Abstract
Enhancers, or cis-regulatory elements, are the principal determinants of spatiotemporal patterning of gene expression. For reasons of clinical and research utility, it is desirable to build customized enhancers that drive novel gene expression patterns, but currently, we largely rely on “found” genomic elements. Synthetic enhancers, assembled from transcription factor binding sites taken from natural signal-regulated enhancers, generally fail to behave like their wild-type counterparts when placed in transgenic animals, suggesting that important aspects of enhancer function are still unexplored. As a step toward the creation of a truly synthetic regulatory element, we have undertaken an extensive structure–function study of an enhancer of the Drosophila decapentaplegic (dpp) gene that drives expression in the developing visceral mesoderm (VM). Although considerable past efforts have been made to dissect the dppVM enhancer, transgenic experiments presented here indicate that its activity cannot be explained by the known regulators alone. dppVM contains multiple, previously uncharacterized, regulatory sites, some of which exhibit functional redundancy. The results presented here suggest that even the best-studied enhancers must be further dissected before they can be fully understood, and before faithful synthetic elements based on them can be created. Implications for developmental genetics, mathematical modeling, and therapeutic applications are discussed.
Introduction
Enhancers were first identified as DNA sequences capable of boosting the rate of transcription at a nearby promoter, and were named for this quantitative property.1,2 It was later discovered that enhancers can also control the spatiotemporal pattern of gene expression within tissues, that they can function at great distances from the promoter being regulated, and that a single gene can harbor many regulatory elements.3 Enhancers are now recognized as principal regulators of gene expression in multicellular eukaryotes.4
As important as enhancers are to organisms for regulating gene expression, they are no less useful to researchers and clinicians who wish to experimentally manipulate gene expression in specific subsets of cells, or restrict the expression patterns of therapeutic transgenes, and many “found” minimally characterized genomic elements lifted directly from native sequences are currently used in a wide range of cell-replacement and gene-therapy approaches for the treatment of disease.5–11 For example, promising strategies for the treatment of Parkinson's and Alzheimer's disease incorporate the use of native enhancer elements, with the goal of promoting neural survival or rescuing neural function.8 In one case, enhancers from the central nervous system–specific intermediate filament gene nestin and the signaling morphogen sonic hedgehog are used as tools to drive embryonic stem cells to differentiate into progenitors of specific subtypes for the purpose of therapeutic transplantation.9 As a second example, approaches to the treatment of myocardial ischemia have also included native tissue–specific enhancers.10 One particularly clever viral gene therapy vector, aptly named the “vigilant vector,” incorporates both a hypoxia response element that upregulates transcription when oxygen levels are low, and a heart-specific promoter isolated from the myosin light chain (MLC-2v).11 The use of tissue or cell type–specific enhancer elements is a prominent theme in treatment strategies for a wide variety of other disease types, including cancer, diabetes, and arthritis.10 In addition, while many advanced inducible gene regulation systems are widely used in a variety of gene therapy applications (including the tetracycline-dependent regulatable gene expression system known as Tet-ON and Tet-OFF), their versatility and wide-spread appeal are in fact a direct result of the inclusion of cell type–specific or organ-specific enhancer/promoters that are still very much required for the specificity of transgene expression.10
As the incorporation of cell type–specific promoters in gene therapy, cell replacement, and tissue engineering strategies becomes more sophisticated, we expect that the need to fine-tune the specificity of these elements and to minimize their size will become more prominent, and the actual limitations of our understanding of enhancer structure and function will become more broadly apparent. We anticipate that the ability to build truly “synthetic” versions of enhancers is a need just on the horizon, and a detailed understanding of how these “found” enhancer sequences function on a base-by-base level will become much more necessary as we attempt to mimic the real complexities of in vivo developmental signaling.
One might expect that, following the principle of combinatorial control, novel gene expression patterns could be generated simply by combining transcription factor (TF) binding sites in synthetic regulatory elements. This can be easily accomplished in the context of cultured cells, where even transfected constructs containing binding sites for a single TF can respond robustly in the presence of that factor.12 However, regulatory sequences inserted into a chromosome, in the context of a multicellular organism, are subject to much tighter restrictions on gene expression, and generally require a surprisingly large number of transcriptional inputs.12,13 In fact, in over 25 years of enhancer research, published reports of robustly active synthetic combinatorial enhancers, built “from scratch” by combining binding sites for multiple TFs, and active in an appropriate pattern in transgenic animals, are exceedingly rare.12–14 As it turns out, real regulatory elements are subject to more complex regulation than typical enhancer models predict. Take, for example, the 480-bp Drosophila even-skipped (eve) stripe 2 enhancer, perhaps the most intensively studied developmental enhancer and, literally, the textbook example of a well-understood cis-regulatory element.15–17 Thirteen regulatory sites, bound by a total of five different TFs, have been identified within the eve stripe 2 enhancer,18,19 and yet of the remaining stretches of sequence between the clusters of known TF binding sites, all are necessary for full enhancer activity in vivo.19 After 20 years of study, the story of this enhancer is still only half-told.
So is eve stripe 2 an unusually baroque element, or is it a fairly typical example of an enhancer? To answer this question, we have built synthetic versions of four previously characterized Drosophila developmental enhancers, all with multiple known direct inputs, by combining the known TF binding sites. We also present the results of an extensive functional analysis of one of these, taken from the Drosophila decapentaplegic (dpp) gene, which encodes a TGF-β/BMP-family ligand and responds to Wnt signaling in the embryonic visceral mesoderm (VM). The goals of this portion of the study are (i) to determine the cis-regulatory complexity of the enhancer and (ii) to build minimal synthetic elements that recapitulate its transcriptional activity in vivo.
