Abstract
The recently determined crystal structure of the human β2-adrenergic (β2AR) G-protein coupled receptor provides an excellent structural basis for exploring β2AR -ligand binding and dissociation process. Based on this crystal structure, we simulated ligand exit from the β2AR receptor by applying the random acceleration molecular dynamics (RAMD) simulation method. The simulation results showed that the extracellular opening on the receptor surface was the most frequently observed egress point (referred to as pathway A) and a few other pathways through inter-helical clefts were also observed with significantly lower frequencies. In the egress trajectories along pathway A, the D192-K305 salt bridge between the extracellular loop 2 (ECL2) and the apex of the transmembrane helix 7 (TM7) was exclusively broken. The spatial occupancy maps of the ligand computed from the 100 RAMD simulation trajectories indicated that the receptor-ligand interactions that restrained the ligand in the binding pocket were the major resistance encountered by the ligand during exit and no second barrier was notable. We next performed RAMD simulations by using a putative ligand-free conformation of the receptor as input structure. This conformation was obtained in a standard MD simulation in the absence of the ligand and it differed from the ligand-bound conformation in a hydrophobic patch bridging ECL2 and TM7 due to the rotation of F193 of ECL2. Results from the RAMD simulations with this putative ligand-free conformation suggest that the cleft formed by the hydrophobic bridge, TM2, TM3 and TM7 on the extracellular surface likely serves as a more specific ligand-entry site and the ECL2-TM7 hydrophobic junction can be partially interrupted upon the entry of ligand that pushes F193 to rotate, resulting in a conformation as observed in the ligand-bound crystal structure. These results may help design β2AR-targeting drugs with improved efficacy as well as understand the receptor subtype-selectivity of ligand binding in the β family of the adrenergic receptors that share almost identical ligand-binding pockets but show notable amino acid sequence divergence in the putative ligand-entry site, including ECL2 and the extracellular end of TM7.
Keywords: β2-adrenergic receptor, ligand, exit pathway, molecular dynamics simulation, conformational change
Introduction
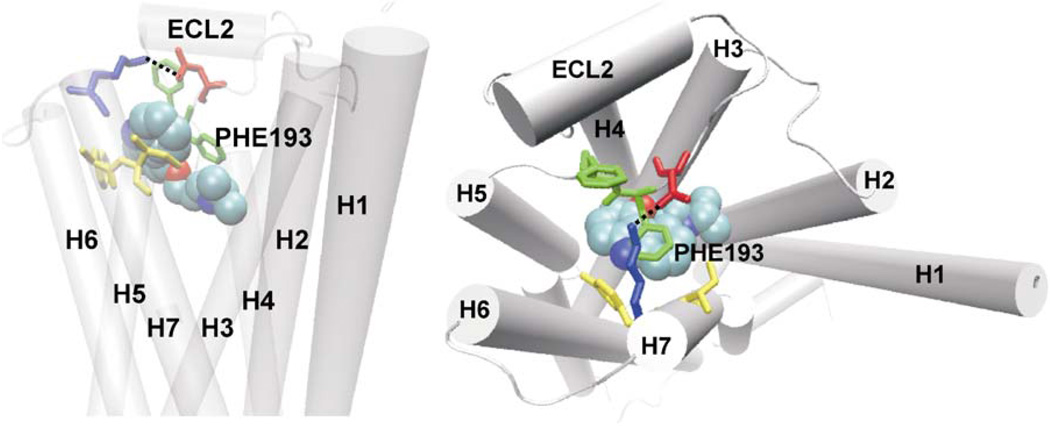
β-adrenergic receptors (βARs) belong to the G-protein coupled receptor (GPCR) super-family and are activated by catecholamine hormones and related molecules. Among the GPCRs that serve as the targets of ca. 50% drugs on the market, βARs represent one of the most important such therapeutic targets. There are three subtypes in the β subfamily: β1, β2, and β3. The inhibitors of βARs, known as β-blockers, are used to treat cardiovascular disorders1 and β2-selective agonists are the mainstay in the treatment of asthma and the chronic obstructive pulmonary disease.2,3 Recently, the crystal structure of the human β2-adrenergic receptor (β2AR) was determined and is one of the two available crystal structures of human GPCRs that bind diffusible ligands.4–8 The β2AR structure shows a high degree of similarity to the rhodopsin structures9,10, consisting of a seven transmembrane helices with connecting loops at the extracellular and the cytoplasmic sides and a partial inverse agonist (carazolol or timotol) bound in the transmembrane region. However, a key difference exits in the second extracellular loop (ECL2) of the β2AR structure compared with the rhodopsin