experiments was performed with a minimum of three different human ASM cell lines.
RESULTS
GRIP-1 Is Essential for GR-Dependent Gene Transcription in ASM Cells
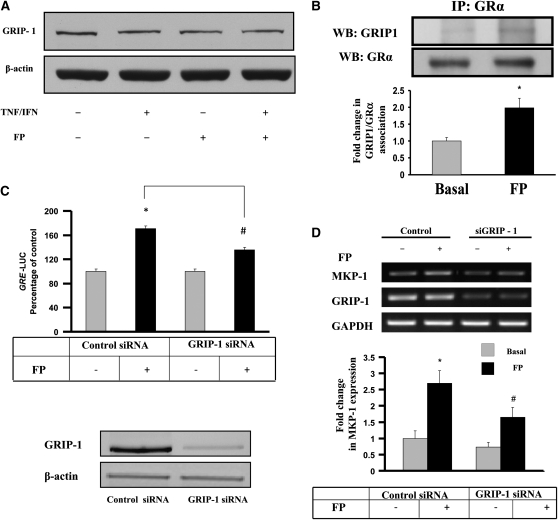
Although GCs suppress activities of some but not all cytokine-induced inflammatory genes (11), the underlying molecular mechanisms appear complex, since GC actions are highly promoter- and cell type–specific (13). Although GRIP-1 is a well-established GR receptor co-activator, its role in GC function is also complex, as it can affect both GR transactivation and transrepression activities (19). As shown in Figure 1A, GRIP-1 was expressed in ASM whole cell extracts, and that expression was unaffected by cytokine and/or steroid treatments. Our finding is in agreement with previous studies showing the conserved expression of GRIP-1 in a variety of smooth muscle cells, including smooth muscle of gastrointestinal and urinary tracts, uterus, epididymis, prostate, and bronchioles of murine tissues (26). Interestingly, we observed a physical interaction between endogenous GR and GRIP-1, and FP treatment significantly enhances this association (1.99 ± 0.3-fold increase over basal) (Figure 1B), suggesting that GRIP-1 recruitment to GR is a prerequisite step to initiate GR transcriptional activity. Interestingly, control immunoprecipitations using nonimmune IgG showed absence of nonspecific binding. Whether GR also binds to other p160 members of the SRC family, as suggested by others (27), remains to be further investigated in ASM. Moreover, we provide the first demonstration of a role of GRIP-1 in GR-dependent gene expression in ASM cells. Indeed, the activity of a reporter construct containing luciferase reporter gene driven by GRE motifs was induced by FP, and GRIP-1 siRNA (but not control siRNA) partially decreased such induction (Figure 1C, top). Of note, specific siRNA GRIP-1 dramatically and efficiently reduced GRIP-1 protein contents (by 91%; Figure 1C, bottom). siRNA-GRIP-1, but not control siRNA, also reduced by more than 50% FP-induced expression of mitogen-activated protein kinase phosphatase 1 (MKP-1) (24, 28, 29) (Figure 1D). This observation provides the first physiologic relevance of GRIP-1 in driving the expression of GC-inducible genes in airway structural cells. This finding is also clinically relevant, as MKP-1 has recently gained a lot of attention as a factor mediating GC inhibition of a number of proasthmatic responses in ASM including expression of inflammatory genes IL-6, GRO-α, and CD38 (24, 28, 29) as well as cell proliferation (30). By regulating the transactivation of GR-inducible proteins (31), our study identifies GRIP-1 as a critical factor in driving the anti-inflammatory actions of GC in ASM.
Figure 1.
