Abstract
Transcriptional regulator protein TM1030 from the hyperthermophile Thermotoga maritima, as well as its complex with DNA, was crystallized at a wide range of temperatures. Crystallization plates were incubated at 4, 20, 37 and 50° C over 3 weeks. The best crystals of TM1030 in complex with DNA were obtained at 4, 20 and 37° C, while TM1030 alone crystallized almost equally well in all temperatures. The crystals grown at different temperatures were used for X-ray diffraction experiments and their structures were compared. Surprisingly, the models of TM1030 obtained from crystals grown at different temperatures are similar in quality. While there are some examples of structures of proteins grown at elevated temperatures in the PDB, these temperatures appear to be underrepresented. Our studies show that crystals of some proteins may be grown and are stable at broad range of temperatures. We suggest that crystallization experiments at elevated temperatures could be used as a standard part of the crystallization protocol.
Keywords: Protein crystallization, TM1030, DNA, transcriptional regulator, crystallization temperature
Introduction
Stable—especially thermostable—enzymes and proteins have great potential for industrial use. They have found application as medicines and as specific catalysts in diagnostic assays, biotransformations, and food processing. 1 Proteins that originate from extremophiles are especially interesting for this role, as they do not require significant amounts of modification in order to preserve their stability and activity in environments that could not be tolerated by most other living organisms. Extremophiles could may classified by temperature, pH or pressure adaptations. 2 For example, the adaptations required for growth at high temperatures include a coordinated set of evolutionary changes affecting nucleic acid thermostability and the stability of codon-anticodon interactions. 3 Growth at high temperatures also requires adaptations that improve protein thermostability. Protein stability is not only a factor that determines its commercial application, but could be also very important in the process of obtaining macromolecular crystals. Most often, the determination of the 3D structure of macromolecules or their complexes by X-ray crystallography is limited by accessibility of well diffracting crystals.
The success in crystallization of biological macromolecules depends on many factors and one of the most significant of them is temperature. 4, 5 Changes in the temperature of crystallization can provide quick and reversible control of the supersaturation level, as the solubility of protein is a function of temperature and may change dramatically even if the temperature change is small. 6, 7 Protein solubility may either increase or decrease as temperature is increased, and this behavior may be significantly altered by both pH and the ionic strength of the solution. 8 Not only can the temperature affect the thermodynamics of the protein solution in terms of the solubility and phase behavior of the solution, but it can also affect the kinetics by which crystals are nucleated and grown affecting crystal size, morphology and quality. Even if initial crystals are obtained, screening a range of different temperatures in addition to sample concentrations, reagent compositions and concentrations, and pH values can increase the probability of producing new or better crystals. Obtaining multiple crystals of a protein from different conditions, especially ones that adopt different crystal forms, increases the probability that one will be more amicable to the methods used to to prepare diffraction experiments, such as co-crystallization, heavy atom derivatization, or cryoprotection.
Despite the fact that small changes in temperature may be a major determinant of the crystallization process, it seems as if relatively little attention has been paid to temperature as a crystallization variable. Most researchers appear to restrict themselves to “room temperature” or to temperatures that can be easily obtained in a laboratory environment, such as 4° C. However, some groups have developed systems to accurately regulate temperature for crystallization experiments. 5-7 Very little has been reported in the literature about crystals grown at elevated temperatures (e.g., above “room temperature”), though apoferritin was reported to produce significantly larger crystals in batch experiments as the temperature of crystallization was increased from 30° to 40° C. 9 In another example, “heat treatment” of a viral protein-ligand solution at 37° C for 5-10 minutes, followed by incubation on ice and crystallization, yielded better-diffracting crystals of the complex. 10
Here we would like to present a study of the crystallization of TM1030, a transcriptional regulator from Thermotoga maritima, a hyperthermophilic bacterium that grows optimally at temperatures of 80°C or higher, as well as co-crystallization of TM1030 with a DNA fragment at a broad range of temperatures of crystallization (here abbreviated Tc).
Materials and Methods
Protein Cloning, Expression and Purification
TM1030 was cloned using the standard protocol developed at the Midwest Center for Structural Genomics. 11 The expression and purification of selenomethionine (Se-Met) incorporated protein was performed according to previously described protocols. 12 The N-terminal His-tag was removed by cleavage with recombinant tobacco etch virus (rTEV) protease, and after cleavage the protein of interest was separated from the His-tagged rTEV and its hydrolyzed tag using cobalt chelate affinity resin (BD Biosciences BD Talon™ Metal Affinity Resin). After the affinity chromatography step, the protein sample was concentrated and further purified using a gel filtration column (HiLoad 6/16 Superdex 200) on an AKTA FPLC system (GE Healthcare). Purified TM1030 used for crystallization was buffer-exchanged into a solution containing 500 mM NaCl and 10 mM HEPES pH 7.5 and concentrated to 9.3 mg/mL.
