Abstract
Bone marrow–derived stromal cells (BMSCs) are defined by their ability to self-renew and differentiate into at least three mesenchymal cell types (bone, adipose, and cartilage). The inability to isolate a reliably efficacious and homogeneous population of early progenitor cells has limited efforts to increase their therapeutic potential. In this study, we focused on identifying protein markers that may be employed to predict the efficacy of a cultured BMSC population. Markers of progenitor status were identified by comparing BMSCs at early and late passage, donor-matched skin fibroblasts, and commercially available dermal fibroblast cell lines. Differentiation potential was determined according to in vitro assays of osteogenesis, adipogenesis, and chondrogenesis. Early-passage BMSCs differentiated into all three lineages, whereas late-passage BMSCs and both fibroblast preparations did not. To identify novel markers of early progenitors, microarray transcript analysis between early-passage BMSCs and fibroblasts was performed. Messenger RNA encoding the cytokine leukemia inhibitory factor (LIF) was identified as differentially expressed. Enzyme-linked immunosorbent assay on conditioned media confirmed that LIF secretion was much higher from early progenitor BMSCs than donor-matched or commercial lines of fibroblasts and dropped with extensive expansion or induction of differentiation. In clonally expanded BMSCs, colonies that retained progenitor status expressed significantly higher levels of LIF than those that failed to differentiate. Our results indicate that LIF expression may represent a marker to quantify the differentiation potential of BMSCs and may be especially suited for the rapid, noninvasive quality control of clinical preparations.
Introduction
Human bone marrow stromal cells (hBMSCs) are nonhematopoietic cells with the capacity to differentiate into at least three mesenchymal tissue lineages: bone, adipose, and cartilage.1–5 hBMSCs are easily obtained from various tissue sources and can be expanded more than one million–fold in culture while retaining their capacity to differentiate.1,6–11 For this reason, they have received wide clinical interest for use in repairing damaged or defective tissue.12 However, the extent of differentiation into the three mesenchymal lineages varies between different preparations of hBMSCs. The main reason for variability is that hBMSC cultures contain a heterogeneous population of cells consisting of tissue progenitor cells and committed or senescent cells that are limited in their capacity to differentiate. The proportion of early progenitors varies as a result of tissue source, cell density, and the number of previous population doublings or passages.
There has therefore been a substantial effort to select and purify hBMSCs with high differentiation potential, and recent reports have described purification protocols based on detection of surface epitopes such as STRO-1, SH2, SH3, and CD49a.13–16 Although used successfully in some cases to isolate the multipotential nonhematopoietic component of human bone marrow, these markers are not always selective for progenitor cells, and expression of these markers frequently occurs on osteoblasts and fibroblasts. This observation therefore effectively dismisses the possibility that these antibodies can be used to enrich the early progenitors of an BMSC population toward homogeneity.17–19 One potential strategy for identification of molecular markers predictive of differentiation potential involves comparison of BMSCs and fibroblasts. Fibroblasts serve as an appropriate negative control, sharing the phenotype and growth characteristics of BMSCs, yet having extremely limited or no potential for differentiation. In a study by Ishii et al.,19 expression profiles of 11 BMSC preparations and 4 fibroblast cell lines were compared. Although numerous distinctions could be made at the mRNA level, only one surface marker was identified, major histocompatibility complex class II receptor. However, there is cause for caution when interpreting these data since in relatively rare cases, some preparations of fibroblasts have the inherent capacity for trans-differentiation or they are simply contaminated by a small amount of BMSCs.20,21 Additional controls are therefore required, and one potential strategy employs the inverse relationship between the level of expansion and the differentiation potential of BMSCs. Unlike minimally expanded BMSCs from early passages, extensively expanded BMSCs from later passages have limited or no differentiation potential.1,2,5,18,19,20
In an attempt to identify robust markers of multipotency in BMSCs, we performed a detailed comparison of minimally expanded BMSCs, late-passage BMSCs, and dermal fibroblasts that were donor matched or acquired commercially. We found that, although the cell preparations were remarkably similar in terms of surface epitopes, maximal secretion of the cytokine leukemia inhibitory factor (LIF) correlated strongly with differentiation potential. LIF is an inflammatory cytokine with multiple biological activities that plays an important role in cell survival, proliferation, and modulation of differentiation.22 We found that LIF expression was high in minimally expanded BMSCs that had a high proportion of early progenitors but fell to barely detectable levels in all control cell preparations. We also found that when early-passage BMSCs were subjected to conditions that induce osteogenic or adipogenic differentiation, LIF secretion was abruptly attenuated. Currently, the role of LIF secretion by BMSCs is unclear, but it may be similar to its effect on murine embryonic, hematopoietic, and neural stem cells in maintaining multipotency.23,24
This manuscript describes a robust, high-fidelity assay for predicting the early progenitor status of BMSC cultures. This quality control assay would be especially useful for large scale current good manufacturing practice (cGMP) compliant preparations of BMSCs for which rapid results are required without disrupting the cultures.
Materials and Methods
Tissue culture
Fresh bone marrow (2 mL) was obtained by posterior iliac crest aspiration from three healthy human donors aged 25 to 32 in accordance with an approved institutional review board (IRB) protocol. The mononuclear fraction of the bone marrow was prepared using discontinuous density gradient centrifugation, and BMSCs were grown from the mononuclear layer as previously described.20,25 The initial plating of the mononuclear cells was designated passage 0. Three skin fibroblast cell lines were obtained from the American Type Culture Collection (ATCC, Manassas, VA) and cultured under the same conditions as BMSCs. CRL1474 was obtained from the skin of a normal 7-year-old male, cell line CRL 2522 was from the foreskin of a normal newborn male, and cell line CRL1502 was derived from the skin of a female 12 weeks post-gestation. For preparation of the donor-matched dermal fibroblasts, 3-mm-diameter skin biopsies were taken from the forearm in accordance with an IRB-approved protocol. The fatty subcutaneous tissue and the epidermal layer were removed and discarded. The dermis was then finely chopped in alpha minimum essential media (αMEM; Invitrogen, Carlsbad, CA) and subjected to digestion with collagenase and dispase (Roche Diagnostics, Indianapolis, IN) for 2 h at 37°C. The digested tissue was then subjected to centrifugation at 500 g for 15 min. The tissue digest was designated passage 0 and cultured thereafter as BMSCs. For expansion and passaging of all preparations used in this study, cells were cultured in complete culture medium (CCM) containing αMEM, 20% (v/v) fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA), 100 U/mL penicillin; 100 μg/mL streptomycin, and 2 mM L-glutamine (Invitrogen) at 37°C in a humidified atmosphere containing 5% (v/v) carbon dioxide (CO2) and frozen at each passage when 70% confluency was reached. For experiments, frozen cell aliquots were thawed at 37°C, plated on 15-cm2 plates, and allowed to recover overnight in the tissue culture incubator. The next day, at 70% confluency (approximately 8,000–12,000 cells/cm2), the cells were lifted with 0.25% (w/v) trypsin and 0.1% (w/v) ethylenediaminetetraacetic acid (Invitrogen) and re-seeded at 100 cm2 on 145-cm2 plates or at 1000 cells/cm2 for assays in a 6-well format (Corning, Pittsburgh, PA). Thereafter, cells were washed with phosphate buffered saline (PBS), and the medium was changed every 2 days. The average number of cell doublings would be predicted to range between six and seven per passage. Aliquots of 1 × 106 cells were frozen in 1 mL of freezing medium containing αMEM with 30% (v/v) FBS and 5% (v/v) dimethyl sulfoxide (Sigma, St. Louis, MO), then slow frozen in an isopropanol bath to −80°C and transferred to the vapor phase of liquid nitrogen for long-term storage. For clonal experiments, single-cell colonies were generated from passage 2 BMSCs and obtained using a Cytomics FC500 flow cytometer (Beckman Coulter, Fullerton, CA). Cells were seeded at 1 cell per well in a 48-well plate and allowed to expand for 1 week in CCM. The cells in the individual wells that grew to confluence were lifted and split into five separate 12-well plates containing CCM, and medium was changed every other day. Cells were expanded for 8 days, at which time medium was collected and cells harvested for LIF enzyme-linked immunosorbent assay (ELISA) from one plate. On day 8, osteogenic and adipogenic differentiation was induced on two plates by replacing the CCM with osteogenic or adipogenic medium. The remaining two plates continued to receive CCM throughout the experiment, serving as controls. Medium was changed every other day until the 21st day, at which time monolayers were fixed and stained with Alizarin red or Oil Red-O.