Materials and Methods
DNA cloning and mutagenesis
All constructs described in this report were subcloned either into the pENTR/D-TOPO plasmid (Invitrogen, Carlsbad, CA) by TOPO cloning or into the pBS-ENTR-TOPO plasmid20 by traditional cloning, and then placed by Gateway recombination cloning into the P-element lacZ reporter vector Ganesh-Z1,20 which inserts pseudo-randomly into the genome as a single copy.
Internal deletions of enhancers were created with overlap extension PCR, or “gene sewing.”21,22 More complex mutations and synthetic enhancer constructs were created with assembly PCR, in which short overlapping oligonucleotides are annealed and assembled into long double-stranded DNAs via PCR.23–25 Detailed protocols are available at sitemaker.umich.edu/barolo/protocols. Full sequences of all wild-type and mutagenized enhancers, synthetic constructs, and PCR primers are available upon request.
Transgenesis
P-element transformation by embryo injection was performed essentially as previously described.26 A slightly modified protocol is available at sitemaker.umich.edu/barolo/protocols. w1118 flies were used for transgenesis. To eliminate the effect of genomic insertion site on gene expression, several independent transgenic lines bearing each construct were examined, and representative results are shown.
Tissue preparation, staining, and microscopy
Staged Drosophila embryos were fixed in formaldehyde and subjected to RNA in situ hybridization with a digoxigenin-labeled lacZ probe, essentially as previously described.27 A slightly modified protocol is available at sitemaker.umich.edu/barolo/protocols. Wing imaginal discs were dissected from crawling third-instar transgenic larvae, fixed in glutaraldehyde, and stained for β-galactosidase activity, as previously described.28 Images of stained embryos and imaginal discs were obtained with an Olympus BX51 microscope and an Olympus DP70 digital camera.
DNA sequence alignment
Orthologous Drosophila genomic sequences were identified by BLAST searches on the DroSpeGe website29 (http://insects.eugenes.org/species). dppVM orthologous sequences were initially aligned, as multiple smaller fragments, with the web-based ClustalW program30 (www.ebi.ac.uk/Tools/clustalw). Default settings were used, with the following exceptions: the gap open penalty was set to 5, and the gap extension penalty was set to 2.5. The ClustalW text output was copied to Microsoft Word, where the subalignments were merged, the alignment was amended by hand in cases where conserved motifs were clearly misaligned, and the large indels in region 4 were removed, where indicated, to conserve space.
Results
In vivo insufficiency of synthetic signal-regulated enhancers
We were curious about the extent to which the activity and pattern of various well-studied enhancers could be explained by the described regulatory inputs. Specifically, we were interested in cases where binding sites for specific regulatory factors had been identified and shown to be essential for the proper regulation of the enhancer, but where the sufficiency of these combined inputs for enhancer activity had not been tested. We built synthetic versions of four Drosophila developmental enhancers, consisting of multimerized binding sites for the known regulatory TFs. The first is the proneural enhancer of the E(spl)m4 gene, which is active in Notch-responsive cells of proneural clusters in larval imaginal discs, and depends on binding sites for Suppressor of Hairless [Su(H)] and Achaete/Scute bHLH proteins for its activation31,32 (Fig. 1A). Our synthetic element, containing multimerized high-affinity Su(H) and Achaete/Scute binding sites, failed to reproduce the E(spl)m4 proneural expression pattern in vivo (Fig. 1B).
FIG. 1.
Synthetic versions of well-studied developmental enhancers fail to recapitulate their activity in vivo. All panels show imaginal discs from larvae carrying lacZ reporter transgenes. Reporter gene expression is visualized by X-gal staining. (A) Proneural expression pattern in the developing wing driven by a Notch-regulated enhancer of the E(spl)m4 gene, which contains binding sites for Achaete/Scute [A] and Su(H) [S]. (B) A synthetic version of the E(spl)m4 enhancer, containing multimerized high-affinity A and S binding sites, does not produce an E(spl)m4-like pattern. (C) Cone cell–specific expression pattern driven by the sparkling (spa) enhancer of the dPax2 gene, which contains binding sites for Lozenge [L], Su(H) [S], and Pointed-P2 [P]. (D) A synthetic enhancer containing high-affinity L, P, and S binding sites does not recapitulate dPax2 expression. (E) A disc-specific enhancer of the dpp gene (dppD), containing binding sites for Engrailed [E] and Ci [C], drives expression in Hedgehog-responding cells of the developing wing. (F) A synthetic version of dppD, containing only the E and C sites in their native spacing, fails to drive gene expression in vivo.
The second enhancer we attempted to re-create was the well-characterized, EGFR- and Notch-regulated sparkling (spa) enhancer of the dPax2 gene, which is directly activated by Su(H), Lozenge, and Pointed-P2 in presumptive cone cells of the developing eye33,34 (Fig. 1C). Again, a synthetic element composed of multiple high-affinity binding sites for these TFs failed to recapitulate the expression of the native enhancer (Fig. 1D). We have subsequently found that a synthetic element in which the TF binding sites are taken directly from the native spa enhancer, and are placed in their native arrangement and spacing, is inactive in vivo (C. Swanson, N. Evans, and S.B., manuscript under review).