Airway smooth muscle (ASM) expresses a functional glucocorticoid receptor (GR) interacting protein 1 (GRIP-1). (A and B) Cells were treated for 6 hours with fluticasone propionate (FP) (100 nM) and/or TNF-α (10 ng/ml) and IFN-γ (500 IU/ml), then lysed and assayed for GRIP-1 by immunoblot analysis (A) or immunoprecipitated with anti-GR antibody, and probed for GRIP-1 by immunoblot analysis (B, top). (B, bottom) Relative change in normalized GRα-bound GRIP-1 in FP-treated cells compared with basal condition (see Materials and Methods).*P < 0.05 compared with untreated cells. (C) ASM cells co-transfected with GRE-luciferase reporter and either GRIP-1 specific or control siRNA were exposed to 100 nM of FP for 6 hours. Luciferase activities were expressed as percentage of control and assessed as described in Materials and Methods (top). In parallel, total cell lysates were assayed for GRIP-1 expression by immunoblotting (bottom, upper gel). The nitrocellulose membrane was then stripped and blotted again for β-actin for equal loading control purpose (bottom, lower gel). *P < 0.05 compared with untreated cells transfected with control siRNA, #P < 0.05 when compared with FP-treated cells transfected with control siRNA. (D) Total mRNA was isolated from cells treated with FP for 6 hours as shown, and subjected to RT-PCR with MKP-1, GRIP-1, and GAPDH primers (top). (D, bottom) Scanning densitometry of three representative gels of MPK-1 normalized to the area density of the corresponding GAPDH control. The results were expressed as fold increase over untreated conditions. *P < 0.05 compared with untreated cells transfected with control siRNA, #P < 0.05 when compared with FP-treated cells transfected with control siRNA.
IRF-1-Induced Transcription Requires GRIP-1
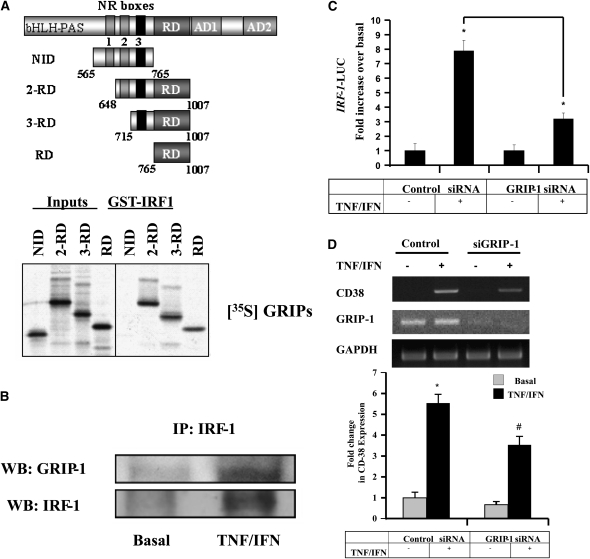
We recently developed a cellular model of steroid insensitivity in which GC-induced suppression of inflammatory genes (CD38, fractalkine) was drastically reduced in the presence of proasthmatic cytokines TNF-α/IFN-γ (8, 14, 25, 32). Cytokine-induced GC insensitivity was mediated, at least in part, by the transcription factor IRF-1 (14) that interfered with GR transactivation activities, although the underlying mechanisms remain unknown. We first examined whether IRF-1 interacts with GRIP-1. Full-length IRF-1 fused to GST was tested for its ability to bind different GRIP-1 derivatives, including: nuclear receptor interaction domain (NID) (aa 565–765), known to bind GR (19, 20); 2-repression domain (RD) (aa 648–1007) containing nuclear receptor (NR) boxes 2 and 3; and the co-repression domain, 3-RD (aa 715–1007) containing NR box 3 and the co-repression domain and RD alone (aa 765–1007) (Figure 2A, top). Interestingly, all but the NID interacted with IRF-1 (Figure 2A, bottom), suggesting a direct GRIP-1:IRF-1 interaction and defining RD as the minimal fragment of GRIP-1 that associates with IRF-1. In addition, the interaction between GRIP-1 and IRF-1 was further confirmed by co-immunoprecipitation using ASM whole cell lysate. Modest interaction was observed in untreated cells, but was dramatically enhanced after cytokine treatment (Figure 2B). Because our in vitro data indicate that cytokine stimulation is not required for the GRIP-1:IRF-1 interaction per se, we speculate that cytokine-dependent IRF-1 accumulation in the nucleus is largely responsible for the relatively greater abundance of GRIP-1:IRF-1 complexes in TNF-α/IFN-γ–treated cells. Indeed, stimulus-dependent nuclear translocation of IRF proteins, including IRF-1, IRF-3, and IRF-7, has been well established (21, 33). Unless cells are exposed to cytokines that lead to IRF accumulation in the nucleus, not much interaction with constitutively nuclear GRIP-1 can be expected under basal conditions in which IRF-1 expression is minimal and mostly in the cytoplasm. It seems, however, unlikely that GRIP-1:IRF-1 in vivo interaction requires GR. Our initial co-imunoprecipitation studies showed no physical interaction between IRF-1 and GR in ASM cells (O. Tliba, unpublished data). Similarly, in macrophages, Reily and coworkers (21), and Ogawa and colleagues (34) were also unable to show any interaction between IRF-3 and GR.