In vitro differentiation
Cells were expanded to 70% to 80% confluency in 6-well plates (10-cm2 surface area, Nunc), and the CCM was replaced with osteogenic medium (CCM supplemented with 1 nM dexamethasone, 50 μM ascorbic acid, and 5 mM β-glycerol phosphate (Sigma)) or adipogenic medium (CCM supplemented with 0.5 μM dexamethasone, 0.5 μM isobutylmethylxanthine, and 50 μM indomethacin (Sigma)). The medium was changed every 2 or 3 days. After 21 days, cultures were fixed for 15 min with 10% (v/v) formalin and stained with appropriate dye. For osteogenic differentiation, calcium deposits were stained with 40 mM Alizarin red S (ARS), pH 4.0. For adipogenic differentiation, lipid droplets were stained with Oil Red-O (0.5% (w/v) in 30% (v/v) isopropanol, ORO). Alkaline phosphatase (ALP) assays were performed as previously described on monolayers using a p-nitrophenol phosphate conversion assay.26 For chondrogenic differentiation, cells were harvested, and micromass pellet cultures were prepared by centrifugation of 2 × 105 cells at 1000 xg for 10 min. Pellets were subsequently cultured for 21 days at 37°C with 5% CO2 in 500 μL of chondrogenic medium containing high-glucose Dulbecco's minimum essential medium (DMEM), 500 ng/mL bone morphogenic protein (BMP)-2, 10 ng/mL transforming growth factor (TGF)-β3 (R&D Systems, Minneapolis, MN), 0.5 μM dexamethasone, 50 μg/mL ascorbate-2-phosphate, 40 μg/mL proline, 100 μg/mL pyruvate (Sigma), and insulin, transferrin, selenium + Premix (Franklin Lakes, NJ). The medium was replaced every 2 to 3 days for 21 days, and then pellets were fixed in formalin, embedded in paraffin, cut into 5-μM sections, and stained with toluidine blue sodium borate. For [35SO42–] incorporation assays, 0.1 μCi/mL of Na2[35SO4] (Amersham Pharmacia, Piscataway, NJ) was added to each medium change. For quantification, the pellets were acid hydrolyzed at 70°C in 6 M hydrochloric acid for 15 h, neutralized with 2 M Tris pH 11.0, and scintillation counted (LS6500, Beckman Coulter, Fullerton, CA).
Surface phenotype analysis according to flow cytometry
Cells were suspended at 3 × 106 cells/mL in PBS. One hundred–μL aliquots containing 3 × 105 cells were incubated in the dark for 20 min at ambient temperature with individual monoclonal antibodies or isotype control antibodies (immunoglobulin (Ig)G1 or IgG2a; DAKO, Fort Collins, CO). Monoclonal antibodies against CD11b, CD14, CD19, CD34, CD36, CD44, CD45, CD49b, CD59, CD79a, CD90, CD105, CD117, CD164, CD166, CD184 (Beckman Coulter), CD29, CD49c, CD49f, CD73a, CD106, CD147, CD271, human leukocyte antigen (HLA)-II HLA-ABC, and LIF (BD Pharmingen, San Jose, CA) were used. After antibody staining, cells were washed three times with PBS, suspended in 1 mL of PBS, and then analyzed using a Cytomics FC500 flow cytometer (Beckman Coulter). The data were interpreted using Cytomics FC500 series CXP software (Beckman Coulter).
Microarray
Total RNA from 1 × 106 cells was isolated from each cell line using an RNA isolation kit (High Pure, Roche, Nutley, NJ) and pooled. Optical density of pooled samples ((BMSC donors A, B, and C) and (fibroblasts cell lines 1052, 1474, and 2522)) was measured to determine RNA concentration and purity and then further confirmed using electrophoresis. Total RNA was used to synthesize double-stranded cDNA (Superscript Choice System; Invitrogen), then purified using phenol/chloroform extraction and concentrated using ethanol precipitation. In vitro transcription was used to produce biotin-labeled cRNA (GeneChip In Vitro Trancription labeling Kit; Affymetrix, Santa Clara, CA). The biotinylated cRNA was then cleaned (RNAeasy Mini Kit; Qiagen, Valencia, CA), fragmented, and hybridized with hybridization controls on the HG-U133 Plus 2.0 microarray chips (Affymetrix). After washing, individual microarray chips were stained with streptavidin–phycoerythrin (Invitrogen), amplified with biotinylated anti-streptavidin (Vector Laboratories, Burlingame, CA), stained with streptavidin–phycoerythrin, and scanned for fluorescence (GeneChip Scanner 3000, Affymetrix) using the GeneChip Operating software 1.0 (Affymetrix). Arrays were checked for quality using spike and housekeeping control signal values and 3′ to 5′ ratios, present call percentage, background signal, and noise levels. The scanned images were then transferred to the dChip program and re-checked for quality. To allow comparisons of different microarrays, one array was chosen as the baseline array (BMSC A, median intensity of 189) against which the other arrays were normalized at the probe intensity level. The dChip program then calculated the model-based expression values using the signal from perfect match (PM) and mismatch (MM) probes. Negative values were assigned a value of 1. Genes were considered significantly different in expression between BMSCs (BMSC A and BMSC B samples) and fibroblasts (FIB A and FIB B samples) if they had at least a two-fold difference (with 90% confidence), were present in both of the samples where the gene was up-regulated, and had a t-test p-value <0.05 between groups (BMSC versus fibroblast). The 382 up-regulated transcripts (257 nonredundant genes based on Gene ID) and 439 down-regulated transcripts (305 nonredundant genes) were used in hierarchical clustering of the samples and genes. The resulting two patterns of expression from the BMSCs and fibroblasts were examined for gene ontology (GO) term enrichment. Next, p-values were calculated for each GO term based on the exact hyper-geometric distribution, to compare the frequencies of GO terms within each pattern with their frequencies across the entire microarray (p-values < 0.01 were considered significant).
Reverse transcriptase polymerase chain reaction analysis
Total RNA was harvested from cells, DNase treated, and quantified. cDNA was synthesized from 2 μg of total RNA (Superscript III, Invitrogen), and 2 μL of resulting cDNA was used as a template for the polymerase chain reaction (PCR) reaction. Reaction conditions were as follows: denaturation at 95°C for 30 s, annealing at 64°C for 30 s, and elongation at 72°C for 30 s for 28 cycles (Platinum PCR supermix, Invitrogen). Primers specific for total LIF, LIF isoforms, LIF receptor, Octamer (OCT)-4, and NANOG have been previously described.27–30 Relative levels of target messenger RNA expression was normalized against glyceraldehyde 3-phosphate dehydrogenase (GAPDH) expression as previously described31 or β-actin (Titan reverse transcriptase (RT)-PCR kit, Roche).