A disc-specific enhancer of the decapentaplegic gene, dppD, responds to Hedgehog signaling in imaginal tissues via Cubitus interruptus (Ci) binding sites, and is directly repressed by Engrailed35 (Fig. 1E). Synthetic enhancer experiments confirm the reported insufficiency of the Ci sites of dppD,35 even when these sites are placed in their native arrangement (Fig. 1F).
The fourth enhancer addressed in this study, dppVM, will be discussed in the following section. In all four cases, the known regulatory inputs, taken together, appear to be insufficient for proper enhancer activity in vivo. This contrasts with the results of numerous reporter gene experiments in transfected cultured cells, in which a small number of binding sites for a single TF (e.g., Su[H] or Ci/Gli binding sites) are often sufficient to drive a strong transcriptional response upon expression of that TF or stimulation of the relevant signaling pathway.12,13 It appears that, at least in these cases, transcriptional activation of a chromatin-embedded gene, under normal cellular conditions, is not simply or easily achieved.
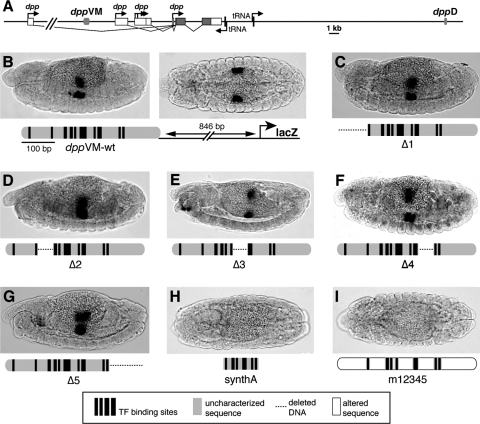
The dppVM enhancer
A 419-bp genomic segment, located 9 kilobases (kb) upstream of the coding sequence of the dpp gene, is capable of driving reporter gene expression in dpp-positive cells in parasegment 7 of the embryonic visceral mesoderm36,37 (Fig. 2B). This enhancer, referred to here as dppVM, contains predicted binding sites for the Hox protein Ultrabithorax (Ubx) and its cofactor Extradenticle (Exd), the Wnt-responsive factor TCF, and the FoxF-related factor Biniou (Bin); all four proteins directly bind to dppVM in vitro.37–39 Ubx and Exd directly activate transcription via dppVM,37,40 while TCF acts as a Wnt-regulated transcriptional switch, activating dpp in Wnt-responding visceral mesoderm cells and repressing dpp in cells not receiving Wnt signaling.38 However, neither ectopic Ubx/Exd nor ectopic Wnt signaling is sufficient to drive enhancer expression outside the visceral mesoderm, and dppVM is not active in other Wnt-responding or Ubx/Exd-expressing cells.37,38,40 Similarly, Bin is required, but is not sufficient, for activation of dpp in the visceral mesoderm.39
FIG. 2.
In vivo functional analysis of the dpp visceral mesoderm enhancer, dppVM. (A) Map of the dpp locus. The dpp transcription unit is depicted as boxes; protein-coding sequence is black. Arrows indicate promoters. Two enhancers of the dpp gene, the imaginal disc enhancer dppD and the visceral mesoderm enhancer dppVM, are shown in gray. (B–I) Whole mount transgenic Drosophila embryos in which lacZ reporter gene expression is detected by RNA in situ hybridization. Anterior is to the left. Black bars indicate known or predicted protein binding sites, gray indicates wild-type uncharacterized sequence, dashed lines indicate deleted sequence, and white (with a black border) indicates sequence that has been altered but not deleted. (B) Embryos carrying the wild-type, 419-bp dppVM in the Ganesh-Z1 vector,30 driving lacZ expression in visceral mesoderm in parasegment 7. Left, lateral view; right, dorsal view; bottom; diagram of enhancer and reporter gene. (C–G) Lateral views of embryos carrying mutant versions of dppVM (Δ1 through Δ5), in which uncharacterized enhancer regions 1 through 5 are deleted, one at a time. No single deletion abolishes enhancer activity. (H) Dorsal view of embryo carrying a “synthetic” 287-bp version of dppVM, in which the functionally significant Ubx, TCF, and Exd sites are placed together, and all other enhancer sequences are deleted. (I) Dorsal view of embryo carrying construct m12345, in which the TF sites are present in their normal arrangement and spacing, and the sequence of regions 1 through 5 is altered.
To test the sufficiency of these regulatory inputs for enhancer activity, we engineered a series of five deletions in sequence of dppVM (named Δ1–Δ5; Fig. 2C–G), which together cover all of the previously uncharacterized sequence of the enhancer. None of these five deletions abolished enhancer activity, even one (Δ2) that removed the only known Bin binding site (Fig. 2D). Of this series of constructs, only Δ3 showed a noticeable difference from the wild-type element, driving a slightly narrower stripe of expression (Fig. 2E, cf. panel B).
Based on these results, it might be concluded that none of the deleted regions is absolutely required for enhancer activity. However, when all five regions were deleted at once, resulting in the minimal “synthetic” construct synthA, enhancer activity in vivo was destroyed (Fig. 2H). To account for the possibility that changes in spacing among TF binding sites are responsible for this failure of activation, we created another synthetic element, m12345, in which regions 1 through 5 were mutated (by changing every other base pair to its noncomplementary transversion: A to C, C to A, G to T, and T to G), but in which the spacing and arrangement of the remaining TF binding sites was normal. This construct was also inactive (Fig. 2I).