Preparation of Sample for Protein-DNA Co-crystallization
An 18-bp palindromic DNA oligomer (5′-TGA CTG ACA TGT CAG TCA-3′) 13 was purchased from IDT and purified by desalting chromatography. 170 nmole of DNA was dissolved in 1 mL of solution containing 500 mM NaCl and 10 mM HEPES pH 7.5. The DNA solution was heated to 94° C for 2 minutes and slowly cooled to 20° C. The protein:DNA complex was prepared by mixing a 2:1 molar ratio of protein to DNA and incubating at 4° C overnight before setting up crystallization trials. Finally the sample was passed through a 0.2 μm filter (Milipore) and then transferred to ice.
Crystallization
Crystallization was performed using the hanging drop vapor diffusion method in Crystool (Qiagen) crystallization plates. Crystallization conditions were screened with the Protein Complex Suite Screen. 14, 15 In the case of the TM1030:DNA complex, drops were created by mixing 1.2 μL of screen solution and 1.2 μL of a solution containing 2:1 mixture of TM1030 (protein forms dimer) and palindromic DNA in 500 mM NaCl and 10 mM HEPES pH 7.5. Identical plates were stored in incubators at 4, 20, 37 and 50° C. Tracking of crystallization experiments and analysis of the results was performed using the Xtaldb crystallization expert system 16. Crystal growth was checked on the 1st, 2nd, 4th, 7th, 14th and 21st day after experiment setup. As a control, crystallizations of either the protein or DNA alone were set up in crystallization experiments using the same conditions. In addition, the original conditions used to produce the crystals of TM1030 alone 12 were repeated at the temperatures used for crystallization of TM1030:DNA complex. Crystals used for diffraction experiments were cryocooled by immersion in liquid N2, using ethylene glycol as cryoprotectant.
Data Collection, Structure Solution and Refinement
Low temperature (100K) data collection was done at the Structural Biology Center 17 at the Advanced Photon Source at Argonne National Laboratory. Data were collected, integrated and scaled with HKL-2000 18. The X-ray structures of TM1030 grown at 4, 37, and 50° C were determined to 2.30 Å, 2.35 Å and 2.30 Å respectively. Structure solution was performed by molecular replacement with MOLREP 19, as incorporated in HKL-3000 20, using the TM1030 structure (PDB id 1z77) as a search model. The protein crystallized in the orthorhombic space group P21212, with one monomer in the asymmetric unit. The refinement was performed using REFMAC 19 and COOT. 21 During the last stages of the refinement TLS was applied. MOLPROBITY 22, PROCHECK 23 and ADIT 24 were used for model validation. A summary of the data collection and refinement statistics are presented in Table 1. The coordinates and structure factors for TM1030 were deposited in the PDB with accession codes 3IH4, 3IH3 and 3IH2.
Table 1.
Summary of X-ray diffraction and structure refinement statistics. Data for the highest-resolution shell are shown in parentheses
| PDB code | 3IH4 | 3IH3 | 3IH2 |
|---|---|---|---|
| Crystallization temperature [°C ] | 4 | 37 | 50 |
| Data collection | |||
| Beamline | 19-ID | 19-ID | 19-BM |
| Wavelength [Å] | 0.9794 | 0.9794 | 0.9790 |
| Unit cell parameters [Å] a, b, c |
55.9, 67.1, 56.2 | 55.9, 67.4, 56.2 | 55.9, 66.9, 55.9 |
| Space group | P21212 | P21212 | P21212 |
| Solvent content [%] | 44 | 44 | 43 |
| Number of protein chains in AU | 1 | 1 | 1 |
| Resolution range [Å] | 50.00-2.30 | 34.20-2.35 | 28.72-2.30 |
| Highest resolution shell [Å] | 2.38-2.30 | 2.39-2.35 | 2.38-2.30 |
| Unique reflection | 9853 | 9334 | 9996 |
| Redundancy | 5.8 (6.0) | 6.4 (6.0) | 6.5 (6.5) |
| Completeness [%] | 99.6 (99.7) | 99.9 (100) | 98.3 (97.8) |
| Rmerge | 0.086 (0.430) | 0.087 (0.512) | 0.067 (0.514) |
| Average I/σ (I) | 39.6 (3.4) | 38.4 (3.2) | 39.9 (3.6) |
| Model and refinement | |||
| R | 0.212 (0.232) | 0.218 (0.282) | 0.224 (0.247) |
| Rfree | 0.273 (0.378) | 0.276 (0.284) | 0.275 (0.309) |
| B from Wilson plot [Å2] | 47.1 | 58.1 | 52.6 |
| Mean B value [Å2] | 39.2 | 33.2 | 30.3 |
| RMS deviation bond lengths [Å] | 0.015 | 0.017 | 0.018 |
| RMS deviation angles [°] | 1.4 | 1.4 | 1.7 |
| Ramachandran plot | |||
| Most favored regions [%] | 98 | 98 | 98 |
| Additional allowed regions [%] | 2 | 2 | 2 |
| Protein residues | 201 | 202 | 201 |
| water molecules | 44 | 29 | 59 |
Crystals of TM1030:DNA complex obtained from condition # 61- 0.1M HEPES pH 7, 20%w/v PEG 8000- of Protein Complex Suite Screen (Qiagen) were used for X-ray diffraction experiments. Prior to data collection, crystal was moved to a cryoprotectant solution consisting of a 2:1 mixture of well solution and ethylene glycol. The complex crystallized in the C2 space group with a = 211.7 Å, b = 44.1 Å, c = 59.6 Å and β = 101.7°. The resulting 2.65 Å data set (completeness 99.2% (99.9%), Rmerge 0.089 (0.458)) was used for structure solution. The structure solution of TM1030:DNA complex was performed using PHASER. 25 It was found that the asymmetric unit contains a dimer of TM1030 and half of the palindromic DNA, with the second half generated by crystallographic symmetry. Initial rigid body refinement with REFMAC gave R and Rfree values of 26% and 32%, respectively. Currently the model of the complex is being refined.