ELISA assays for human LIF, apolipoprotein E, and osteoprotegrin
Medium levels of osteoprotegrin (OPG) were measured using ELISA as previously described.26 Apolipoprotein E (ApoE) was measured using 50 A-G1b as a capture antibody and 50H-G1a as the detection antibody (Academy Bio-Medical, Houston, TX). To measure levels of soluble LIF, MAB250, a capture antibody, was used in combination with biotinylated BAF250 detection antibody (R&D Systems) followed by incubation with streptavidin horseradish peroxidase (Pierce, Rockford, IL). Plates were developed by adding 3, 3′, 5, 5′-tetramentylbenzidine (Pierce) and stopped with the addition of 2 N sulfuric acid. Absorbance was read at 450 nm and background corrected (FLUOstar, BMG Labtech, Durham, NC). ApoE (Calbiochem, San Diego, CA), OPG, and LIF (R&D Systems) concentrations were calculated against reference standards. To normalize protein secretion per cell, cells were lifted and counted using CyQuant cell proliferation assay (Invitrogen). The average was taken from three separate experiments, each with n = 12.
Statistical analysis
All analyses were performed using the Student t-test, as indicated for each respective data set. Statistical significance was defined as p < 0.05. Error bars represent standard deviations.
Results
The ex vivo differentiation potential of expanded BMSCs and dermal fibroblasts
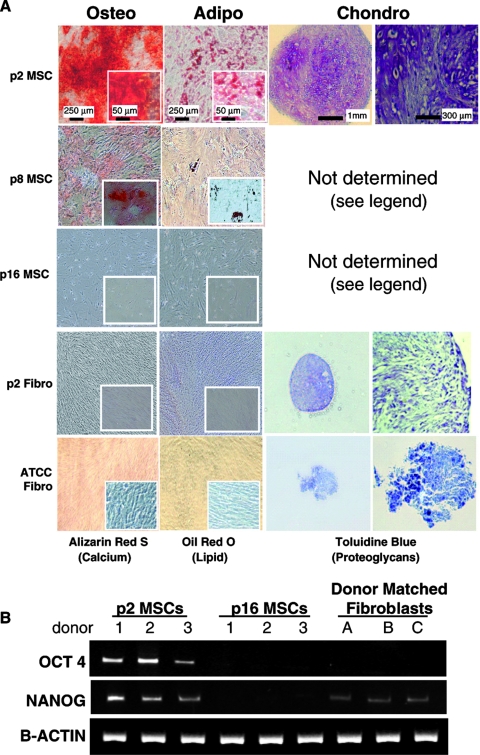
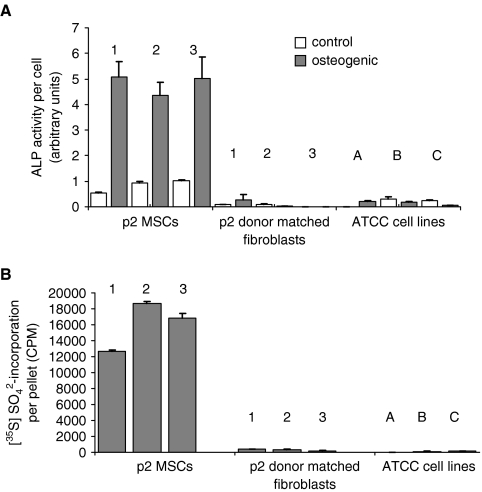
To identify markers of early progenitor markers expressed by BMSCs, a comparative study was first performed to fully characterize the early-passage BMSCs and control cell lines. The progenitor status of passage 2 (p2) BMSCs from three donors was compared with those of four types of control cell lines with little or no predictable differentiation potential. These controls were: extensively expanded passaged, donor-matched BMSCs at p8 and p16, the respective donor-matched dermal fibroblasts at p2, and three commercially acquired dermal fibroblast cell lines. Osteogenic, adipogenic, and chondrogenic differentiation was induced for 21 days. As expected, the p2 BMSCs readily differentiated into osteoblasts, adipocytes, and chrondrocytes, as defined by extensive ARS, ORO, and toluidine blue staining (Fig. 1A).The control cell lines fared poorly in all three assays. In all but a single case, the ARS, ORO, and proteoglycan staining was sporadic or absent (Fig. 1A). The exception was one of the donor-matched fibroblast preparations, which produced an ARS-stained mineralization matrix upon osteogenic stimulus, although adipogenic and chondrogenic assays were both negative (data not shown). Chondrogenesis assays were attempted for p8 and p16 BMSC cultures, but the cells failed to adhere and form micromasses (Fig. 1A). RT-PCR was conducted on p2 and p16 BMSCs and donor-matched fibroblasts to determine the relative expression levels of NANOG and OCT-4, two transcription factors found to play a central role in the regulation of pluripotency and self-renewal. As shown in Figure 1B, early-passage BMSCs highly expressed OCT-4 and NANOG, whereas the p16 BMSCs had no detectable levels of OCT-4 or NANOG mRNA. The donor-matched fibroblast had no detectable levels of OCT-4 message, although they expressed lower levels of NANOG than the p2 BMSCs. To further confirm that the early-passage BMSCs were distinct in their differentiation potential, semiquantitative chondrogenic and osteogenic assays were then performed (Fig. 2). Quantitative osteogenic assays based on ALP activity (Fig. 2A) and chondrogenic assays based on incorporation of radioactive sulfate into proteoglycans (Fig. 2B) were positive for p2 BMSCs but negative for the fibroblasts (including the fibroblasts from the donor that appeared to differentiate in the ARS-based assays). These data confirmed, at the population level, that the minimally expanded BMSCs had a large proportion of progenitor cells and readily differentiated into osteoblasts, adipocytes, and chondrocytes, whereas the fibroblasts and late-passage BMSCs exhibited limited (in the case of p8 cells) or no (in the case of p16 cells and fibroblasts) potential for differentiation. The dermal fibroblast lines acquired from ATCC contrasted most with the p2 BMSCs in that they exhibited no differentiation whatsoever. It was therefore confirmed that the four control cell lines were a valid bank of negative controls for the identification of markers that predict early progenitor status in BMSCs.
FIG. 1.
(A) Micrographs of bone marrow–derived stromal cells (BMSCs), donor-matched skin fibroblasts, and American Type Culture Collection (ATCC) acquired fibroblasts after differentiation into adipocytes, chondrocytes, and osteoblasts. Bone differentiation was detected using Alarizan Red, fat differentiation using Oil Red-O, and cartilage differentiation using toluidine blue. Passage 2 (p2) BMSCs stained strongly positive for all three lineages, whereas p8 and p16 BMSCs and skin fibroblasts stained weakly or negative. Passage 8 and p16 BMSCs consistently failed to form a condensed micromass pellet and were therefore omitted from the panel. Micrographs presented for one of the three cell preparations tested per group. (B) Reverse transcriptase polymerase chain reaction assays for expression of Octamer (OCT)-4 and NANOG by p2 and p16 BMSCs and donor-matched fibroblasts.
FIG. 2.
Quantitative assays of osteogenic and chondrogenic differentiation of bone marrow–derived stromal cells (BMSCs), donor-matched fibroblasts and American Type Culture Collection (ATCC) cell lines. (A) BMSCs (donors 1, 2, and 3), donor-matched fibroblasts (donors 1, 2, and 3), and ATCC fibroblasts (cell lines 1052, 1474, and 2522) were cultured in osteogenic medium (gray bars) or complete culture medium (open bars) for 10 days before alkaline phosphatase (ALP) assays. All BMSC donors had significantly greater ALP activity, but no significant activity was observed in any of the fibroblast cell lines. (B) Incorporation of sulfate was measured over 14 days in chondrogenic conditions. After the assay, the pellets were hydrolyzed, and incorporation of sulfate into proteoglycans was measured. Only the p2 BMSCs significantly incorporated sulfate into their extracellular matrix.