Figure 2.
GRIP-1 is novel co-activator in IRF-1 regulation of gene transcription in ASM cells. (A, top panel) A diagram of GRIP-1 derivatives tested for binding to GST-IRF-1 including NID, 2-RD, 3-RD, and RD alone, as indicated. (A, bottom panel) Mapping of the interacting surfaces on GRIP-1. In vitro–produced GRIP-1 derivatives from the top panel in A were tested for their ability to interact with GST-IRF-1. (B) Cells stimulated with TNF-α (10 ng/ml) and IFN-γ (500 IU/ml) for 6 hours, as shown, were lysed, immunoprecipitated with anti–IRF-1 antibody, and assayed for GRIP-1 by immunoblot analysis. (C) ASM cells co-transfected with IRF-1-luciferase reporter construct and either 100 nM of GRIP-1 siRNA or control siRNA were exposed to TNF-α (10 ng/ml) and IFN-γ (500 IU/ml) for 6 hours. The results were expressed as fold increase over basal. The luciferase activity was assessed as described in Materials and Methods. *P < 0.05 compared with untreated cells transfected with siRNA control. (D) In parallel, total mRNA was subjected to RT-PCR with CD38, GRIP-1, and GAPDH primers (top). (D, bottom) Scanning densitometry of CD38 gels performed exactly as in Figure 1C. *P < 0.05 compared with untreated cells transfected with control siRNA, #P < 0.05 when compared with FP-treated cells transfected with control siRNA.
We next investigated whether GRIP-1 recruitment to IRF-1 is functional, in which case manipulating GRIP-1 levels would modulate IRF-1 transactivation activities. The use of silencing strategies demonstrated that the siRNA to GRIP-1 (but not siRNA controls) reduced by more than 57% TNF-α/IFN-γ–induced IRF-1–dependent reporter gene activity (Figure 2C). GRIP-1 depletion also abrogated the expression of CD38, an IRF-1–dependent gene in ASM cells (14) as shown by the 47% reduction of cytokine-induced CD38 mRNA up-regulation (Figure 2D). Although SRC family members interact with other transcription factors and regulators such as AP-1, serum response factor (SRF), NF-κB, CREB, and as muscle-specific factor, myocyte enhancer factor 2C (MEF2C) (20, 31, 35–37), our findings are the first to demonstrate that GRIP-1 also interacts with IRF-1 and modulates IRF-1 transactivation activities. Together, these observations suggest that IRF-1–induced transcription requires GRIP-1 and raise the hypothesis that an excess of IRF transcription factors could potentially affect GC function by competing with GR for GRIP-1 required for GC transcriptional functions.