structures. Unlike in the rhodopsin structures in which ECL2 forms a β-hairpin, folding down into the transmembrane core and thus covers the ligand-binding pocket, ECL2 in β2AR forms a short α-helix displaced away from the ligand-binding pocket, exhibiting a partial opening on the extracellular surface. This opening appears to serve as a ligand entrance to the binding pocket that is deeply buried in the transmembrane region. The ligand, carazolol (PDB entry 2RH1) sits at the bottom of the binding pocket, ca. 13.0Å deep from the putative entrance and is flanked by two aromatic residues at the extracellular top of the binding pocket: F193 of ECL2 and Y308 of the transmembrane helix 7 (TM7). Right above the binding-site crevice is a salt bridge of D192-K305 between ECL2 and the apex of TM7 that divides the extracellular opening into two small areas. In the binding pocket, carazolol formed close interactions with several polar transmembrane residues, D113(TM3), N312(TM7), Y316(TM7), S203(TM5) and S207(TM5), and the interactions include three hydrogen bonds between its alkylamine N atom with D113 and N312 and the alcohol O atom with N312. The crystal structure (see Figure 1) shows a static view of β2AR-ligand bound state but lacks information about how a ligand diffuses in and out of the receptor or any conformational changes of the receptor accompanying ligand binding and dissociation. These questions are of importance as understanding the routes and mechanisms of ligand entry and exit may help to design drugs with higher efficacy and longer duration of action.
Figure 1.
The crystal structure of the β2AR-carazolol complex structure. Left: the β2AR receptor is represented by cartoon and the seven transmembrane helices are labeled. Carazolol is represented by van der Waals spheres. Residues Asp192 and Phe193 of ECL2, and Lys305 and Tyr308 of TM7 are represented by stick and in different colors. The salt bridge between Asp192 and Lys305 is represented by dash line, which partially covers the extracellular opening and divides it into two small areas. Right: carazolol formed close interactions with several polar transmembrane residues D113 (TM3), N312 (TM7), Y316(TM7), S203(TM5) and S207(TM5). Hydrogen bonds are represented by dash lines.
In addition, because the residues lining the ligand-binding pockets in the β-family of adrenergic receptors share high sequence similarity among the β1, β2 and β3 subtypes, differences in the mechanisms of ligand entry and exit may play a role in receptor sub-type selectivity of ligand binding. In the β2AR crystal structure with carazolol bound (PDB entry 2RH1), the residues within 5.0Å distance of carazolol are identical to the corresponding residues in β1 and β3, except for Y308, which is a phenylanaline in both β1 and β3, and T195 being a serine in β3. However, the extracellular surface residues, in particular in the region around the putative entrance, show much less conservation, which may introduce differences in ligand entry and exit.
Compared to the transmembrane core region, the extracellular loops are less known with respect to their structural and functional roles in GPCR-ligand binding and activation. The recent studies showed that in addition to its best-known role of ligand recognition, the ECL2 was found to be critical to ligand-binding kinetics due to its conformational flexibility11 and its interactions with the proximal helices contributed to the stability of the inactive states of GPCRs.12,13 These findings may have general implications to the GPCRs beyond those studied in the experiments.