Analysis of cell-surface antigens
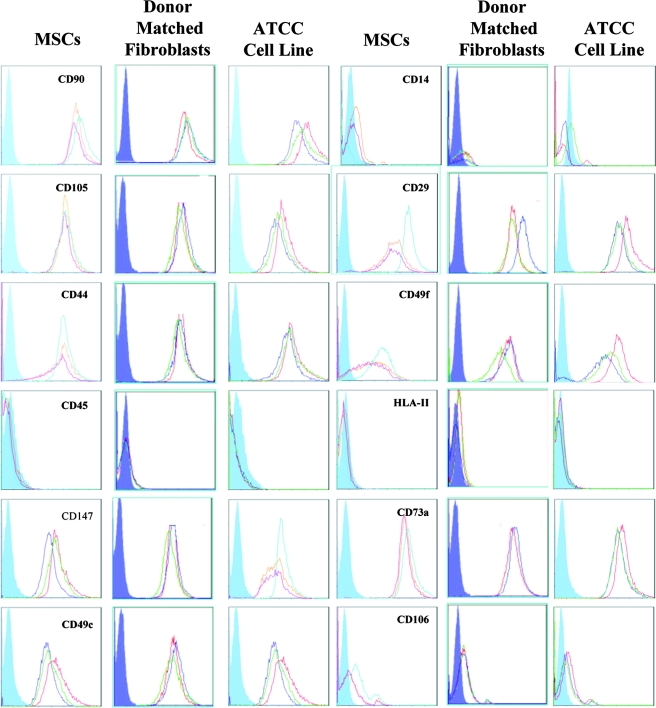
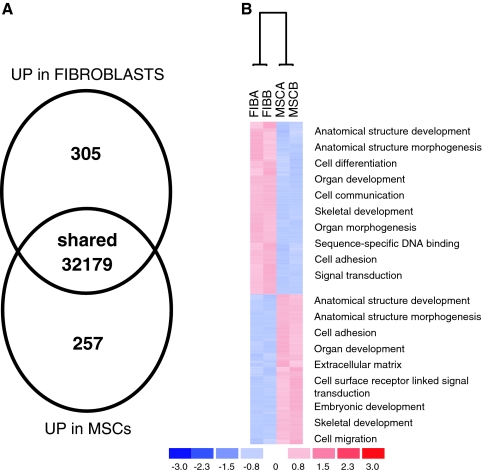
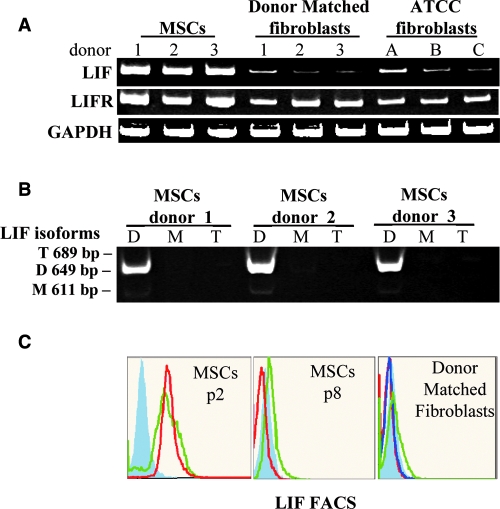
The cell-surface-antigen profile of BMSCs and fibroblasts was examined with the objective of identifying surface markers that were unique to BMSCs. An initial screen of 24 commonly employed surface markers demonstrated that the BMSCs and fibroblasts cell lines were not significantly different. All were negative for CD11b, CD14, CD19, CD34, CD36, CD45, CD79a, CD117, CD184, CD271, and HLA-II; positive for CD29, CD44, CD49b, CD49c, CD49f, CD59, CD73a, CD90, CD105, CD147, CD166, and HLA-ABC; and dim for CD106 (Fig. 3). In an attempt to detect previously unidentified multipotentiality markers, microarray analysis was performed to compare gene expression profiles between BMSCs and fibroblasts. RNA from p2 BMSCs and the ATCC-derived skin fibroblast cell lines were compared because the ATCC-derived cell lines did not detectably differentiate. The RNA from three p2 BMSC donors and three ATCC fibroblast lines were pooled before probe synthesis. Genes that were more up-regulated in BMSCs than in fibroblasts with a signal-log ratio greater than 2 were regarded as potential markers of multipotency. The mRNA samples were remarkably similar, with only a handful of differentiated genes detectable (Fig. 4A). Upon analysis of the gene ontology tags for the differentially expressed genes, it could be seen that BMSCs conspicuously up-regulated genes responsible for embryonic development and migration, whereas the fibroblasts seemed to preferentially express adhesion- and differentiation-related genes (Fig. 4b). Candidate genes that met our stringency parameters included LIF-1, NOTCH 3, and INT α-10, with LIF having the highest up-regulated signal-log ratio of 3.9 (see supplemental data, available online at www.liebertonline.com/ten). Although there were some cell-surface antigens that appeared differentially regulated at the level of mRNA, flow cytometry and protein detection assays demonstrated that all membrane-localized candidates had unpredictable protein presentation at the membrane. We therefore decided to focus our efforts on LIF, because it represented the most differentially transcribed secreted protein on the panel as well as possessing a role in a number of developmental processes, hematopoiesis, and bone metabolism. To confirm the microarray analysis, RT-PCR for total LIF mRNA was performed on all of the p2 BMSCs and fibroblast cell lines. In all instances, LIF mRNA levels were lower in fibroblast cells lines than in the p2 BMSCs. In addition, LIF receptor mRNA was present at equivalent levels in all cultures, suggesting that the LIF-responsive pathways were present in all cell types (Fig. 5A). Because variant LIF transcripts exist, as well as the common mRNA (termed LIF-D), we examined expression of LIF-M (encoding a LIF molecule confined to the extracellular matrix) and LIF-T (a putative intracellular LIF molecule).27,28 When assayed using RT-PCR, only the conventional LIF-D transcript was detected (Fig. 5B) in p2 BMSCs. There were no detectable levels of variant LIF-M and LIF-T transcripts in any of the control cell lines and barely detectable levels of LIF-D mRNA. Fluorescence-activated cell sorting analysis of fixed and permeablized cell preparations confirmed that LIF protein levels correlated with the RT-PCR data (Fig. 5C).
FIG. 3.
Fluorescence-activated cell sorting analysis of bone marrow–derived stromal cells (BMSCs) and skin fibroblasts presenting 12 of the 24 surface antigen profiles tested. Closed blue lines represent isotype controls, and open colored lines represent the three donors tested per group. Inspection of the traces and software analysis demonstrated that the expression profiles were virtually identical. ATCC, American Type Culture Collection.
FIG. 4.
Microarray analysis of pooled bone marrow–derived stromal cells (BMSC) mRNA versus pooled fibroblast mRNA. (A) Summary of up-regulated genes in BMSCs and fibroblasts. (B) Hierarchical clustering of up-regulated genes in BMSCs and fibroblasts with gene ontology tags. Supplemental data for this figure is available online at www.liebertonline.com/ten.
FIG. 5.
Assays of leukemia inhibitory factor (LIF) expression. (A) Reverse transcriptase polymerase chain reaction (RT-PCR) was performed on the microarrray samples to confirm presence of total LIF mRNA. In all instances, LIF mRNA was lower in the fibroblast cell lines than in the early-passage bone marrow–derived stromal cells (BMSCs). In contrast, mRNA expression of the LIF receptor was equivalently expressed by all three cell lines. (B) RT-PCR for three variant LIF transcripts: D, M, and T. In early-passage BMSCs, only the LIF-D transcript was detected. (C) Fluorescence-activated cell sorting analysis on fixed cells for intracellular LIF was positive for passage 2 (p2) BMSCs (n = 2). Donor-matched fibroblasts (n = 3) and p8 BMSCS (n = 2) were negative. Closed blue lines represent isotype controls, and open colored lines represent the donors tested.