Recruitment of GRIP-1 by IRF-1 Mediates Cytokine-Induced GR Dysfunction
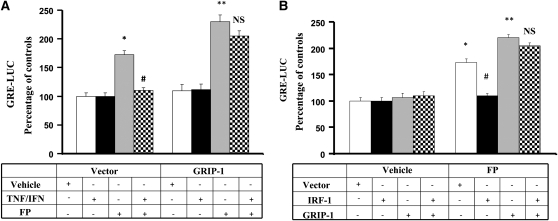
We previously showed that inflammatory cytokines (TNF-α and IFN-γ) promote steroid insensitivity in ASM cells by inhibiting GR transactivation activity, an effect mediated by IRF-1 (14). Interestingly, GR competition for GRIP-1 was shown to be critical in antagonizing IRF-3–regulated transcription in macrophages (21). We therefore examined whether the presence of an excess of GRIP-1 would restore cell sensitivity to steroids diminished by cytokine treatment. As reported previously (14), treatment of ASM cells with TNF-α/IFN-γ combination strongly suppressed FP-dependent gene expression as shown by the reduced FP-induced GRE-luciferase reporter activity in vector-transfected cells (Figure 3A). This suppression, however, was no longer observed in cells overexpressing GRIP-1 (Figure 3A). Similarly, when steroid insensitivity was induced by overexpressing IRF-1, increased levels of GRIP-1 restored GR reporter activity in IRF-1–transfected cells (Figure 3B). Of note, GRIP-1 overexpression dramatically and efficiently increased GRIP-1 protein contents (by 2.2-fold; data not shown). These findings suggest that inflammatory cytokines promote GC insensitivity via activation of IRF-1 that associates GRIP-1 and sequesters GRIP-1 away from GR (Figure 4). This represents a potential molecular mechanism underlying steroid insensitivity induced by inflammatory cytokines. Whether GRIP-1 acts directly or indirectly through the recruitment of additional co-regulators to overcome IRF-1 regulation of GR signaling needs further investigation. Further, since GRIP-1 depletion inhibits only partially FP-transactivation activities (Figures 1C and 1D), additional studies are also needed to examine the possible redundant and/or overlapping functions of other SRC family members and specifically define whether other p160 co-factors drive GR transcription and whether such pathways contribute to the anti-inflammatory actions of GCs in ASM cells. Importantly, while the expression of IRF-1 has been demonstrated in the airways of patients with asthma cca38), whether the expression of IRF-1 or GRIP-1 is altered in GC-resistant individuals with asthma remains to be determined.
Figure 3.
GRIP-1 overexpression rescues GR transcriptional activation from cytokine-/IRF-1–dependent inhibition. (A) ASM cells co-transfected with GRE-luciferase reporter and either 2 μg of full-length GRIP-1 or pcDNA3 were exposed to TNF-α (10 ng/ml) and IFN-γ (500 IU/ml) for 6 hours and/or FP (100 nM) added 2 hours before. The luciferase activity was determined as described in Materials and Methods. (B) ASM cells co-transfected with GRE-luciferase reporter construct and 2 μg of full-length GRIP-1 and/or 2 μg of full-length IRF-1 were exposed to FP (100 nM) for 6 hours, and luciferase activity was determined as described in Materials and Methods. Results are presented as % of control and were representative of three separate transfections. *P < 0.05 compared with untreated cells transfected with pcDNA3; **P < 0.01 compared with untreated cells transfected with pcDNA3; #P < 0.05 when compared with FP-treated cells transfected with pcDNA3 (NS, not significant).
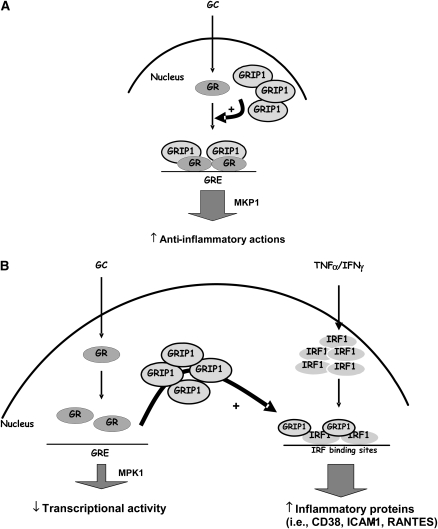
Figure 4.