To explore these questions, we first simulated the exit of carazolol from β2AR’s binding pocket in a solvated phospholipid bilayer environment by applying the random acceleration molecular dynamics (RAMD) method.14–19 In the RAMD simulations, a small randomly oriented force was applied to the center of mass of carazolol to accelerate its dynamics of motions. With the extra random force, the carazolol molecule has an enhanced tendency to break the interactions that restrain it in the binding pocket and exit in a computationally feasible time. Unlike methods such as “steered molecular dynamics”20 which simulates a ligand exiting from the binding pocket along a pre-defined deterministic path, RAMD does not assume the path and allows the ligand to “find” its way out, providing a realistic way to evaluate plausible pathways. The broken interactions along the path of ligand exit may indicate the resistance to ligand dissociation as well as the obstacles for ligand entry. By analyzing 100 egress simulation trajectories, we found that carazolol predominantly exited from the binding pocket through the opening on the extracellular surface via diverse routes and the exit caused the breakage of the hydrogen bond between D192 and K305. The simulations trajectories were further analyzed by computing the ligand spatial occupancy maps to obtain a statistical view on the distribution and frequency of ligand’s transient stay within the receptor before it reached the egress point, which together with structural inspection of the trajectories elucidated the ligand exit mechanisms. We then investigated the conformational flexibility of the β2AR receptor in the presence as well as the absence of carazolol by applying standard MD simulations for a total of over 120 ns trajectories. The most significant conformational change was observed in F193 of ECL2, which rotated to point toward TM7 and formed tight packing with the D192-K305 salt bridge in a carazolol-absent simulation. This conformational change created a hydrophobic cluster bridging ECL2 and TM7. On the assumption that this conformation represents the ligand-free state of the receptor, we performed another 100 RAMD simulations to probe the pathways of ligand entry and the role of the ECL2-TM7 hydrophobic bridge.
Results
Exit from the carazolol-bound crystal structure
Two sets of RAMD simulations were performed to investigate carazolol exit from the crystal structure. The first set of 60 simulations was carried out with four different magnitudes of accelerations 0.20, 0.23, 0.25, 0.30 kcal/Å·g along with different random number generation seeds. The second set of 40 simulations was carried out with the smallest acceleration used in the first set. The observations from these two sets of simulations were statistically summarized and compared in Table 1. Complete exit from the receptor was observed in all 60 simulations of the first set and 38 of 40 simulations of the second set. All the 100 simulation trajectories were visualized in Figure 2 by showing the path of the center of mass of carazolol. The exit pathways can be classified into six pathways based on the point of egress: the extracellular surface opening (pathway A, i.e. the putative entrance), the cleft between TM4 and TM5 (pathway B), the cleft between TM5 and TM6 (pathway C), the cleft between TM1 and TM2 (pathway D), the cleft between TM1 and TM7 (pathway E) and the cleft between TM6 and TM7 (pathway F).
Table 1.
Statistical summary of the RAMD simulations of the carazolol-bound crystal structure. The results are separately listed from 60 simulations with 4 different accelerations and 40 simulations with the smallest acceleration. The latter results are in the gray-shaded rows.
| Pathway | Egress point | Number of trajectories |
Acceleration (kcal/Å·g) |
Trajectory length (ps) |
|---|---|---|---|---|
| A | Extracellular opening | 41 (19a, 21b) | 0.20, 0.23, 0.25, 0.30 | 22–976 |
| 28 (11a, 17 b) | 0.20 | 22–928 | ||
| B | H4–H5 cleft | 16 | 0.20, 0.23, 0.25, 0.30 | 16–878 |
| 5 | 0.20 | 144–596 | ||
| C | H5–H6 cleft | 2 | 0.30, 0.30 | 90, 104 |
| 1 | 0.20 | 152 | ||
| D | H1–H2 cleft | 1 | 0.20 | 500 |
| 1 | 0.20 | 930 | ||
| E | H1–H7 cleft | 0 | ||
| 2 | 0.20 | 564,1000 | ||
| F | H6–H7 cleft | 0 | ||
| 1 | 0.20 | 438 | ||
| No egress | 0 | |||
| 2 | 0.20 | 1000 |
carazolol passed out of the receptor from the “left” side (sub-pathway A1) and the “right” side(sub-pathway A2) of the D192-K305 salt bridge, respectively.