Analysis of PDB Deposits
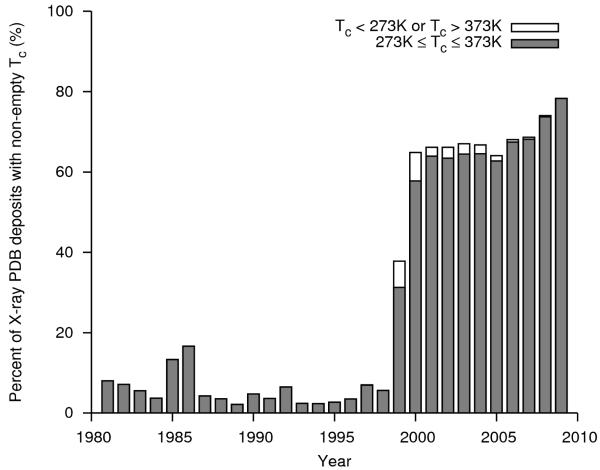
The temperature of crystallization (Tc) is reported in the _exptl_crystal_grow.temp data item in the mmCIF representations of Protein Data Bank (PDB) entries 26, 27 structures, in Kelvins. The local MYPDB database 28, which normalizes and curates data from the PDB and builds them into a relational database, was extended to extract the reported value of Tc from the _exptl_crystal_grow.temp mmCIF 29 data item for each structure. The Biological Macromolecular Crystallization Database (BMCD) contains manually curated crystallization parameters for a set of crystal structures, using information derived both from the PDB and from structure citations in the published literature. 14 BMCD revision 4.02, which contains information on 14372 crystal entries, was queried through its web interface (http://xpdb.nist.gov:8060/BMCD4/) and the reported Tc for each structure was extracted. The values of Tc in MYPDB and the BMCD for the structures that report crystal unit cell parameters (e,g, those solved by X-ray or neutron diffraction) were used to calculate the distribution by year Fig. 3 and the distribution by reported Tc in Fig. 4 and Tables 3 and 4. For purposes of simplicity in Tables 3 and 4, we convert from units of Kelvin to degrees Centigrade by subtracting 273.0° C, rather than 273.15° C.
Figure 3.
Percentage of PDB deposits with a non-empty value for the _exptl_crystal_grow.temp data item (reported temperature of crystallization or Tc) in the mmCIF representation of each structure, grouped by the year of deposition. The gray portion of each bar represents the percentage of structures where 273K ≤ Tc ≤ 373K, and the white portion represents the percentage of structures where Tc lay outside this range.
Figure 4.
Histograms of the reported Tc for all structures that reported that parameter to the PDB (upper graph; 31884 structures) or were recorded in the BMCD (lower graph; 8587). On both graphs, the boxes marked * collect all structures with reported Tc < 0° C. The boxes marked ** collect all structures with reported Tc ≥ 38° C (311 K).
Table 3.