LIF secretion as a marker of early progenitor status in BMSCs
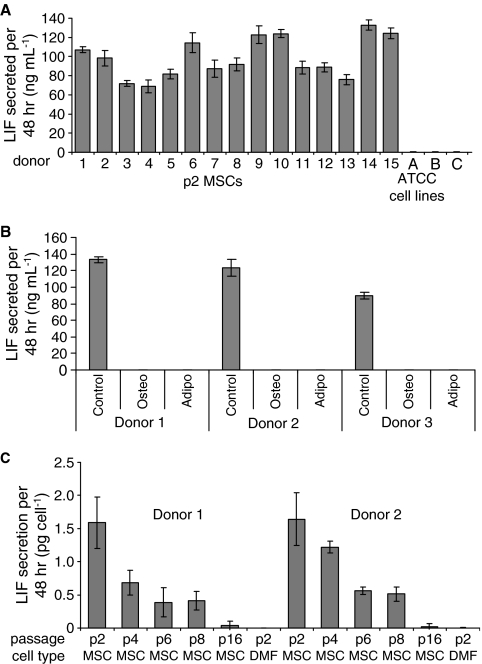
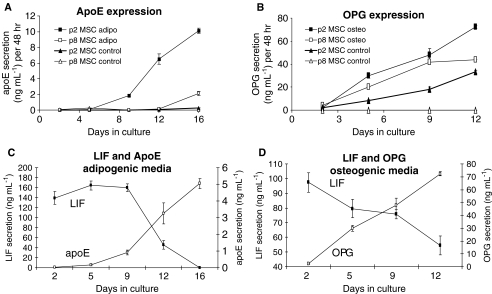
To further examine the potential of LIF as a predictor of early progenitor status in BMSCs, the level of LIF secretion was measured using ELISA assays. In initial experiments, p2 BMSCs and fibroblasts were cultured until 60% to 70% confluency was obtained. From these experiments, we observed that all 15 p2 BMSC donors secreted LIF during the log phase of growth, in contrast to ATCC-derived (Fig. 6A) and donor-matched (Fig. 6C) fibroblasts, which produced barely detectable levels. In the next series of experiments, we examined the effect of passaging on LIF secretion by BMSCs. We observed the maximal secretion of LIF by BMSCs at p2 (Fig. 6C), and levels of LIF secretion gradually decreased to zero as the BMSCs were passaged from p2 to p4 to p6 to p8, to p16 (Fig. 6C). To account for variations in the proliferative capacity of BMSCs at various passages, LIF secretion was normalized to cell number. The results indicate that LIF expression was once again coincident with the presence of progenitor cells, because late-passage BMSCs lost their differentiation potential (Fig. 1). We next examined the effects of BMSC differentiation on LIF secretion. It was hypothesized that committed BMSCs should attenuate expression of LIF as they lose progenitor status through differentiation. Passage 2 BMSCs were incubated with osteogenic or adipogenic differentiation medium for 14 days. The medium conditioned between day 12 and 14 was recovered for LIF analysis. Consistent with the hypothesis that early progenitor BMSCs express LIF, we observed complete down-regulation of LIF secretion upon differentiation of BMSCs (Fig. 6B). We next investigated the kinetics of LIF down-regulation during differentiation. To evaluate the differentiation process without significantly disrupting the cultures, we assayed secreted markers of osteogenesis (OPG) and adipogenesis (ApoE). To validate the use of these proteins as differentiation markers, we compared their expression from p2 and p8 BMSCs in osteogenic and adipogenic assays. As expected, in osteogenic assays, OPG secretion gradually increased from both p2 and p8 BMSCs, but remained lower from p8 BMSCs (Fig. 7B). Similarly, ApoE expression increased in p2 BMSCs, but was much lower in p8 BMSCs (Fig. 7A). For assays of LIF expression, p2 BMSCs were transferred to osteogenic and adipogenic medium for 12 and 16 days, respectively. Media were changed every 2 to 4 days, and conditioned medium was retained for ELISA measurements. For all of the BMSC donors examined, OPG levels rose steadily after 2 days and were accompanied by a steady drop in LIF expression (Fig. 7D). In adipogenic assays, we observed a drop in LIF secretion with up-regulation of ApoE at approximately day 5. Secretion of LIF dropped significantly at day 9 and was undetectable by day 16 (Fig. 7C). The results indicate that LIF expression drops as BMSCs commit to differentiation, further confirming the supporting role of LIF as an indicator of early progenitor status in of BMSCs.
FIG. 6.
Enzyme-linked immunosorbent assay for leukemia inhibitory factor (LIF) secretion by bone marrow–derived stromal cells (BMSCs), donor-matched fibroblasts and American Type Culture Collection (ATCC) fibroblasts (A) LIF secretion by 15 passage 2 (p2) BMSCs donor and ATCC fibroblasts during the log phase of growth. (B) LIF secretion by confluent BMSCs after incubation for 21 days in control medium (complete culture medium), osteogenic differentiation medium (OSTEO), or adipogenic differentiation medium (ADIPO). High LIF secretion was confined to early-passage BMSCs that were not subjected to differentiation medium. (C) LIF secretion by BMSCs at p2–16 and p2 donor-matched skin fibroblasts (DMF) during the log phase of growth.
FIG. 7.
Enzyme-linked immunosorbent assay for leukemia inhibitory factor (LIF), apolipoprotein (apo)E, and osteoprotegrin (OPG) secretion by differentiating bone marrow–derived stromal cells (BMSCs) (A) ApoE secretion by confluent passage 2 (p2) and p8 BMSCs undergoing adipogenic differentiation or in the presence of control medium. (B) OPG secretion by confluent p2 and p8 BMSCs undergoing osteogenic differentiation or in the presence of control medium. (C) Time course comparison of the secretion of apoE and LIF by cultures of p2 BMSCs differentiating into adipocytes. (D) Time course comparison of the expression of OPG and LIF by cultures of p2 BMSCs differentiating into osteoblasts.
LIF secretion and clonally expanded BMSCs
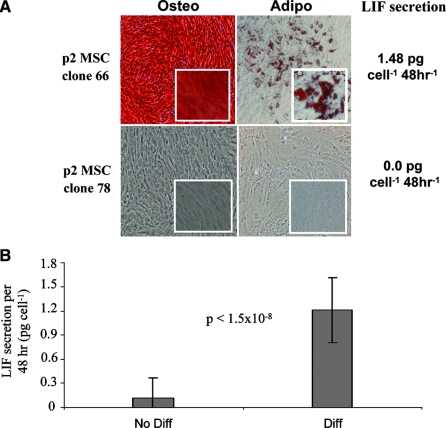
BMSC cultures are a heterogeneous population and thus contain cells with variable proliferative and differentiation potential. We sought to examine whether LIF expression defined BMSCs that could differentiate into both the osteogenic and adipogenic lineage or was associated with cells that could be directed to just one lineage. Single p2 BMSCs from two donors were allowed to establish clonally derived cultures. Resultant cultures (∼60) were then passaged into multiple cultures, re-expanded, and subjected to osteogenic or adipogenic differentiation. Before and during the differentiation assays, LIF levels were assayed from control cultures that did not receive differentiation stimulus. Upon staining, we found only two groups of cells: those that did not differentiate to either lineage and those that formed both osteoblasts and adipocytes (Fig. 8A). Cells that retained bi-directional differentiation potential expressed significantly higher levels of LIF than those that failed to differentiate (Fig. 8B), and of the colonies that failed to differentiate, most did not express LIF at all. Likewise, all BMSCs that differentiated expressed at least 0.8 pg/cell per 48 h, suggesting that, at the single-cell level, BMSCs can maintain their progenitor status or lose the ability to differentiate entirely.
FIG. 8.