Later experiments in which the GC content of mutated enhancer sequences is preserved (see Fig. 4) indicate that it is not merely the local GC content (and therefore the DNA rigidity) of these regions that is important, but rather the specific sequence within these regions. This is at least consistent with the possibility that dppVM harbors functionally important binding sites for as-yet-unknown regulatory factors.
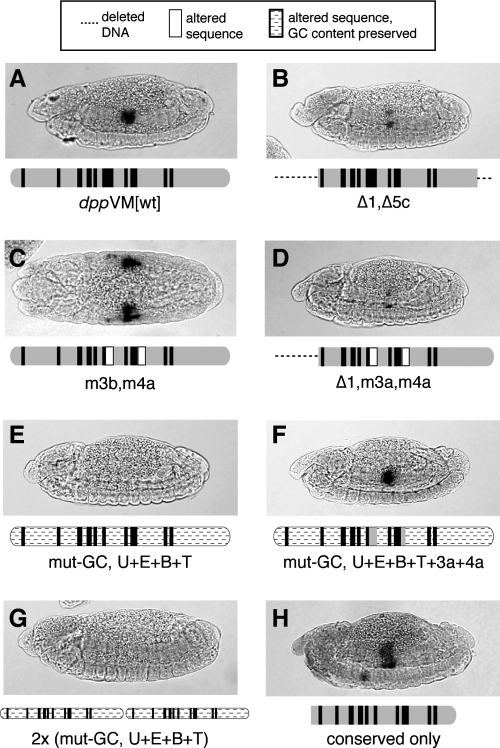
FIG. 4.
Combinatorial mutations reveal functional redundancy in novel regulatory sequences within dppVM. Embryos are stained and presented as in Figure 2. (A) Wild-type dppVM. (B) Lateral view of embryo carrying a mutated dppVM lacking region 1 and the 3′ third of region 5 (region 5c). (C) Dorsal view of embryo carrying an enhancer in which the ATGYTGCA repeats (region 3b) and similar sequences (region 4a) have been altered. (D) Combining the deletion of region 1 with mutations in regions 3b and 4a causes a significant loss of enhancer activity. (E) A mutated enhancer in which all known or predicted binding sites (Ubx + Exd + Bin + TCF) are preserved, but all other sequences are altered such that the wild-type GC content is maintained. (F) A construct similar to the previous, but with the addition of the wild-type ATGYTGCA repeats (3b) and related sequences (4a). Enhancer activity is restored. (G) Two tandem copies of the construct shown in panel E are insufficient for enhancer activity. (H) A construct containing only the most highly conserved sequences from dppVM shows visceral mesoderm activity.
Taken together, these results suggest that additional regulatory sequences beyond the known TF sites are required, but that there is functional redundancy among these sequences such that no single subelement is essential. The proposal of functional redundancy is further supported by the fact that the Bin site is essential in the context of a 261-bp truncated version of the enhancer that extends only to within region 4,22 but that site is not essential in the 419-bp fragment (Fig. 2D). We therefore conclude that the 3′ end of the enhancer, a region that contains no known or predicted Bin binding sites, harbors a regulatory site or sites that are functionally redundant with the Bin site in region 2.
Evolutionary dynamics of regulatory sequences
Comparing orthologous sequences from related species allows us to observe how regulatory DNA changes over time. Evolutionary sequence alignments can also be useful in enhancer dissection, since functionally critical regulatory sequences are often, though not always,41 relatively well conserved. Thanks to the recent sequencing of the genomes of twelve Drosophila species, the evolution of the dppVM enhancer within that genus can be visualized (Fig. 3). As with many aligned enhancer sequences, the following features are observed:
Known regulatory binding sites are generally well conserved (more so in dppVM than in many other enhancers, in our experience).
Such sites are often (but not always) clustered within small islands of highly conserved sequence, within which there is little or no change in binding site spacing.
The linear order of conserved sequence islands is maintained throughout the lineage.
Uncharacterized sequences (i.e., those lacking known TF binding sites) range from rapidly evolving to very well conserved.
FIG. 3.
Evolutionary alignment of the sequence of dppVM across 12 Drosophila species: mel, D. melanogaster; sec, D. sechellia; sim, D. simulans; yak, D. yakuba; ere, D. erecta; ana, D. ananassae; pse, D. pseudoobscura; per, D. persimilis; wil, D. willistoni; vir, D. virilis; moj, D. mojavensis; gri, D. grimshawi. Dashes indicate gaps added to align orthologous sequences. Known and predicted protein binding sites are labeled and highlighted in black; the number indicates position, given in base pairs from the 5′ end of the enhancer. Regions 1 through 5 are indicated with arrows. ATGYTGCA repeats are in bold and underlined. Bases identical to the D. melanogaster sequence are shaded gray. Numbers after the sequences represent the length of the sequences, measured from the PstI site to the XbaI site.