List of structures in the PDB with reported Tc ≥ 311K. n indicates the number of structures at a given Tc value. The rows in italics denote structures with reported Tc values at or above the boiling point of water
| Tc (K) | Tc (°C) | n | PDB identifiers |
|---|---|---|---|
| 311 | 38 | 3 | 2I9F,2FTS,2FU3 |
| 312 | 39 | 2 | 1PEB,1XMK |
| 313 | 40 | 10 | 1WTN,1WTM,1B7Y,1B70,1V7S,1V2L,1PS5,2D4K,3C2J,1YEM |
| 314 | 41 | 1 | 1S2W |
| 315 | 42 | 7 | 1SX5,1QAL,1TQE,1QAF,3FHJ,1D2R,3FI0 |
| 316 | 43 | 7 | 1R12,1NR0,1R15,2G5F,1R16,1R0S,2NTN |
| 318 | 45 | 5 | 1YXI,1JZN,1L0S,1PK3,353D |
| 318.15 | 45.15 | 2 | 1RB8,1M06 |
| 319 | 46 | 1 | 1OMI |
| 320 | 47 | 22 | 1JEY,3B7Q,1R65,1OM3,1K7Y,2HDV,1Z40,1JEQ,1OP5,1JRQ, 1Q6K,2QTV,1OP3,1K98,1EUN,3B74,3B7N,3B7Z,2F68,2F6A, 2AUX,2AUZ |
| 321 | 48 | 4 | 2FP7,2FOM,1G9U,1SJM |
| 322 | 49 | 2 | 1X7O,1X7P |
| 323 | 50 | 66 | 2GW5,1SNR,2GW1,1F9Z,1MIJ,1OYW,1R3F,1FA5,1Q0C,1Q5M, 2B61,2AEB,2B9A,1R3E,1I9B,1G4U,1JW4,1SEG,1SN2,1SN0,1T9J, 1SN5,2AZ5,2GDR,2H13,2GHJ,1QC6,1T9I,1MNV,1MV8,1DFA, 2HWO,1Z32,2HWP,1Z63,1I2M,1ZLT,1YV0,1MUU,2EDM,1NZV, 3EG9,1JW5,3EGD,2OKY,3EGX,2Q3Z,1MFZ,1Q0O,2H14,1M5X, 3E7O,1NW1,3EFO,1M54,1NZL,1PW6,1NM9,1Z6A,2ED6,1Z5Z, 2ID5,2HOX,2HOR,1OQO,1OQX |
| 333 | 60 | 8 | 2I2E,2I2A,2I2D,2I29,2I2C,2I1W,2I2B,2I2F |
| 334 | 61 | 1 | 1JJE |
| 335 | 62 | 1 | 1P2F |
| 353 | 80 | 2 | 1V9G,1G7R |
| 370 | 97 | 1 | 2HU4 |
| 373 | 100 | 9 | 1SKQ,2FZ9,2QTR,2GME,2NYC,2GMM,1I4P,3BQ4,2NYE |
| 378 | 105 | 1 | 3DDH |
| 392 | 119 | 2 | 1OXH,1OX0 |
| 393 | 120 | 3 | 1NWK,2E9T,1LJL |
| 394 | 121 | 1 | 1MIQ |
| 395 | 122 | 2 | 1N2V,1ZWY |
| 398 | 125 | 5 | 1S5M,1S5N,1XQK,1XQL,1N9E |
| 410 | 137 | 4 | 1TN5,1Q5I,1TN0,1Q5J |
| 497 | 224 | 1 | 2FJT |
| 589 | 316 | 2 | 2GVD,2GVZ |
| 777 | 504 | 1 | 2I89 |
Table 4.
List of structures in the BMCD with reported Tc ≥ 311K. n indicates the number of structures with a given Tc value. The rows in italics represent structures with reported Tc values at or above the boiling point of water
| Tc (K) | Tc (°C) | n | PDB identifiers |
|---|---|---|---|
| 312 | 39 | 1 | 1PEB |
| 313 | 40 | 2 | 1PS5,1VZL |
| 314 | 41 | 1 | 1S2W |
| 315 | 42 | 4 | 1D2R,1QAF,1QAL,1SX5 |
| 316 | 43 | 5 | 1NR0,1R0S,1R12,1R15,1R16 |
| 318 | 45 | 2 | 1JZN,1L0S |
| 318.15 | 45.15 | 2 | 1M06,1RB8 |
| 320 | 47 | 10 | 1EUN,1JEQ,1JRQ,1K7Y,1K98,1OM3,1OP3,1OP5,1Q6K,1R65 |
| 321 | 48 | 2 | 1G9U,1SJM |
| 323 | 50 | 33 | 1DFA,1F29,1FA5,1G4U,1I2H,1I9B,1JW4,1JW5,1M54,1M5X, 1MFZ,1MIJ,1MNV,1MVV,1MV8,1NM9,1NW1,1NZL,1NZV, 1OYW,1PW6,1Q0C,1Q0O,1R3E,1R3F,1SEG,1SN0,1SN2,1SN5, 1SNR,1SY6,1T9I,1T9J |
| 334 | 61 | 1 | 1JJE |
| 373 | 100 | 1 | 1SKQ |
| 392 | 119 | 2 | 1OX0,10XH |
| 393 | 120 | 1 | 1NWK |
| 395 | 122 | 1 | 1N2V |
| 398 | 125 | 3 | 1N9E,1S5M,1S5N |
Information from the Prokaryotic Growth Temperature database (PGTdb; http://pgtdb.csie.ncu.edu.tw/) was used to correlate the distribution of the temperature of crystallization of a protein to the temperature of optimal growth of its source organism. 30 The PGTdb contains information on 1086 eubacterial and archaebacterial species, divided into 4 categories based on their growth temperature optima: psychrophilic (<20°C), mesophilic (20-45°C), thermophilic (45-80°C), and hyperthermophilic (>80°C). The list of 31884 structures with a reported Tc in the PDB was filtered, by creating a subset of structures that (a) contained only one source organism—e.g., a single unique value in the list of _entity_src_gen.pdbx_host_org_ncbi_taxonomy_id mmCIF data items—and (b) had a record in the PGTdb that matched that source organism. Structures from eukaryotes and bacteria without growth temperature data in the PGTdb were excluded. The subset of structures was then divided into different groups by reported Tc and by growth temperature category, as shown in Table 5.