Clonal analysis of differentiation potential and leukemia inhibitory factor (LIF) secretion. (A) Representative micrographs of clonally derived bone marrow–derived stromal cell (BMSC) cultures after differentiation into osteoblasts and adipocytes. Clone 66 stained strongly for both lineages and secreted an average of 1.48 pg of LIF per cell. In contrast, clone 78 stained negative for both lineages and had no detectable levels of LIF. (B) LIF secretion from 62 clonally derived cultures. Forty-five clones stained negative for both osteogenic and adipogenic differentiation (No Diff), and 17 clones stained positive for both lineages (Diff). Average LIF expression before differentiation was 0.12 and 1.21 pg/cell per 48 hours, respectively.
Discussion
Despite the increasing interest in the potential therapeutic applications of BMSCs, there are major obstacles to their processing. It has been demonstrated that the standard cultures of BMSCs contain a heterogeneous population of cells, varying in their capacity to differentiate.18 Currently, the most favored method for BMSC expansion is at low monolayer density, which reduces contact-induced differentiation.18 Irrespective of the care taken to expand BMSCs, the cells can dramatically lose their capacity to differentiate even after relatively short periods of expansion. This is probably due to dilution of the early progenitor cells with partially or fully committed precursors. There is therefore a need for an assay that rapidly and robustly predicts the potential efficacy of a given BMSC culture without the necessity for time-consuming and often subjective assays of differentiation. Equipped with such a tool, investigators have the ability to identify and quantify the early progenitor status in BMSC cultures and improve current BMSC culture strategies.
In an attempt to identify the most robust markers of differentiation potential, we carefully designed a comparative study consisting of early-passage BMSCs and a panel of negative controls. The controls consisted of the appropriately passaged donor-matched dermal fibroblasts, late-passage BMSCs, and ATCC-derived fibroblast cell lines. We applied in vitro differentiation assays to osteoblasts, chondrocytes, and adipocytes to validate our sources of early progenitor BMSCs and negative controls. All of the controls had barely detectable or absent potential for transdifferentiation, with the exception of one of the donor-matched fibroblast preparations. The cells from this donor reproducibly differentiated into mineralizing osteoblasts but failed to elicit a positive response in the ALP assays that define early, premineralizing osteoblasts. The cells also failed to produce adipocytic cultures and chondrocyte pellets, suggesting that their transdifferentiation potential was confined to one particular osteogenic pathway that probably bypasses the immature osteogenic stage. Although the physiological relevance of such a phenomenon is therefore questionable, differentiation by fibroblasts has been documented in the literature,21,32,33 demonstrating the need for validation of fibroblast controls and alternative negative controls, such as late-passage BMSCs.
Initially, we set out to compose a detailed profile of surface epitopes currently favored for the identification and characterization of BMSCs. We found no differences in the expression of 24 of the most commonly used markers when fibroblasts and p2 BMSCs were compared. Not surprisingly, all cultures were negative for hematopoietic markers. Markers that are commonly used for proving the identity of BMSCs, especially CD105, CD49, and CD90, were expressed in fibroblasts, demonstrating that these markers are limited in utility.
Comparative transcriptome analysis using the Affymetrix microarray revealed that BMSCs and fibroblasts are strikingly similar, with only a handful of differentially expressed genes between them. The microarray data predicted that some surface epitopes may be useful for direct selection of early progenitor BMSCs, but when analyzed using fluorescence-activated cell sorting, levels of surface protein were variable (see supplemental data). It was therefore decided to examine transcripts that encoded secreted proteins. One of the most prominent of this group of genes was LIF. LIF is a member of the interleukin (IL)6-type cytokine family comprising IL6, IL11, oncostatin M, ciliary neurotrophic factor, cardiotrophin-1, and cardiotrophin-like cytokine.34,35 In addition to their function in inflammation and immunity, these cytokines also play a crucial role in hematopoiesis, tissue regeneration, embryonic development, osteogenesis, and adipogenesis. LIF transduces its signal via sequestration of a monomer of the IL6 receptor (GP130) and the specific LIF receptor (LIFR).36 The resultant complex transmits conformational changes into the cell, leading to a sequence of phosphorylation events and the recruitment of Janus kinase type 3 (Jak3). Jak3 is transiently immobilized at the membrane, where it activates members 1, 3, and 5 of the signal transducer and activation of transcription (STAT) family of transcription factors.35,37 The presence of the LIFR transcript in BMSCs suggests that there is potential for LIF-induced autocrine modulation of the Jak/STAT pathway.
Although the role of LIF secretion by human BMSCs is poorly understood, LIF seems to have a negative effect on differentiation by stromal progenitors. Malaval and colleagues, who demonstrated that chronic LIF exposure could inhibit osteogenic differentiation by rat calvarial progenitor cells38–43 and that dexamethasone41,42 or a dominant negative form of LIF40,43 could antagonize this effect, first reported this. LIF has also been shown to inhibit adipogenesis by a multipotent murine bone marrow–derived cell line.44 Based on the available literature, the effects of LIF on adult stromal progenitor cells appear to mimic the effects of LIF on murine embryonic stem cells, inhibiting inappropriate differentiation while supporting proliferation.45 LIF has been reported to stimulate proliferation of human adipose–derived BMSCs and multipotential adult progenitor cells in vitro.46,47 Although our donor pool had a limited age range (24–46), we found that the proliferative and differentiation capacity of the BMSCs did not appreciably vary with age at early passage. Levels of LIF secretion, which was maintained between 86.4 and 166.16 ng/mL per 48 h, reflected this. Our results are similar to those of Igarashi et al.,48 who examined the effect of age on gene expression by iliac crest–derived bone marrow stromal cells. Samples were obtained from seven young donors (aged 18–39) and LIF secretion was compared with that of seven older donors (aged 53–81). No significant difference in the expression of LIF or differentiation potential was observed between these groups. Control fibroblasts in the study expressed negligible levels of LIF. Therefore, our observations and those of Igarashi et al. strongly suggest that LIF could be of great importance in clinical studies for patients of various ages.
In contrast with some studies, we failed to detect any monopotent cells. In the two key studies that document their existence, their detection was probably dependent on the culture conditions. Muraglia et al.49 employed an insulin-mediated adipogenic protocol, whereas our experiments employed the protocol of Digirolamo et al.,50 which is based on pharmaceutical manipulation of peroxisome proliferator-activated receptors gamma activity and cyclic 3',5'-adenosine monophosphate levels.1 We propose that the adipogenic medium used in this study is more powerful in inducing the adipogenic phenotype, accounting for the detection of more adipogenic clones than in the Maraglia study.50 Indeed, Digirolamo demonstrated that all of their early-passage clones had adipogenic and osteogenic potential; only when the cells were cultured to senescence did they exhibit exclusively osteogenic potential. The BMSC cultures in this study were cultured and tested for differentiation potential within 28 days, a shorter duration than in the Muriglia and Digirolamo studies. It is likely that we would have detected clones with only osteogenic potential if we had increased the number of cell doublings.
In the future, LIF secretion may prove to have an essential role in the growth and maintenance of BMSCs, accounting for its presence in the conditioned medium. The data presented here demonstrate that detection of LIF secretion using ELISA assay represents a robust predictor and indicator of early progenitor status in adult bone marrow stromal cells. Measurement of this parameter is rapid and noninvasive to the culture, suggesting that it may be an invaluable future tool for the production of high-quality BMSCs for clinical trial purposes.
Supplementary Material
Acknowledgments
We thank Bindiya Patel and Dr. Bruce Bunnell (Center for Gene Therapy, Tulane University Health Sciences Center, New Orleans, LA.) for their helpful contribution with Oct-4 assays. The work was supported in part by National Institutes of Health Grants HL075161-01, DK071780, and P20RR020152–1 from National Center For Research Resources and the Louisiana Gene Therapy Research Consortium.