Figure 2.
Paths of the center of mass of carazolol in the RAMD trajectories starting from the ligand-bound crystal structure. Left: the first set of 60 simulations with four different acceleration magnitudes; Right: the second set of 40 simulations with the smallest acceleration used in the first set. The color scale is from yellow (start) to blue (end). The starting conformation of the receptor is shown in cartoon. For clarity, the second extracellular loop of residues 173 to 196 and the solvated lipid bilayer environment are not shown. In both sets of the simulations, the extracellular surface opening (pathway A) was the most frequently observed egress point, and the TM4–TM5 cleft (pathway B) was the second.
Extracellular surface opening - pathway A
The egress from the extracellular opening (pathway A) was observed in 41 of 60 egress trajectories in the first set and 28 of 38 egress trajectories (no egress in 2 simulations) in the second set, being the most frequently observed egress pathway in both simulation sets. This indicates that the putative entrance is also the most likely ligand exit pathway. Furthermore, by using the D192-K305 salt bridge as a division line, pathway A can be classified into two sub-pathways A1 and A2, in which the surface opening of sub-pathway A1 is on the “left” side of the salt bridge, formed by TM5, TM6 and TM7 and sub-pathway A2 is the “right” cleft formed by TM2, TM3 and TM7 (see Figure 1 for the location of the D192-K305 salt bridge). These two sub-pathways were observed to have practically equal frequency to be used as an exit pathway. Figure 3 shows the snapshots of two egress trajectories along these two sub-pathways. The ligand egress along pathway A started with the upward tilting of the carbazole ring head and proceeded with the further ascent of the ring system into either the “left” cleft or the “right” cleft, while the alkylamine-alcohol tail was tethered by the hydrogen bonds with D113 (TM3) and N312 (TM7). Once these hydrogen bonds were broken, the ligand exited the receptor quickly. This exit mechanism also resulted in the observation that the heterocyclic ring of carazolol passed out first in all egress trajectories.
Figure 3.
Ligand exit mechanisms along the extracellular surface opening (pathway A). Snapshots of two egress trajectories in which carazolol passed out of the protein from the right side (upper panel, sub-pathway A2) and the left side (lower panel, sub-pathway A1) of the D192-K305 salt bridge (not shown). The protein is shown in transparent cartoon. Carazolol is shown in stick surrounded by its molecular surface presentation and the initial bound position is colored yellow. The side chain of the protein residues D113 (red, TM3), N312 (green, TM7), Y316 (green, TM7) are shown in stick. The red balls represent water molecules at 3.0Å distance of carazolol and the three protein residues. Hydrogen bonds between carazolol and protein residues are shown in dash lines, which were the major resistance encountered by the ligand during exit. Two movies showing the complete egress trajectories are available in Supporting Information (Movies 1–2).