A model illustrating the role of IRF-1 in modulating steroid responsiveness in ASM cells. (A) In steroid-sensitive conditions, GC promotes GRIP-1 recruitment to GR at the GREs such as those of the MKP-1 promoter, stimulating its expression, thereby contributing to the anti-inflammatory actions of GC. (B) In steroid-resistant conditions, cytokine treatment leads to enhanced expression of IRF-1, which would deplete GRIP-1 from GR complex, thereby reducing the transcription of GR-dependent genes such as MKP-1 and promoting the expression of IRF-1–dependent pro-inflammatory genes such as CD38. GC, glucocorticoid; GRE, glucocorticoid responsive elements; GR, glucocorticoid receptor.
CONCLUSIONS
This study reveals a potential mechanism for the mutual antagonism observed between IRF-1 and GR in their ability to activate gene transcription. As a shared co-factor required for maximal gene activation by both transcription factors, limiting the amount of GRIP-1 available to GR in the presence of high level of IRF-1 would dramatically affect GC function. Our study also shows that the induction of anti-inflammatory proteins such as MKP-1 by GCs represents a GRIP-1–dependent response. In inflammatory diseases such as asthma in which structural cells such as ASM are exposed to multiple proinflammatory factors, an abnormal increase in IRF-1 levels would not only lead to increased IRF-1–dependent inflammatory proteins but would also impair tissue steroid responsiveness by decreasing GC function.
Acknowledgments
The authors thank Su Gu for his excellent technical assistance and Mary McNichol for assistance in the preparation of the manuscript.
This work was supported by National Institutes of Health grant R00 HL089409-03 (OT), American Lung Association grant RG-49342-N (OT), and R01 HL-064063 (Y.A.). O.T. is a Parker B. Francis Fellow in Pulmonary Research.
Originally Published in Press as DOI: 10.1165/rcmb.2009-0239RC on October 5, 2009
Conflict of Interest Statement: None of the authors has a financial relationship with a commercial entity that has an interest in the subject of this manuscript.
References
- 1.Adcock IM, Barnes PJ. Molecular mechanisms of corticosteroid resistance. Chest 2008;134:394–401. [DOI] [PubMed] [Google Scholar]
- 2.Adcock IM, Ford PA, Bhavsar P, Ahmad T, Chung KF. Steroid resistance in asthma: mechanisms and treatment options. Curr Allergy Asthma Rep 2008;8:171–178. [DOI] [PubMed] [Google Scholar]
- 3.Gross KL, Lu NZ, Cidlowski JA. Molecular mechanisms regulating glucocorticoid sensitivity and resistance. Mol Cell Endocrinol 2009;300:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Michetti P, Mottet C, Juillerat P, Pittet V, Felley C, Vader JP, Gonvers JJ, Froehlich F. Severe and steroid-resistant Crohn's disease. Digestion 2007;76:99–108. [DOI] [PubMed] [Google Scholar]
- 5.Pujols L, Mullol J, Picado C. Alpha and beta glucocorticoid receptors: relevance in airway diseases. Curr Allergy Asthma Rep 2007;7:93–99. [DOI] [PubMed] [Google Scholar]
- 6.Kam JC, Szefler SJ, Surs W, Sher ER, Leung DY. Combination IL-2 and IL-4 reduces glucocorticoid receptor-binding affinity and T cell response to glucocorticoids. J Immunol 1993;151:3460–3466. [PubMed] [Google Scholar]
- 7.Almawi WY, Lipman ML, Stevens AC, Zanker B, Hadro ET, Strom TB. Abrogation of glucocorticoid-mediated inhibition of T cell proliferation by the synergistic action of IL-1, IL-6, and IFN-gamma. J Immunol 1991;146:3523–3527. [PubMed] [Google Scholar]