References
- 1.Gregory C.A. Prockop D.J. Spees J.L. Non-hematopoietic bone marrow stem cells: molecular control of expansion and differentiation. Exp Cell Res. 2005;306:330. doi: 10.1016/j.yexcr.2005.03.018. [DOI] [PubMed] [Google Scholar]
- 2.Friedenstein A.J. Chailakhyan R.K. Latsinik N.V. Panasyuk A.F. Keiliss-Borok I.V. Stromal cells responsible for transferring the microenvironment of the hemopoietic tissues. Cloning in vitro and retransplantation in vivo. Transplantation. 1974;17:331. doi: 10.1097/00007890-197404000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Pereira R.F. Halford K.W. O'Hara M.D. Leeper D.B. Sokolov B.P. Pollard M.D. Bagasra O. Prockop D.J. Cultures of adherent cells from marrow can serve as long-lasting precursor cells for bone, cartilage and lung in irradiated mice. Proc Natl Acad Sci U S A. 1995;19:4857. doi: 10.1073/pnas.92.11.4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pittenger M.F. Mackay A.M. Beck S.C. Jaiswal R.K. Douglas R. Mosca J.D. Moorman M.A. Simonetti D.W. Craig S. Marshak D.R. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- 5.Sekiya I. Vuoristo J.T. Larson B.L. Prockop D.J. In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis. Proc Natl Acad Sci U S A. 2002;99:4397. doi: 10.1073/pnas.052716199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sakaguchi Y. Sekiya I. Yagishita K. Ichinose S. Shinomiya K. Muneta T. Suspended cells from trabecular bone by collagenase digestion become virtually identical to mesenchymal stem cells obtained from marrow aspirates. Blood. 2004;104:2728. doi: 10.1182/blood-2003-12-4452. [DOI] [PubMed] [Google Scholar]
- 7.Yoshimura H. Muneta T. Nimura A. Yokoyama A. Koga H. Sekiya I. Comparison of rat mesenchymal stem cells derived from bone marrow, synovium, periosteum, adipose tissue, and muscle. Cell Tissue Res. 2006;372:449. doi: 10.1007/s00441-006-0308-z. [DOI] [PubMed] [Google Scholar]
- 8.Sakaguchi Y. SekiyaI I. Yagishita K. Muneta T. Comparison of human stem cells derived from various mesenchymal tissues: superiority of synovium as a cell source. Arthritis Rheum. 2005;52:2521. doi: 10.1002/art.21212. [DOI] [PubMed] [Google Scholar]
- 9.De Bari C. Dell'Accio F. Tylzanowski P. Luyten F.P. Multipotent mesenchymal stem cells from adult human synovial membrane. Arthritis Rheum. 2001;44:1928. doi: 10.1002/1529-0131(200108)44:8<1928::AID-ART331>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 10.Zuk P.A. Zhu M. Ashjian P. De Ugarte D.A. Huang J.I. Mizuno H. Alfonso Z.C. Fraser J.K. Benhaim P. Hedrick M.H. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miura M. Gronthos S. Zhao M. Lu B. Fisher L.W. Robey P.G. Shi S. SHED: stem cells from human exfoliated deciduous teeth. Proc Natl Acad Sci U S A. 2003;100:5807. doi: 10.1073/pnas.0937635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prockop D.J. Marrow stromal cells as stem cells for non-hematopoietic tissues. Science. 1997;276:711. doi: 10.1126/science.276.5309.71. [DOI] [PubMed] [Google Scholar]
- 13.Simmons P.J. Torok-Storb B. Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1. Blood. 1991;78:55. [PubMed] [Google Scholar]
- 14.Gronthos S. Franklin D.M. Leddy H.A. Robey P.G. Storms R.W. Gimble J.M. Surface protein characterization of human adipose tissue-derived stromal cells. J Cell Physiol. 2001;189:54. doi: 10.1002/jcp.1138. [DOI] [PubMed] [Google Scholar]
- 15.Haynesworth S.E. Baber M.A. Caplan A.I. Cell surface antigens on human marrow-derived mesenchymal cells are detected by monoclonal antibodies. Bone. 1992;13:69. doi: 10.1016/8756-3282(92)90363-2. [DOI] [PubMed] [Google Scholar]
- 16.Letchford J. Cardwell A.M. Stewart K. Coogans K.K. Cox J.P. Lee M. Beresford J.N. Perry M.J. Welham M.J. Isolation of C15: a novel antibody generated by phage display against mesenchymal stem cell-enriched fractions of adult human marrow. J Immunol Methods. 2006;308:124. doi: 10.1016/j.jim.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Carver W. Molano I. Reaves T.A. Borg T.K. Terracio L. Role of the alpha 1 beta 1 integrin complex in collagen gel contraction in vitro by fibroblasts. J Cell Physiol. 1995;165:425. doi: 10.1002/jcp.1041650224. [DOI] [PubMed] [Google Scholar]
- 18.Gregory C.A. Ylostalo J. Prockop D.J. Adult bone marrow stem/progenitor cells (MSCs) are pre-conditioned by micro-environmental “niches” in culture: A two-stage hypothesis for regulation of MSC fate. Sci STKE. 2005;294:pe37. doi: 10.1126/stke.2942005pe37. [DOI] [PubMed] [Google Scholar]
- 19.Ishii M. Koike C. Igarashi A. Yamanaka K. Pan H. Higashi Y. Kawaguchi H. Sugiyama M. Kamata N. Iwata T. Matsubara T. Nakamura K. Kurihara H. Tsuji K. Kato Y. Molecular markers distinguish bone marrow mesenchymal stem cells from fibroblasts. Biochem Biophys Res Commun. 2005;332:297. doi: 10.1016/j.bbrc.2005.04.118. [DOI] [PubMed] [Google Scholar]
- 20.Sekiya I. Larson B.L. Smith J.R. Pochampally R. Cui J.G. Prockop D.J. Expansion of human adult stem cells from bone marrow stroma: conditions that maximize the yields of early progenitors and evaluate their quality. Stem Cells. 2002;20:530. doi: 10.1634/stemcells.20-6-530. [DOI] [PubMed] [Google Scholar]
- 21.Sabatini F. Petecchia L. Tavian M. Jodon de Villeroché V. Rossi G.A. Brouty-Boyé D. Human bronchial fibroblasts exhibit a mesenchymal stem cell phenotype and multilineage differentiating potentialities. Lab Invest. 2005;85:962. doi: 10.1038/labinvest.3700300. [DOI] [PubMed] [Google Scholar]
- 22.Heinrich P.C. Behrmann I. Haan S. Hermanns H.M. Müller-Newen G. Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374:1. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verfaillie C. McGlave P. Leukemia inhibitory factor/human interleukin for DA cells: a growth factor that stimulates the in vitro development of multipotential human hematopoietic progenitors. Blood. 1991;77:263. [PubMed] [Google Scholar]
- 24.Carpenter M.K. Cui X. Hu Z.Y. Jackson J. Sherman S. Seiger A. Wahlberg L.U. In vitro expansion of a multipotent population of human neural progenitor cells. Exp Neurol. 1999;158:265. doi: 10.1006/exnr.1999.7098. [DOI] [PubMed] [Google Scholar]
- 25.Colter D.C. Class R. DiGirolamo C.M. Prockop D.J. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from bone marrow. Proc Natl Acad Sci U S A. 2000;97:3213. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gunn W.G. Conley A. Deininger L. Olson S.D. Prockop D.J. Gregory C.A. A crosstalk between myeloma cells and marrow stromal cells stimulates production of DKK1 and interleukin-6: a potential role in the development of lytic bone disease and tumor progression in multiple myeloma. Stem Cells. 2005;24:986. doi: 10.1634/stemcells.2005-0220. [DOI] [PubMed] [Google Scholar]