The enhancer is subject to many “indels” (relative insertions and deletions), causing the orthologs to vary dramatically in size (from 418 to 1425 bp, a 3.4-fold range), and resulting in variable distances between neighboring islands of conserved sequence. As in many other enhancers, the distance between closely spaced binding sites (such as Ubx294 and Exd305) is sometimes well conserved, which could reflect important pairwise interactions between neighboring TFs or their co-factors. By contrast, there appears to be little selection pressure maintaining the relative spacing of these small clusters of TF binding sites (see regions 1 through 5), which could in turn suggest a lack of rigid structure within the enhancer as a whole. It is also possible, however, that the enhancer and its bound regulators do form a precise three-dimensional structure, and that “spacer” sequences are merely “looped out,” rendering their length relatively unimportant. The generally compact size of known enhancers, along with studies of functional interactions among TFs, suggests that there are usually upper limits of binding site spacing within an enhancer, beyond which the activities of its bound regulators fail to cohere into a unified signal to the promoter. This may not always be the case, however,42 and it is possible that our current views on enhancer size are skewed by sample bias, both in the design of experiments and in the publication of results.
Another notable sequence feature is the presence of well-conserved repeats at the 5′ end of region 3, first noticed by Sun et al.37 This repeated octamer sequence, which we define as ATGYTGCA (where Y = C or T), occurs as two overlapping repeats, followed by another sequence with a one-base mismatch, in the 5′ end of region 3 (called region 3a). Of the dppVM orthologs in the 12 sequenced Drosophila species, all contain from one to four instances of this octamer (bold and underlined in Fig. 3). In four species, octamers are found both in region 3a and at the 5′ end of region 4 (region 4a). We investigated the possible function of these sequences in further mutagenesis experiments (below).
Combinatorial mutations reveal novel, functionally redundant activities
Certain previously uncharacterized sequences within dppVM (i.e., those sequences for which no binding factor has been identified) are highly conserved throughout the genus Drosophila, which could reflect an important functional role for those sites. However, as we have shown (Fig. 1), none of these novel conserved sites is necessary for enhancer function in the context of the full-length dppVM element. For example, a sequence that we called region 5c, at the 3′ end of region 5, is highly conserved (Fig. 3), but is not necessary for dppVM enhancer activity (Fig. 2G). However, we found that combining the deletion of region 5c with the deletion of region 1 caused a severe reduction in reporter gene expression (Fig. 4B, cf. panel A), despite the fact that both sequences are dispensable when removed individually. We conclude that these regulatory regions, though dissimilar at the sequence level, are functionally redundant.
We also investigated the functional role of the ATGYTGCA octamer repeats found in region 3a (and in some species, also in region 4a). Mutations in region 3a do not abolish dppVM activity in vivo,21 nor does a combination of mutations in regions 3a and 4a (Fig. 4C). When these two mutations were combined with the deletion of region 1, however, enhancer activity was nearly completely lost (Fig. 4D). Here again, we observed functional redundancies among regulatory sequences with little or no sequence similarity.
Two routes to a near-minimal functional enhancer
As a starting point in our next attempt to create a functional but minimal version of the dppVM enhancer, we built a mutant element in which all of the known or predicted Ubx, Exd, Bin, and TCF sites are maintained, and are placed in their native spacing, but the interstitial sequences are mutated such that the GC content of each region is preserved (every other base altered; A to T, T to A, C to G, and G to C). This construct (mut-GC, U + E + B + T) was inactive in vivo (Fig. 4E). However, restoring regions 3a and 4a to the construct (mut-GC, U + E + B + T + 3a + 4a) largely, though not completely, rescued its activity (Fig. 4F).
Interestingly, creating a tandem duplication of the construct mut-GC, U + E + B + T failed to rescue its activity (Fig. 4G). The fact that adding a second copy of each previously identified regulatory binding site did not reconstitute enhancer activity, while adding regions 3a and 4a did (Fig. 4F), suggests that these novel regulatory sites may contribute a qualitatively distinct activity that is essential for enhancer function, and that cannot be substituted with additional binding sites for the known regulatory factors.
A second approach to building a functional minimal enhancer took advantage of evolutionary conservation data. We created a 287-bp synthetic element containing only the most highly conserved sequences, with all other sequences deleted. This construct (dppVM-conserved only) drove reporter gene expression in the proper pattern, though not at levels equal to the wild-type element (Fig. 4H). This is consistent with the proposal, stated above, that spacing among islands of conserved sequence does not play a critical role in the function of this enhancer, although it may make a minor contribution with respect to proper quantitative levels of gene expression.
Toward a truly minimal, yet functional, synthetic enhancer
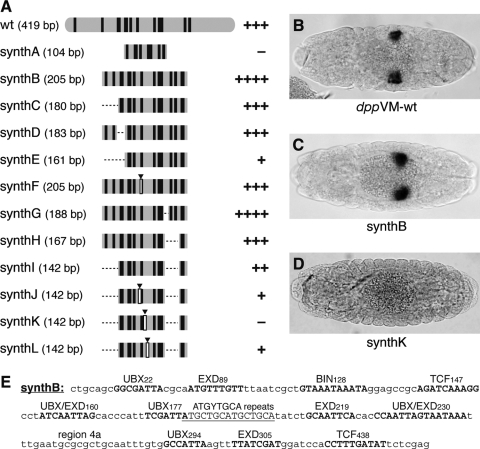
The previous section describes two methods by which we created functional (if not full-strength) elements lacking a large proportion of the sequence of the native enhancer. Building on these findings, we then created an even smaller element (called synthB), containing only those sequences shared by both mut-GC, U + E + B + T + 3a + 4a and dppVM-conserved only, with the addition of a second, highly conserved TCF site located just 3′ of dppVM (see Fig. 3 for sequence). The 205-bp synthB element (diagrammed in Fig. 5A; sequence given in panel E) activated gene expression in the proper pattern, and even more intensely than the wild-type 419-bp enhancer (Fig. 5C; cf. panel B).