D192-K305 salt bridge
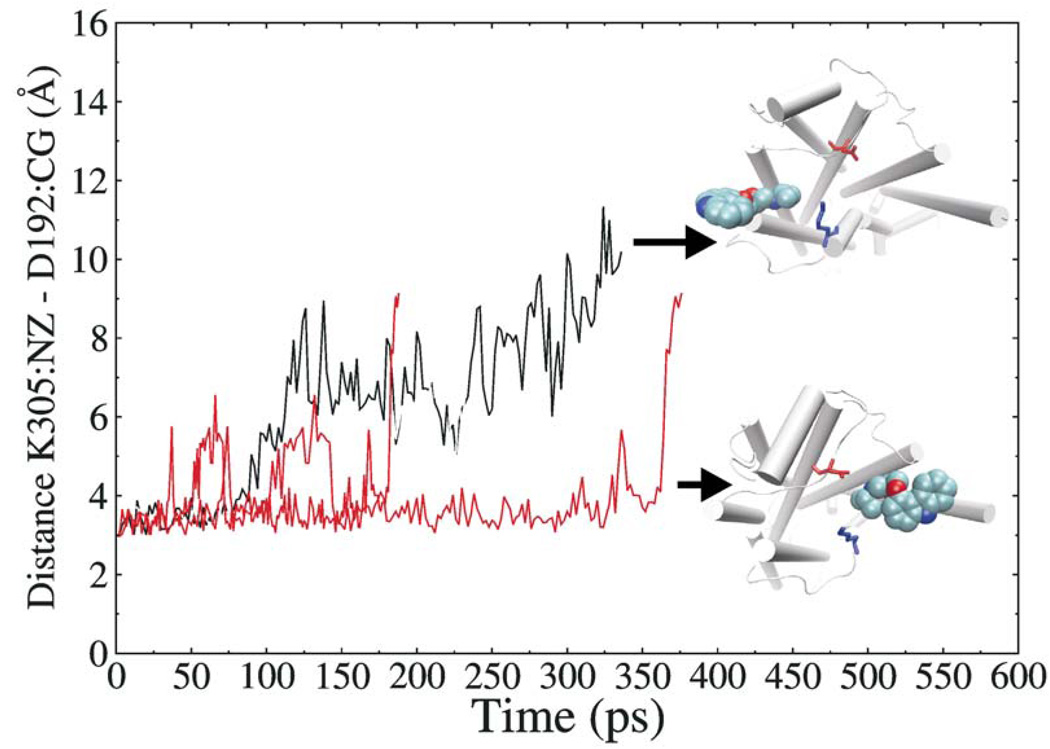
Another significant observation was that egress along pathway A exclusively interrupted the D192-K305 salt bridge (see Figure 4). In most trajectories, the breakage of the salt bridge was due to the direct collision with carazolol at the last moment of exiting the receptor, and in a few trajectories, the salt bridge was already lost at the early stage of egress. These results can indicate the low stability of the salt bridge in the ligand egress process and that the salt bridge may represent a last obstacle influencing ligand passage along the extracellular opening.
Figure 4.
Variation of the D192-K305 salt bridge distance as a function of time in two egress trajectories along pathway A (sub-pathways A1 and A2, respectively).
Inter-helical clefts - pathways B – F
Despite the presence of the extracellular opening and the closure of the transmembrane helix bundle, carazolol was observed to pass out of the protein from inter-helical clefts in 19 egress trajectories in the first simulation set (pathways B, C and D) and 10 egress trajectories in the second simulation set (pathways B, C, D, E, and F). While pathways D–F are rather rarely observed, pathway B (TM4–TM5 cleft) is statistically significant, accounting for 27% egress trajectories of the first simulation set and 20% of the second simulation set. The egress opening along pathway B was made by transiently breaking the inter-helical hydrophobic interactions due to the stretching and bumping of the ligand’s heterocycle ring head. In our previous study on retinal channeling in the inactivated bovine rhodopsin,18 the TM4–TM5 cleft was also identified as one of the major egress pathways of retinal. In both β2AR and rhodopsin structures, the ring systems of the ligands are located proximal to TM4 and TM5. Therefore, the TM4–TM5 cleft may present a relatively straight way for the ligands to exit the binding pocket.
In addition, the major difference in the results from the two simulation sets lie in the additional pathways E (TM1–TM7 cleft) and F (TM6–TM7 cleft) found only in the second set. This was most likely due to the larger numbers of simulations with the small acceleration used in the second set, which allowed the ligand to take time to explore less straight pathways.