Table 5.
Number of structures in the PDB derived from mesophilic or extremophilic prokaryotes (as identified by the Prokaryote Growth Temperature database), grouped by reported temperature of crystallization (Tc) in the PDB. The numbers in parentheses indicate the number of structures in a given growth category and Tc range as a percentage of all structures crystallized in the given Tc range
| Tc range (°C) | Psychrophilic | Mesophilic | Thermophilic | Hyper- thermophilic |
Total |
|---|---|---|---|---|---|
| 0 ≤ Tc < 20 | 74 (2.3%) |
2373 (73.5%) |
514 (15.9%) |
267 (8.3%) |
3228 |
| 20 ≤ Tc < 38 | 66 (1.0%) |
4317 (66.6%) |
1519 (23.4%) |
577 (8.9%) |
6479 |
| 38 ≤ Tc < 100 | 0 (0.0%) |
34 (70.8%) |
11 (22.9%) |
3 (6.2%) |
48 |
Results and Discussion
Se-Met Derivative of TM1030
Crystallization of Se-Met protein was performed using hanging-drop vapor diffusion method and crystallization conditions (solution # 95 of Hampton Research’s Index Screen: 0.1 M KSCN and 30 %w/v polyethylene glycol monomethyl ether 2000), as described previously. 12 Identical plates were stored at 4, 20, 37 and 50° C, and at all temperatures, crystals suitable for X-ray diffraction experiments were obtained (Fig. 1). Structures of TM1030 from 4, 37, and 50° C have the same overall conformation and the refined models are of a quality comparable to those obtained from crystals grown at 20° C. 12 After superposition of Cα carbons r.m.s.d. values between structures range from 0.2 to 0.4 Å. Table 1 (see also Supporting Figure 1) shows that the mean B-factors of the structures decrease as the crystallization temperatures of the corresponding crystals increase. This may indicate that TM1030 is more ordered at higher temperatures that are closer to its physiological conditions. Such results suggest that a wide range of temperature could be used for crystallization of TM1030 and crystallization at temperatures above 20° C can also result in diffraction quality crystals. In our study we were not able to crystallize the protein in higher temperature mainly due to the limitations of the incubators that were used for crystallization experiments.
Figure 1.
The Se-Met derivative of TM1030 crystallized at different temperatures. All pictures were recorded at similar magnification and grown from the same solution (# 95 of Hampton Research’s Index Screen).
TM1030:DNA Complex
Identical crystallization screens were set up of the TM1030:DNA complex at four different temperatures and was incubated for three weeks. This work demonstrated how screening temperature and conditions could affect and enhance the results obtained from the Protein Complex Suite Screen (PCSS). 15 The complex crystallized in one condition at 4° C, four conditions at 20° C, four conditions at 37° C and three conditions at 50° C. The number of days it took to obtain crystals in different crystallization conditions are given in Table 2. The crystallization experiment results are summarized in Figures 1 and 2. The best crystals were observed at 4° C, 20° C and 37° C. Screen condition # 61: 0.1 M HEPES pH 7.0, 20 %w/v PEG8000 was especially productive, as high-quality crystals were obtained at four different temperatures. Within 2 days the results shown in Table 2 were obtained for the complex in screen conditions # 9, 33, 35, 61, and 66. After two weeks crystals were observed in conditions # 37 and 61, and after three weeks in condition # 93. pH 7 proved effective in conditions # 61, 66, and 93. In different conditions the additives PEG4000 (conditions # 29, 33, 35, and 37) and HEPES (conditions # 37, 61, and 66) increased the number of crystals produced. Crystal size also varied with solution conditions, with the largest crystals obtained at temperatures of 4°, 20°, and 37° C, and pH values of 6.5, 7, 7.5, and 8.
Table 2.