FIG. 5.
Toward a functional “synthetic” visceral mesoderm enhancer. (A) Diagrams (to scale) of a series of constructs (synthA through synthL) containing subsets of sequences from dppVM. Arrowheads indicate sites of deletions, relative to constructs shown above. Strength of reporter gene expression in visceral mesoderm is summarized on the right, where “ + + + ” indicates wild-type expression levels and “–” indicates no detectable expression. (B–D) Dorsal views of embryos carrying constructs dppVM-wt, synthB, and synthK. (E) Annotated sequence of the synthB construct.
At 205 bp, synthB is half the length of the native enhancer it is derived from. We made several attempts to further reduce the size of our synthetic element (synthC through synthL), but found that nearly all of the regulatory sequences in synthB make a significant contribution to its activity in vivo (Fig. 5A). Only region 4a could be removed without affecting gene expression levels (synthG). The lower size limit for a full-strength synthetic dppVM element, therefore, appears to be roughly half the length of the natural enhancer.
Several of our synthetic enhancer experiments provide evidence for the functional significance of the octamer repeats (Fig. 5A; synthF, -K, and -L), at least in the context of a minimal enhancer construct. However, as mentioned above, the octamers are not necessary for the activity of the full-length dppVM. This could be due to functional redundancy between the octamers and sequences within dppVM that were excluded from the synthetic constructs.
Discussion
Building enhancers from spare parts
We have demonstrated that dppVM cannot in fact be reduced to a functional synthetic element composed simply of all of the currently characterized regulatory sites. We have also shown this to be true of three other well-characterized, signal-regulated developmental enhancers: a proneural enhancer of the E(spl)m4 gene, an eye enhancer of the dPax2 gene, and an imaginal disc enhancer of dpp (Fig. 1). As previously discussed, the extensively dissected eve stripe 2 enhancer also cannot be similarly reduced.19 Our attempt to further simplify and reduce the dppVM enhancer has revealed the presence of multiple unknown functionally redundant regulatory elements in addition to binding sites for the four known regulators, and our goal of generating a truly “synthetic” functional enhancer remains, to date, unachieved. It is possible that the requirement of a unexpectedly large number of transcriptional inputs reflects a requirement for a minimum number of activating TFs, each of which contributes a small amount of “activation activity,” before a cumulative threshold is crossed and activation can occur. Alternatively, it may be that different activating TFs recruit different biochemical activities to the enhancer (such as chromatin remodeling, DNA looping, subnuclear localization of the locus, etc.), all of which must act in concert, or sequentially, to stimulate the promoter. In addition to the well-known classes of cell type–specific TFs, which often recruit chromatin-remodeling cofactors, there may also be more general enhancer-binding factors with different biochemical properties still awaiting discovery.
Insights from “promoter bashing”
It is important to emphasize how substantially this work contradicts the general perception of our level of understanding of enhancer structure and function. The conventional wisdom among many researchers is that, with the discovery of combinatorial control, the enhancer is essentially “solved,” and the rest is mere detail. As we have demonstrated here, there is still much of interest to learn, even at the level of cataloging and characterizing regulatory DNA sites. Simplified cartoon models of enhancer function may have led to a mistaken perception that “promoter bashing,” as mutagenesis of reporter genes is sometimes referred called, has reached the end of its usefulness, and that in silico or systems-biology approaches will inevitably render small-scale functional studies unnecessary. As this work has demonstrated, not only does much remain to be discovered at the functional level, but much further effort will be required before fully understood synthetic constructs can be designed that can mimic the real complexities of in vivo developmental signaling. Such capabilities are becoming increasingly more desirable for the development of more sophisticated therapeutic tissue repair and regeneration strategies. In addition, the results presented here have potential implications for the development of mathematical models of the in vivo activity of complex regulatory pathways. Models relying on a simple Boolean logic of cis-regulatory input may, in the end, be insufficient to explain natural enhancers—or at the very least, the number of required terms may be greater than expected.43,44
Although we cannot yet successfully recapitulate existing expression patterns without including “mystery” sequences (i.e., sequences without known direct binding factors), results such as those presented here still present significant opportunities for further discovery in molecular biology. For example, by identifying and characterizing the unknown regulators of dppVM, particularly the repeated octamer, we may discover new regulatory factors that play significant roles in the development of the tissues where those enhancers are active. Further analyses of the other enhancer elements examined here have already begun to yield results with interesting implications for developmental genetics, including evidence suggesting that a subelement of the sparkling enhancer is specifically required for action at a distance from the promoter (C. Swanson, N. Evans, and S.B., manuscript under review), and evidence for the special importance of low-affinity TF binding sites in signal-responsive enhancers (D. Parker and S.B., manuscript in preparation). We hope and expect that once the regulatory inputs of a few test enhancers have been comprehensively identified and characterized, this function-based reverse-engineering approach will lead to a better understanding of mechanisms of transcriptional control, an improved ability to mathematically model and predict in vivo transcriptional networks, and, ultimately, the advancement of therapeutic transgene strategies via the creation of custom-built cell– and tissue–specific enhancers.