- 8.Clarke D, Damera G, Sukkar MB, Tliba O. Transcriptional regulation of cytokine function in airway smooth muscle cells. Pulm Pharmacol Ther 2009;22:436–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goleva E, Kisich KO, Leung DY. A role for STAT5 in the pathogenesis of IL-2-induced glucocorticoid resistance. J Immunol 2002;169:5934–5940. [DOI] [PubMed] [Google Scholar]
- 10.Spahn JD, Szefler SJ, Surs W, Doherty DE, Nimmagadda SR, Leung DY. A novel action of IL-13: induction of diminished monocyte glucocorticoid receptor-binding affinity. J Immunol 1996;157:2654–2659. [PubMed] [Google Scholar]
- 11.Tliba O, Panettieri RA Jr. Noncontractile functions of airway smooth muscle cells in asthma. Annu Rev Physiol 2009;71:509–535. [DOI] [PubMed] [Google Scholar]
- 12.Hirst SJ, Lee TH. Airway smooth muscle as a target of glucocorticoid action in the treatment of asthma. Am J Respir Crit Care Med 1998;158:S201–S206. [DOI] [PubMed] [Google Scholar]
- 13.Tliba O, Amrani Y, Panettieri RA Jr. Is airway smooth muscle the “missing link” modulating airway inflammation in asthma? Chest 2008;133:236–242. [DOI] [PubMed] [Google Scholar]
- 14.Tliba O, Damera G, Banerjee A, Gu S, Baidouri H, Keslacy S, Amrani Y. Cytokines induce an early steroid resistance in airway smooth muscle cells: novel role of interferon regulatory factor-1. Am J Respir Cell Mol Biol 2008;38:463–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroger A, Koster M, Schroeder K, Hauser H, Mueller PP. Activities of IRF-1. J Interferon Cytokine Res 2002;22:5–14. [DOI] [PubMed] [Google Scholar]
- 16.Nakao F, Ihara K, Kusuhara K, Sasaki Y, Kinukawa N, Takabayashi A, Nishima S, Hara T. Association of IFN-gamma and IFN regulatory factor 1 polymorphisms with childhood atopic asthma. J Allergy Clin Immunol 2001;107:499–504. [DOI] [PubMed] [Google Scholar]
- 17.Mamane Y, Heylbroeck C, Genin P, Algarte M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene 1999;237:1–14. [DOI] [PubMed] [Google Scholar]
- 18.Yamada K, Elliott WM, Hayashi S, Brattsand R, Roberts C, Vitalis TZ, Hogg JC. Latent adenoviral infection modifies the steroid response in allergic lung inflammation. J Allergy Clin Immunol 2000;106:844–851. [DOI] [PubMed] [Google Scholar]
- 19.Rogatsky I, Luecke HF, Leitman DC, Yamamoto KR. Alternate surfaces of transcriptional coregulator GRIP1 function in different glucocorticoid receptor activation and repression contexts. Proc Natl Acad Sci USA 2002;99:16701–16706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rogatsky I, Zarember KA, Yamamoto KR. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J 2001;20:6071–6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reily MM, Pantoja C, Hu X, Chinenov Y, Rogatsky I. The GRIP1:IRF3 interaction as a target for glucocorticoid receptor-mediated immunosuppression. EMBO J 2006;25:108–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Panettieri RA, Murray RK, DePalo LR, Yadvish PA, Kotlikoff MI. A human airway smooth muscle cell line that retains physiological responsiveness. Am J Physiol 1989;256:C329–C335. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa K, Yokosawa H. Degradation of transcription factor IRF-1 by the ubiquitin-proteasome pathway: the C-terminal region governs the protein stability. Eur J Biochem 2000;267:1680–1686. [DOI] [PubMed] [Google Scholar]
- 24.Issa R, Xie S, Khorasani N, Sukkar M, Adcock IM, Lee KY, Chung KF. Corticosteroid inhibition of growth-related oncogene protein-alpha via mitogen-activated kinase phosphatase-1 in airway smooth muscle cells. J Immunol 2007;178:7366–7375. [DOI] [PubMed] [Google Scholar]