Summary of crystallization conditions. The different conditions that produced crystals are listed, along with the number of days it to produce crystals at each temperature
| PCSS# | Crystallization conditions | Days to crystallize at temperature [°C] |
|||
|---|---|---|---|---|---|
| 4 | 20 | 37 | 50 | ||
| 9 | 0.2 M NaCl, 0.1 M MES pH 6.0, 20%w/v PEG2000 MME | - | - | 2 | - |
| 29 | 0.1 M Tris pH 8.0, 20%w/v PEG4000 | - | 7 | - | - |
| 33 | 0.1 M sodium cacodylate pH 5.5, 25% w/v PEG4000 | - | - | - | 2 |
| 35 | 0.1 M sodium cacodylate pH 6.5, 25% w/v PEG4000 | - | 2 | 2 | - |
| 37 | 0.2 M NaCl, 0.1M HEPES pH 7.5, 25% w/v PEG4000 | - | 14 | - | - |
| 61 | 0.1M HEPES pH 7.0, 20% w/v PEG8000 | 14 | 4 | 4 | 2 |
| 66 | 0.1 M HEPES pH 7.0, 15% w/v PEG20000 | - | - | 2 | - |
| 93 | 0.1 M imidazole pH 7.0, 50% MPD | - | - | - | 21 |
Figure 2.
Crystals of the TM1030:DNA complex obtained at different temperatures. A) PCSS #61, 20° C; B) PCSS #61, 37° C; C) PCSS #61, 50° C; D) PCSS #35, 20° C; E) PCSS #35, 37° C; F) PCSS #33, 50° C. All pictures were recorded at similar magnification levels.
Crystallization Temperatures – Analysis of the PDB and BMCD
The original PDB flat-file format contained a means to represent information about crystallization conditions, including Tc. However, this was done via means of an arbitrary-text COMMENT data field in the PDB file header, and often Tc was omitted from this comment (Fig. 3). In contrast, the mmCIF format contains a specific data item to represent the temperature of crystallization in Kelvins. With the advent of the mmCIF format, coupled with improved tools for upload of structures to the PDB such as ADIT, the percentage of structures that reported Tc leapt from about 2-15% in 1981-1998 to 65-78% in 2000-2009 (Fig. 3).
However, not all of the PDB structures that report Tc report plausible values for that parameter. The vast majority of protein crystallizations are carried out either via batch or diffusion methods, both of which require that a protein be dissolved (along with other components) in a liquid solvent, which in virtually all cases is water. Thus reported Tc values where water is not a liquid at atmospheric pressure (i.e. Tc < 273K or Tc > 373K, Figs. 3 and 4) are suspicious, and are likely to be erroneous. Indeed, a brief sampling of literature citations of structures with implausible reported Tc values, showed that most of the publications either directly disagreed with the Tc values in their PDB deposits or did not mention the Tc at all (data not shown). In addition, the majority of Tc values < 273K have values like 4, 20 or 25, strongly suggesting that the depositors thought that the parameter was specified in units of °C instead of K. The fraction of suspicious Tc values was greatest in 2000, comprising 7% of all deposited crystal structures, and decreased in subsequent years (Fig. 3). However, as recently as 2008, 17 structures were deposited to the PDB reporting suspicious Tc values. We suggest that the crystallization parameters, such as crystallization temperature, protein concentration, crystallization method, composition of precipitating buffer along with its pH, should be obligatory in all newly deposited structures. We also suggest that some rudimentary constraints be placed on all parameters like the crystallization temperature to exclude physically impossible values. Application of simple validation tools would make the PDB a more reliable source of data on protein crystallization.
The majority of protein crystallization data in the PDB reports Tc values of either 20°C or 25° C, and a significant number were reported to be 4° C (Fig. 4). Structures in the PDB with Tc ≥ 38° C (311 K) are listed in Table 4. In other words, most experiments were performed either at “room temperature” or in a standard refrigerator or cold-room (or in incubators that replicated those temperatures). Very few structures reported Tc values at temperatures above “room temperature” (e.g., > 26° C). Among crystallization experiments at these elevated temperatures, small clusters of Tc values were seen at 30°, 37° (physiological temperature), or 50° C. 731 structures have Tc values less than 0° C. There are 31 structures with reported Tc values at or above 100° C, and 13 structures with 60° ≤ Tc < 100° C. None of the structures with reported Tc ≥ 60°C could be verified by their citations in the published literature. One curious example is that of the structure of Thermotoga maritima DrrB (1P2F), which is reported to crystallize at 62° C (Table 3), but was described in the literature as being crystallized at 37° C and then transferred to 25° C after crystals had formed (note that 37+25 = 62). 31
A survey of the BMCD, which is reported to contain crystallization information for PDB deposits manually curated by comparison to literature citations, displayed a very similar distribution of reported Tc values (Fig. 4). Curiously, the BMCD also contains a small fraction of reported Tc’s with suspicious values, though the fraction is smaller for that of the whole PDB (Fig. 4). In addition to containing 87 structures with Tc values below 0° C, the BMCD data set contains 10 structures with reported Tc ≥ 100° C (373 K), all of which were found to disagree with the temperatures reported in the corresponding published literature citations.