Acknowledgments
This research was supported, in part, by NIH grant GM076509. We thank Trish Hinrichs and Zeeshaan Bhaati for laboratory assistance. S.B. thanks the members of his lab, especially Paulette Ference and David S. Parker, and also thanks Jim Posakony, Ken Cadigan, Deneen Wellik, Ben Novitch, Doug Engel, and Tom Glaser, as well as many members of their laboratories, for valuable discussions.
References
- 1.Banerji J. Rusconi S. Schaffner W. Expression of a β-globin gene is enhanced by remote SV40 DNA sequences. Cell. 1981;27:299. doi: 10.1016/0092-8674(81)90413-x. [DOI] [PubMed] [Google Scholar]
- 2.Moreau P. Hen R. Wasylyk B. Everett R. Gaub M.P. Chambon P. The SV40 72 base repair repeat has a striking effect on gene expression both in SV40 and other chimeric recombinants. Nucleic Acids Res. 1981;9:6047. doi: 10.1093/nar/9.22.6047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atchison M.L. Enhancers: mechanisms of action and cell specificity. Annu. Rev. Cell Biol. 1988;4:127. doi: 10.1146/annurev.cb.04.110188.001015. [DOI] [PubMed] [Google Scholar]
- 4.Maston G.A. Evans S.K. Green M.R. Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:29. doi: 10.1146/annurev.genom.7.080505.115623. [DOI] [PubMed] [Google Scholar]
- 5.Brand A.H. Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 6.Richards C.A. Austin E.A. Huber B.E. Transcriptional regulatory sequences of carcinoembryonic antigen: identification and use with cytosine deaminase for tumor-specific gene therapy. Hum. Gene Ther. 1995;6:881. doi: 10.1089/hum.1995.6.7-881. [DOI] [PubMed] [Google Scholar]
- 7.Phillips M.I. Tang Y.L. Genetic modification of stem cells for transplantation. Adv. Drug Deliv. Rev. 2008;60:160. doi: 10.1016/j.addr.2007.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korecka J.A. Verhaagen J. Hol E.M. Cell-replacement and gene-therapy strategies for Parkinson's and Alzheimer's disease. Regen. Med. 2007;2:425. doi: 10.2217/17460751.2.4.425. [DOI] [PubMed] [Google Scholar]
- 9.Andersson E. Tryggvason U. Deng Q. Friling S. Alekseenko Z. Robert B. Perlmann T. Ericson J. Identification of intrinsic determinants of midbrain dopamine neurons. Cell. 2006;124:393. doi: 10.1016/j.cell.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 10.Goverdhana S. Puntel M. Xiong W. Zierger J.M. Barcia C. Curtin J.F. Soffer E.B. Mondkar S. King G.D. Hu J. Sciascia S.A. Candolfi M. Greengold D.S. Lowenstein P.R. Castro M.G. Regulatable gene expression systems for gene therapy applications: progress and future challenges. Curr. Gene Ther. 2006;6:421. doi: 10.1016/j.ymthe.2005.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Phillips M.I. Tang Y. Schmidt-Ott K. Qian K. Kagiyama S. Vigilant vector: heart-specific promoter in an adeno-associated virus vector for cardioprotection. Hypertension. 2002;39:51. doi: 10.1161/hy0202.103472. [DOI] [PubMed] [Google Scholar]
- 12.Barolo S. Posakony J.W. Three habits of highly effective signaling pathways: principles of transcriptional control by developmental cell signaling. Genes Dev. 2002;16:1167. doi: 10.1101/gad.976502. [DOI] [PubMed] [Google Scholar]
- 13.Barolo S. Transgenic Wnt/TCF pathway reporters: all you need is Lef? Oncogene. 2006;25:7505. doi: 10.1038/sj.onc.1210057. [DOI] [PubMed] [Google Scholar]
- 14.Guss K.A. Nelson C.E. Hudson A. Kraus M.E. Carroll S.B. Control of a genetic regulatory network by a selector gene. Science. 2001;292:1164. doi: 10.1126/science.1058312. [DOI] [PubMed] [Google Scholar]
- 15.Alberts B. Johnson A. Lewis J. Raff M. Roberts K. Walter P. 4th. New York: Garland Science; 2002. Molecular Biology of the Cell; pp. 408–410. [Google Scholar]
- 16.Wolpert L. 2nd. Oxford: Oxford University Press; 2002. Principles of Development; pp. 168–169. [Google Scholar]
- 17.Gilbert S. 8th. Sunderland, MA: Sinauer; 2006. Developmental Biology; pp. 278–279. [Google Scholar]
- 18.Small S. Blair A. Levine M. Regulation of even-skipped stripe 2 in the Drosophila embryo. EMBO J. 1992;11:4047. doi: 10.1002/j.1460-2075.1992.tb05498.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Andrioli L.P. Vasisht V. Theodosopoulou E. Oberstein A. Small S. Anterior repression of a Drosophila stripe enhancer requires three position-specific mechanisms. Development. 2002;129:4931. doi: 10.1242/dev.129.21.4931. [DOI] [PubMed] [Google Scholar]
- 20.Swanson C.I. Hinrichs T. Johnson L.A. Zhao Y. Barolo S. A directional recombination cloning system for restriction- and ligation-free construction of GFP, DsRed, and lacZ transgenic Drosophila reporters. Gene. 2008;408:180. doi: 10.1016/j.gene.2007.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Horton R.M. Cai Z. Ho S.N. Pease L.R. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. BioTechniques. 1990;8:528. [PubMed] [Google Scholar]