- 25.Tliba O, Cidlowski JA, Amrani Y. CD38 expression is insensitive to steroid action in cells treated with tumor necrosis factor-alpha and interferon-gamma by a mechanism involving the up-regulation of the glucocorticoid receptor beta isoform. Mol Pharmacol 2006;69:588–596. [DOI] [PubMed] [Google Scholar]
- 26.Puustinen R, Sarvilinna N, Manninen T, Tuohimaa P, Ylikomi T. Localization of glucocorticoid receptor interacting protein 1 in murine tissues using two novel polyclonal antibodies. Eur J Endocrinol 2001;145:323–333. [DOI] [PubMed] [Google Scholar]
- 27.Xu J, Li Q. Review of the in vivo functions of the p160 steroid receptor coactivator family. Mol Endocrinol 2003;17:1681–1692. [DOI] [PubMed] [Google Scholar]
- 28.Kang BN, Jude JA, Panettieri RA Jr, Walseth TF, Kannan MS. Glucocorticoid regulation of CD38 expression in human airway smooth muscle cells: role of dual specificity phosphatase 1. Am J Physiol Lung Cell Mol Physiol 2008;295:L186–L193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quante T, Ng YC, Ramsay EE, Henness S, Allen JC, Parmentier J, Ge Q, Ammit AJ. Corticosteroids reduce IL-6 in ASM cells via up-regulation of MKP-1. Am J Respir Cell Mol Biol 2008;39:208–217. [DOI] [PubMed] [Google Scholar]
- 30.Burgess JK, Lee JH, Ge Q, Ramsay EE, Poniris MH, Parmentier J, Roth M, Johnson PR, Hunt NH, Black JL, et al. Dual ERK and phosphatidylinositol 3-kinase pathways control airway smooth muscle proliferation: differences in asthma. J Cell Physiol 2008;216:673–679. [DOI] [PubMed] [Google Scholar]
- 31.Leo C, Chen JD. The SRC family of nuclear receptor coactivators. Gene 2000;245:1–11. [DOI] [PubMed] [Google Scholar]
- 32.Sukkar MB, Issa R, Xie S, Oltmanns U, Newton R, Chung KF. Fractalkine/CX3CL1 production by human airway smooth muscle cells: induction by IFN-gamma and TNF-alpha and regulation by TGF-beta and corticosteroids. Am J Physiol Lung Cell Mol Physiol 2004;287:L1230–L1240. [DOI] [PubMed] [Google Scholar]
- 33.Tliba O, Tliba S, Da Huang C, Hoffman RK, DeLong P, Panettieri RA Jr, Amrani Y. Tumor necrosis factor alpha modulates airway smooth muscle function via the autocrine action of interferon beta. J Biol Chem 2003;278:50615–50623. [DOI] [PubMed] [Google Scholar]
- 34.Ogawa S, Lozach J, Benner C, Pascual G, Tangirala RK, Westin S, Hoffmann A, Subramaniam S, David M, Rosenfeld MG, et al. Molecular determinants of crosstalk between nuclear receptors and toll-like receptors. Cell 2005;122:707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen SL, Dowhan DH, Hosking BM, Muscat GE. The steroid receptor coactivator, GRIP-1, is necessary for MEF-2C-dependent gene expression and skeletal muscle differentiation. Genes Dev 2000;14:1209–1228. [PMC free article] [PubMed] [Google Scholar]
- 36.Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev 1998;12:3343–3356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gehin M, Mark M, Dennefeld C, Dierich A, Gronemeyer H, Chambon P. The function of TIF2/GRIP1 in mouse reproduction is distinct from those of SRC-1 and p/CIP. Mol Cell Biol 2002;22:5923–5937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sampath D, Castro M, Look DC, Holtzman MJ. Constitutive activation of an epithelial signal transducer and activator of transcription (STAT) pathway in asthma. J Clin Invest 1999;103:1353–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]