To determine if there was a correlation between the reported temperature of crystallization of a protein and the optimum growth temperature of the source organism, the subset of structures derived from prokaryotes identified as mesophiles or extremophiles by the PGTdb were grouped by temperature and by growth temperature category (Table 5). For all three Tc categories—low (Tc < 20° C), medium (20°C ≤ Tc < 38° C), and high (Tc ≥ 38° C)—the distributions of source organism by growth category were very similar. In fact, the percentage of proteins crystallized at medium Tc from thermophiles is about the same as the corresponding percentage of proteins crystallized at high Tc from thermophiles. A similar observation may be made of hyperthermophiles (Table 5).
Conclusions
Temperatures of crystallization (Tc) above “room temperature” (e.g., > 26° C) appear to be underrepresented in the PDB. As well, there appears to be no significant difference in the distribution of structures of thermophilic and hyperthermophilic proteins at medium vs. high Tc values—i.e., thermophiles and hyperthermophiles are not overrepresented at higher Tc. It is unclear if higher Tc values are more rarely used by experimenters or if the overall rate of success for high Tc experiments is lower, either for mesophiles or thermophiles. By definition, PDB structures represent only successful crystallization experiments, and we are unable to measure the relative rate of success at different temperatures directly. However, a small number of successful crystallizations, including the crystallization of the TM1030:DNA complex described in this work, have been shown to crystallize at temperatures as high as 50° C.
We also note that there are some issues with the crystallization temperature data reported in both the PDB and the BMCD. Many temperatures that are reported for PDB structures are implausible at best, and in many cases directly contradict the experimental methods described in the primary literature citations. It seems that it is necessary to restrict values for the _exptl_crystal_grow.temp mmCIF data item to the range of 273 K to 373 K during the deposition process. If a macromolecular crystal is in fact grown at a temperature outside of that range, the _exptl_crystal_grow.temp_details data item may (and should) be used to describe such an atypical crystallization experiment.
Our results show that some proteins may be crystallized in much higher temperatures than are typically sampled. Moreover, in the case of the TM1030:DNA complex, it was demonstrated that quality of the crystallographic data and subsequent refined structure do not depend on the crystallization condition. The current results show that higher temperatures may be used not only for crystallization of proteins alone but also for complexes with DNA fragments. It may be important that TM1030, a protein from a hyperthermophilic organism, is able to crystallize well at elevated temperatures, as it is a likely assumption that the protein has better solubility behavior at those temperatures. Although proteins from (hyper-)thermophilic organisms are not seen more frequently in crystals grown at high Tc, it is not clear if this is significant given the very small number of crystals with high Tc values overall.
In all cases, temperature should be a variable explored in the process of crystallization optimization. The current work shows that for proteins from both mesophilic and thermophilic organisms, the range of temperatures explored should include those above room temperature.
Supplementary Material
Acknowledgments
The authors would like to thank Alexander Wlodawer, Andrzej Joachimiak and the members of the Structural Biology Center at the Advanced Photon Source and the Midwest Center for Structural Genomics for help and discussions. The work described in the paper was supported by NIH PSI grants GM62414 and GM074942. The results shown in this report are derived from work performed at Argonne National Laboratory, at the Structural Biology Center of the Advanced Photon Source. Argonne is operated by University of Chicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357.
References
- (1).Lehmann M, Pasamontes L, Lassen SF, Wyss M. The consensus concept for thermostability engineering of proteins. Biochim. Biophys. Acta. 2000;1543:408–415. doi: 10.1016/s0167-4838(00)00238-7. [DOI] [PubMed] [Google Scholar]
- (2).Razvi A, Scholtz JM. Lessons in stability from thermophilic proteins. Protein Sci. 2006;15:1569–78. doi: 10.1110/ps.062130306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Basak S, Ghosh TC. On the origin of genomic adaptation at high temperature for prokaryotic organisms. Biochem. Biophys. Res. Commun. 2005;330:629–32. doi: 10.1016/j.bbrc.2005.02.134. [DOI] [PubMed] [Google Scholar]
- (4).Boistelle R, Astier JP. CRYSTALLIZATION MECHANISMS IN SOLUTION. J. Cryst. Growth. 1988;90:14–30. [Google Scholar]
- (5).Budayova-Spano M, Dauvergne F, Audiffren M, Bactivelane T, Cusack S. A methodology and an instrument for the temperature-controlled optimization of crystal growth. Acta Crystallogr. Sect. D. 2007;63:339–47. doi: 10.1107/S0907444906054230. [DOI] [PubMed] [Google Scholar]