- 22.Ge L. Rudolph P. Simultaneous introduction of multiple mutations using overlap extension PCR. BioTechniques. 1997;22:28. doi: 10.2144/97221bm03. [DOI] [PubMed] [Google Scholar]
- 23.Dillon P.J. Rosen C.A. A rapid method for the construction of synthetic genes using the polymerase chain reaction. BioTechniques. 1990;9:298. [PubMed] [Google Scholar]
- 24.Prodromou C. Pearl L.H. Recursive PCR: a novel technique for total gene synthesis. Protein Eng. 1992;5:827. doi: 10.1093/protein/5.8.827. [DOI] [PubMed] [Google Scholar]
- 25.Stemmer W.P.C. Crameri A. Ha K.D. Brennan T.M. Heyneker H.L. Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene. 1995;164:49. doi: 10.1016/0378-1119(95)00511-4. [DOI] [PubMed] [Google Scholar]
- 26.Rubin G.M. Spradling A.C. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 27.Tautz D. Pfeifle C. A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma. 1989;98:81. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 28.Romani S. Campuzano S. Macagno E.R. Modolell J. Expression of achaete and scute genes in Drosophila imaginal discs and their function in sensory organ development. Genes Dev. 1989;3:997. doi: 10.1101/gad.3.7.997. [DOI] [PubMed] [Google Scholar]
- 29.Gilbert D.G. DroSpeGe: rapid access database for new Drosophila species genomes. Nucleic Acids Res. 2007;35:D480. doi: 10.1093/nar/gkl997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chenna R. Sugawara H. Koike T. Lopez R. Gibson T.J. Higgins D.G. Thompson J.D. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bailey A.M. Posakony J.W. Suppressor of Hairless directly activates transcription of Enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 32.Nellesen D.T. Lai E.C. Posakony J.W. Discrete enhancer elements mediate selective responsiveness of Enhancer of split complex genes to common transcriptional activators. Dev. Biol. 1999;213:33. doi: 10.1006/dbio.1999.9324. [DOI] [PubMed] [Google Scholar]
- 33.Fu W. Duan H. Frei E. Noll M. shaven and sparkling are mutations in separate enhancers of the Drosophila Pax2 homolog. Development. 1998;125:2943. doi: 10.1242/dev.125.15.2943. [DOI] [PubMed] [Google Scholar]
- 34.Flores G.V. Duan H. Yan H. Nagaraj R. Fu W. Zou Y. Noll M. Banerjee U. Combinatorial signaling in the specification of unique cell fates. Cell. 2000;103:75. doi: 10.1016/s0092-8674(00)00106-9. [DOI] [PubMed] [Google Scholar]
- 35.Müller B. Basler K. The repressor and activator forms of Cubitus interruptus control Hedgehog target genes through common generic Gli-binding sites. Development. 2000;127:2999. doi: 10.1242/dev.127.14.2999. [DOI] [PubMed] [Google Scholar]
- 36.Masucci J.D. Hoffmann F.M. Identification of two regions from the Drosophila decapentaplegic gene required for embryonic midgut development and larval viability. Dev. Biol. 1993;159:276. doi: 10.1006/dbio.1993.1240. [DOI] [PubMed] [Google Scholar]
- 37.Sun B. Hursh D.A. Jackson D. Beachy P.A. Ultrabithorax protein is necessary but not sufficient for full activation of decapentaplegic expression in the visceral mesoderm. EMBO J. 1995;14:520. doi: 10.1002/j.1460-2075.1995.tb07028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang X. van Beest M. Clevers H. Jones T. Hursh D.A. Mortin M.A. decapentaplegic is a direct target of dTcf repression in the Drosophila visceral mesoderm. Development. 2000;127:3695. doi: 10.1242/dev.127.17.3695. [DOI] [PubMed] [Google Scholar]
- 39.Zaffran S. Küchler A. Lee H.H. Frasch M. biniou (FoxF), a central component in a regulatory network controlling visceral mesoderm development and midgut morphogenesis in Drosophila. Genes Dev. 2001;15:2900. doi: 10.1101/gad.917101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stultz B.G. Jackson D.G. Mortin M.A. Yang X. Beachy P.A. Hursh D.A. Transcriptional activation by extradenticle in the Drosophila visceral mesoderm. Dev. Biol. 2006;290:482. doi: 10.1016/j.ydbio.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 41.Wittkopp P.J. Evolution of cis-regulatory sequence and function in Diptera. Heredity. 2006;97:139. doi: 10.1038/sj.hdy.6800869. [DOI] [PubMed] [Google Scholar]
- 42.Halfon M.S. (Re)modeling the transcriptional enhancer. Nat. Genet. 2006;38:1102. doi: 10.1038/ng1006-1102. [DOI] [PubMed] [Google Scholar]
- 43.Howard M.L. Davidson E.H. cis-Regulatory control circuits in development. Dev. Biol. 2004;271:109. doi: 10.1016/j.ydbio.2004.03.031. [DOI] [PubMed] [Google Scholar]
- 44.Wang Z. Wei G. Liu D. Liang C. Unraveling the world of cis-regulatory elements. Med. Bio. Eng. Comput. 2007;35:709. doi: 10.1007/s11517-007-0195-9. [DOI] [PubMed] [Google Scholar]