- 27.Knight D.A. Lydell C.P. Zhou D. Weir T.D. Robert Schellenberg R. Bai T.R. Leukemia inhibitory factor (LIF) and LIF receptor in human lung. Am J Respir Cell Mol Biol. 1999;20:834. doi: 10.1165/ajrcmb.20.4.3429. [DOI] [PubMed] [Google Scholar]
- 28.Hisaka T. Desmouliere A. Taupin J.L. Daburon S. Neaud V. Senant N. Blanc J.F. Moreau J.F. Rosenbaum J. Expression of leukemia inhibitory factor (LIF) and its receptor gp190 in human liver and in cultured human liver myofibroblasts. Cloning of new isoforms of LIF mRNA. Comp Hepatol. 2004;263:10. doi: 10.1186/1476-5926-3-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jungwoon L. Hye Kyoung K. Jeung-Yon R. Yong-Mahn H. Jungho K. The human OCT-4 isoforms differ in their ability to confer self-renewal. J Biol Chem. 2006;281:33554. doi: 10.1074/jbc.M603937200. [DOI] [PubMed] [Google Scholar]
- 30.Takahashi K. Tanabe K. Ohnuki M. Narita M. Ichisaka T. Tomoda K. Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:5. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 31.Gregory C.A. Singh H. Perry A.S. Prockop D.J. The Wnt signaling inhibitor dickkopf-1 is required for reentry into the cell cycle of human adult stem cells from bone marrow. J Biol Chem. 2003;278:28067. doi: 10.1074/jbc.M300373200. [DOI] [PubMed] [Google Scholar]
- 32.Ishii G. Ito TK. Aoyagi K. Fujimoto H. Chiba H. Hasebe T. Fujii S. Nagai K. Sasaki H. Ochiai A. Presence of human circulating progenitor cells for cancer stromal fibroblasts in the blood of lung cancer patients. Stem Cells. 2007;26:1469. doi: 10.1634/stemcells.2006-0449. [DOI] [PubMed] [Google Scholar]
- 33.Sudo K. Kanno M. Miharada K. Ogawa S. Hiroyama T. Saijo K. Nakamura Y. Mesenchymal progenitors able to differentiate into osteogenic, chondrogenic, and/or adipogenic cells in vitro are present in most primary fibroblast-like cell populations. Stem Cells. 2007;25:1610. doi: 10.1634/stemcells.2006-0504. [DOI] [PubMed] [Google Scholar]
- 34.Heinrich P.C. Behrmann I. Haan S. Hermanns H.M. Muller-Newen G. Schaper F. Principles of interleukin (IL)-6-type cytokine signalling and its regulation. Biochem J. 2003;374(Pt 1):1. doi: 10.1042/BJ20030407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stahl N. Boulton T.G. Farruggella T. Ip N.Y. Davis S. Witthuhn B.A. Quelle F.W. Silvennoinen O. Barbieri G. Pellegrini S. Association and activation of Jak-Tyk kinases by CNTF-LIF-OSM-IL-6 beta receptor components. Science. 1994;263:92. doi: 10.1126/science.8272873. [DOI] [PubMed] [Google Scholar]
- 36.Bravo J. Heath J.K. Receptor recognition by gp130 cytokines. EMBO J. 2000;19:2399. doi: 10.1093/emboj/19.11.2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hermanns H.M. Radtke S. Haan C. Schmitz-Van de Leur H. Tavernier J. Heinrich P.C. Behrmann I. Contributions of leukemia inhibitory factor receptor and oncostatin M receptor to signal transduction in heterodimeric complexes with glycoprotein 130. J Immunol. 1999;163:6651. [PubMed] [Google Scholar]
- 38.Malaval L. Gupta A.K. Aubin J.E. Leukemia inhibitory factor inhibits osteogenic differentiation in rat calvaria cell cultures. Endocrinology. 1995;136:411. doi: 10.1210/endo.136.4.7895651. [DOI] [PubMed] [Google Scholar]
- 39.Malaval L. Aubin J.E. Biphasic effects of leukemia inhibitory factor on osteoblastic differentiation. Cell Biochem. 2001;S36:63. doi: 10.1002/jcb.1086. [DOI] [PubMed] [Google Scholar]
- 40.Malaval L. Liu F. Vernallis A.B. Aubin J.E. GP130/OSMR is the only LIF/IL-6 family receptor complex to promote osteoblast differentiation of calvaria progenitors. J Cell Physiol. 2005;204:585. doi: 10.1002/jcp.20312. [DOI] [PubMed] [Google Scholar]
- 41.Malaval L. Gupta A.K. Liu F. LIF, but not IL-6, regulates osteoprogenitor differentiation in rat calvaria cell cultures: modulation by dexamethasone. J Bone Miner Res. 1998;13:175. doi: 10.1359/jbmr.1998.13.2.175. [DOI] [PubMed] [Google Scholar]
- 42.Liu F. Aubin J.E. Malaval L. Expression of leukemia inhibitory factor (LIF)/interleukin-6 family cytokines and receptors during in vitro osteogenesis: differential regulation by dexamethasone and LIF. Bone. 2002;31:212. doi: 10.1016/s8756-3282(02)00806-2. [DOI] [PubMed] [Google Scholar]
- 43.Vernallis A.B. Hudson K.R. Heath J.K. An antagonist for the leukemia inhibitory factor receptor inhibits leukemia inhibitory factor, cardiotrophin-1, ciliary neurotrophic factor, and oncostatin M. M J Biol Chem. 1997;272:26947. doi: 10.1074/jbc.272.43.26947. [DOI] [PubMed] [Google Scholar]
- 44.Gimble J.M. Wanker F. Wang C.S. Bass H. Wu X. Kelly K. Yancopoulos G.D. Hill M.R. Regulation of bone marrow stromal cell differentiation by cytokines whose receptors share the gp130 protein. Cell Biochem. 1994;54:122. doi: 10.1002/jcb.240540113. [DOI] [PubMed] [Google Scholar]
- 45.Pease S. Braghetta P. Gearing D. Grail D. Williams R.L. Isolation of embryonic stem (ES) cells in media supplemented with recombinant leukemia inhibitory factor (LIF) Dev Biol. 1990;141:344. doi: 10.1016/0012-1606(90)90390-5. [DOI] [PubMed] [Google Scholar]
- 46.Song H.Y. Jeon E.S. Jung J.S. Kim J.H. Oncostatin M induces proliferation of human adipose tissue-derived mesenchymal stem cells. Int J Biochem Cell Biol. 2005;37:2357. doi: 10.1016/j.biocel.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 47.Jiang Y. Jahagirdar B.N. Reinhardt R.L. Schwartz R.E. Keene C.D. Ortiz-Gonzalez X.R. Reyes M. Lenvik T. Lund T. Blackstad M. Du J. Aldrich S. Lisberg A. Low W.C. Largaespada D.A. Verfaillie C.M. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature. 2002;418:41. doi: 10.1038/nature00870. [DOI] [PubMed] [Google Scholar]
- 48.Igarashi A. Segoshi K. Sakai Y. Pan H. Kanawa M. Higashi Y. Sugiyama M. Nakamura K. Kurihara H. Yamaguchi S. Tsuji K. Kawamoto T. Kato Y. Selection of common markers for bone marrow stromal cells from various bones using real-time RT-PCR: effects of passage number and donor age. Tissue Eng. 2007;13:2405. doi: 10.1089/ten.2006.0340. [DOI] [PubMed] [Google Scholar]
- 49.Muraglia A. Cancedda R. Quarto R. Clonal mesenchymal progenitors from human bone marrow differentiate in vitro according to a hierarchical model. J Cell Sci. 2000;113:1161. doi: 10.1242/jcs.113.7.1161. [DOI] [PubMed] [Google Scholar]
- 50.Digirolamo C. Stokes D. Colter D. Phinney D.G. Class R. Prockop D.J. Propagation and senescence of human marrow stromal cells in culture: a simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107:275. doi: 10.1046/j.1365-2141.1999.01715.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.