- (6).Bartling K, Sambanis A, Rousseau RW. Multiwell microbatch crystallization on a thermal gradient. Cryst. Growth Des. 2005;5:1559–1564. [Google Scholar]
- (7).Berg M, Urban M, Dillner U, Muhlig P, Mayer G. Development and characterization of temperature-controlled microreactors for protein crystallization. Acta Crystallogr. Sect. D. 2002;58:1643–8. doi: 10.1107/s0907444902012726. [DOI] [PubMed] [Google Scholar]
- (8).McPherson A. Crystallization of Biological Macromolecules. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, New York: 1999. [Google Scholar]
- (9).Bartling K, Sambanis A, Rousseau RW. Dependence of apoferritin crystal growth on temperature and cadmium concentration. Cryst. Growth Des. 2007;7:569–575. [Google Scholar]
- (10).Hassell AM, An G, Bledsoe RK, Bynum JM, Carter HL, 3rd, Deng SJ, Gampe RT, Grisard TE, Madauss KP, Nolte RT, Rocque WJ, Wang L, Weaver KL, Williams SP, Wisely GB, Xu R, Shewchuk LM. Crystallization of protein-ligand complexes. Acta Crystallogr. Sect. D. 2007;63:72–9. doi: 10.1107/S0907444906047020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Zhang RG, Skarina T, Katz JE, Beasley S, Khachatryan A, Vyas S, Arrowsmith CH, Clarke S, Edwards A, Joachimiak A, Savchenko A. Structure of Thermotoga maritima stationary phase survival protein SurE: a novel acid phosphatase. Structure. 2001;9:1095–106. doi: 10.1016/s0969-2126(01)00675-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Koclega KD, Chruszcz M, Zimmerman MD, Cymborowski M, Evdokimova E, Minor W. Crystal structure of a transcriptional regulator TM1030 from Thermotoga maritima solved by an unusual MAD experiment. J. Struct. Biol. 2007;159:424–32. doi: 10.1016/j.jsb.2007.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Subramanian SK, Godzik A. personal communication [Google Scholar]
- (14).Gilliland GL, Tung M, Ladner JE. The Biological Macromolecule Crystallization Database: crystallization procedures and strategies. Acta Crystallogr. Sect. D. 2002;58:916–20. doi: 10.1107/s0907444902006686. [DOI] [PubMed] [Google Scholar]
- (15).Radaev S, Sun PD. Crystallization of protein-protein complexes. J. Appl. Crystallogr. 2002;35:674–676. [Google Scholar]
- (16).Zimmerman MD, Chruszcz M, Koclega KD, Otwinowski Z, Minor W. The Xtaldb system for project salvaging in high-throughput crystallization. Acta Crystallogr. Sect. A. 2005;61:c178–c179. [Google Scholar]
- (17).Rosenbaum G, Alkire RW, Evans G, Rotella FJ, Lazarski K, Zhang RG, Ginell SL, Duke N, Naday I, Lazarz J, Molitsky MJ, Keefe L, Gonczy J, Rock L, Sanishvili R, Walsh MA, Westbrook E, Joachimiak A. The Structural Biology Center 19ID undulator beamline: facility specifications and protein crystallographic results. J. Synchr. Radiat. 2006;13:30–45. doi: 10.1107/S0909049505036721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- (19).Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. Sect. D. 1997;53:240–55. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- (20).Minor W, Cymborowski M, Otwinowski Z, Chruszcz M. HKL-3000: the integration of data reduction and structure solution--from diffraction images to an initial model in minutes. Acta Crystallogr. Sect. D. 2006;62:859–66. doi: 10.1107/S0907444906019949. [DOI] [PubMed] [Google Scholar]
- (21).Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. Sect. D. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- (22).Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–50. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- (23).Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- (24).Yang H, Guranovic V, Dutta S, Feng Z, Berman HM, Westbrook JD. Automated and accurate deposition of structures solved by X-ray diffraction to the Protein Data Bank. Acta Crystallogr. Sect. D. 2004;60:1833–9. doi: 10.1107/S0907444904019419. [DOI] [PubMed] [Google Scholar]
- (25).McCoy AJ. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr. Sect. D. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- (27).Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–42. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Zheng H, Chruszcz M, Lasota P, Lebioda L, Minor W. Data mining of metal ion environments present in protein structures. J. Inorg. Biochem. 2008;102:1765–76. doi: 10.1016/j.jinorgbio.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Bourne PE, Berman HM, McMahon B, Watenpaugh KD, Westbrook JD, Fitzgerald PM. Macromolecular Crystallographic Information File. Methods Enzymol. 1997;277:571–90. doi: 10.1016/s0076-6879(97)77032-0. [DOI] [PubMed] [Google Scholar]
- (30).Huang SL, Wu LC, Liang HK, Pan KT, Horng JT, Ko MT. PGTdb: a database providing growth temperatures of prokaryotes. Bioinformatics. 2004;20(2):276–8. doi: 10.1093/bioinformatics/btg403. [DOI] [PubMed] [Google Scholar]
- (31).Robinson VL, Wu T, Stock AM. Structural analysis of the domain interface in DrrB, a response regulator of the OmpR/PhoB subfamily. J Bacteriol. 2003;185(14):4186–94. doi: 10.1128/JB.185.14.4186-4194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.