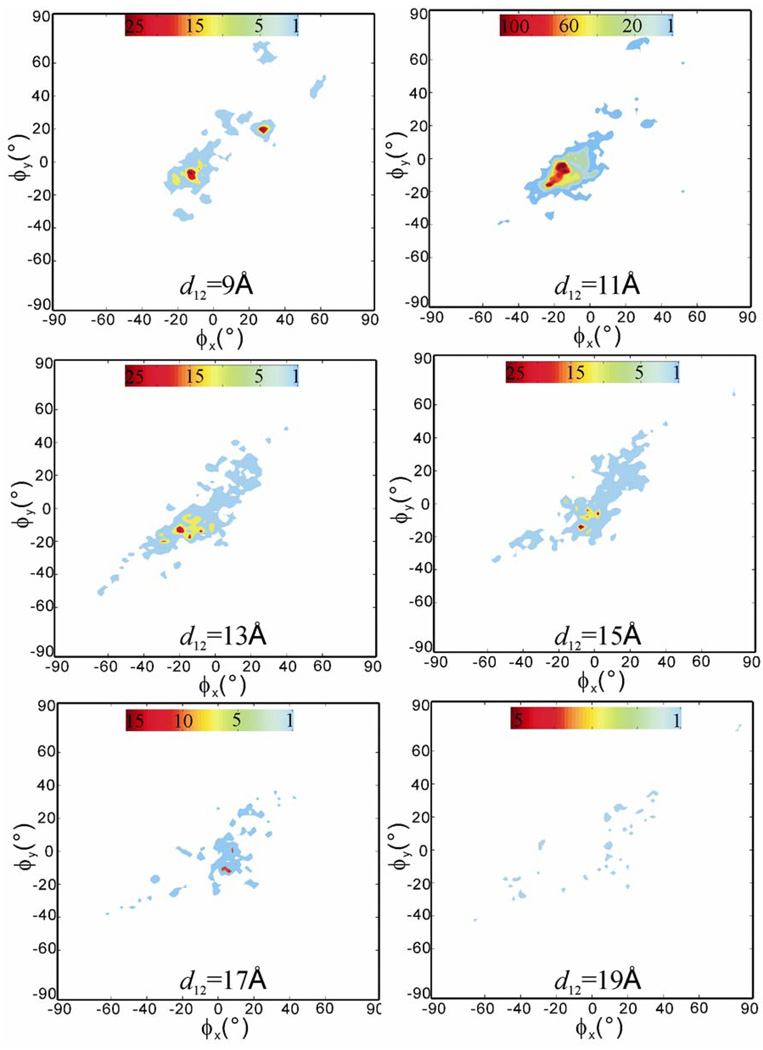
Spatial occupancy maps of the ligand
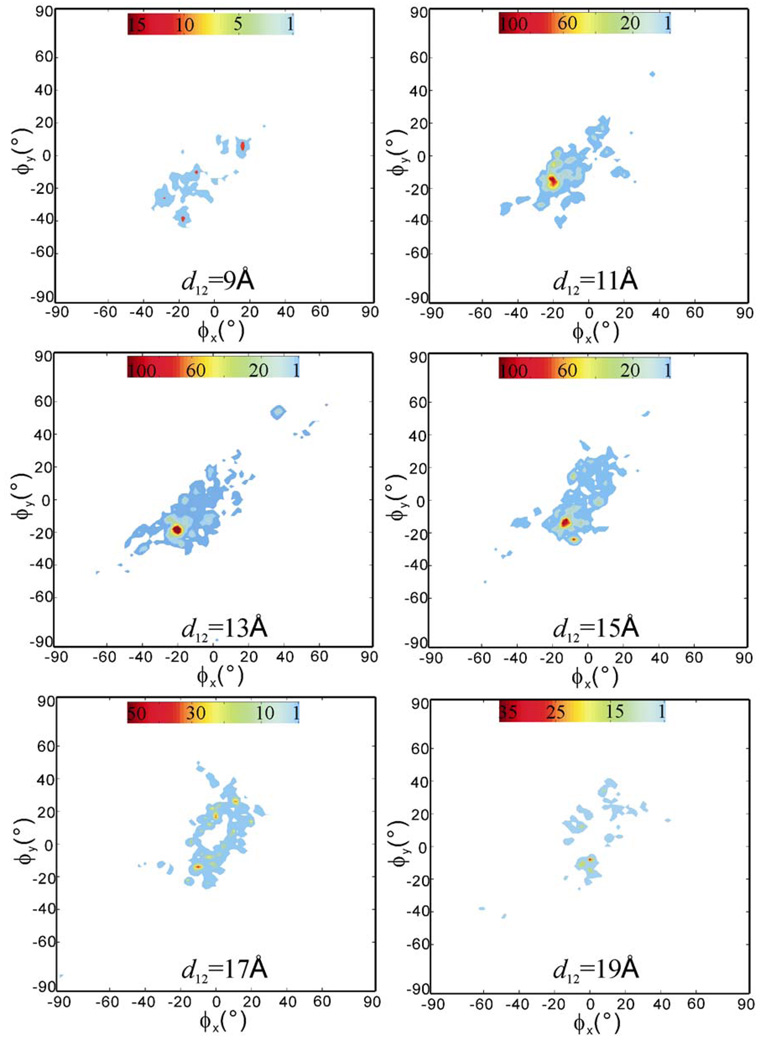
To obtain a statistical view on the distribution and frequency of ligand’s transient stay within the receptor during the RAMD simulations, we computed the spatial occupancy maps of the ligand as a function of the receptor-ligand center-to-center distance d12. The occupancy maps are two-dimensional population histograms defined by two azimuth angles, the ϕx, ϕy angles that show the relative positions of the ligand in the receptor structure (see the Method section for the definition of the ϕx, ϕy angles). All 100 RAMD trajectories were used. Figure 5 shows the maps computed at d12 =9, 11, 13, 15, 17 and 19Å with an allowance of ±1.0Å, in which 11Å is close to the initial receptor-ligand distance of 11.6Å and 19Å is the farthest distance that the ligand can move away while still staying in the receptor structure. We can see that the space traversed by the ligand, i.e, the colored regions in the maps of Figure 5, was rather limited, which reflects the fact that the movement of the ligand was confined within the helical bundle of the receptor structure. The high occupancy region, colored red with small negative ϕx, ϕy angles in the map at d12 = 11 Å indicates the most stayed region, which is close to the initial bound position. With the increase of d12, i.e. the moving up of the ligand toward the extracellular surface opening, the occupancy clouds became wider and less dense and no comparable maximum was observed in the maps at d12 = 13 to 19 Å. These results indicate that the ligand-receptor interactions that restrained the ligand in the binding pocket were the major resistance encountered by the ligand during exit and no second barrier was notable. The interactions were mainly the hydrogen bonds formed between D113 (TM3) and N312 (TM7) and the alkylamine-alcohol tail of the ligand. At the early stage of egress, the carbazole ring head either stretched away or bended toward its alkylamine-alcohol tail, resulted in the change of mass center distance d12 while the tail was still tethered by the hydrogen bonds in the position close to the initial bound position. This tethering effect was reflected in the minor maximum areas in the maps of d12=9, 13, and 15 Å, and the structural states can be observed in Figure 3. However, the low occupancy clouds spread in the maps of d12=13–19 Å indicate that the exit routes are diverse.
Figure 5.
The computed spatial occupancy maps of the ligand from 100 RAMD trajectories as a function of receptor-ligand center-to-center distance d12 = 9, 11, 13, 15, 17, and 19Å. The starting structure is the ligand-bound crystal structure. Red means high occupancy and blue low occupancy. Note: each map has its own color scale.
Formation of a hydrophobic patch bridging ECL2 and TM7 in the absence of carazolol
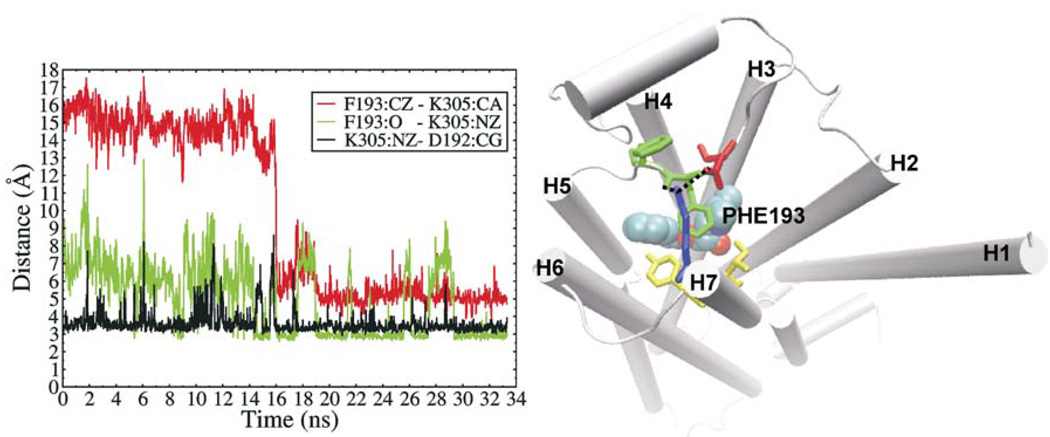
Conformational dynamics of the β2AR structure were investigated by the standard MD simulations in the presence as well as the absence of carazolol. In the two simulations starting from the carazolol-bound crystal structure, which lasted for 28.2 ns and 28.4 ns, respectively, carazolol remained bound in the binding pocket and the receptor structure did not show notable change. The backbone root-mean-square deviations (RMSD) of the receptor from the crystal structure were lower than 2.0Å. However, we found that the D192 - K305 salt bridge opened and reformed spontaneously and frequently during the simulations (see Supporting Information Figure S1). This dynamic nature of the salt bridge would be favorable for ligand exit as its breakage has been observed in all egress trajectories along the predominant pathway (pathway A in Table 1). In one of the two MD simulations in the absence of carazolol, F193 was observed to undergo a dramatic conformational and rotameric state change after about 16 ns. It rotated out toward TM7 such that its phenyl ring formed face-to-face hydrophobic packing with the D192 - K305 salt bridge that has shifted toward TM4 in response, and the backbone oxygen atom of F193 formed a hydrogen bond with K305. This constituted a hydrophobic bridge between ECL2 and TM7, and the neighboring F194 of ECL2 and Y308 and I309 of TM7 further extended this hydrophobic patch (see Figure 6). This new conformation remained throughout the rest 18 ns simulation. In the other carazolol-absent simulation, F193 was observed stayed in the middle way of rotating toward TM7, but returned back to the original conformation after ca.1.5 ns (see Supporting Information Figure S2). In Figure 6, we can also see that since the rotation of F193 at 16 ns, the distance fluctuation of the D192 - K305 salt bridge became much smaller, indicating the stabilization of the salt bridge by the hydrophobic packing with F193. Although the hydrophobic cluster bridge does not show steric clash with carazolol after superimposed on the carazolol-bound structure, it presents as a cap on the ligand-binding pocket and is located on the path of ligand entry/exit from the extracellular opening to the binding pocket. In the carazolol-bound crystal structure, the χ angles of F193 were χ1=62.76, χ2=62.75, respectively, which do not correspond to any of the four rotamers in the penultimate rotamer library.21 It is likely that this unusual rotameric state was due to the binding of carazolol. In the simulated conformation saved at 20 ns and shown in Figure 6, besides backbone rotation, the χ angles of F193 changed to χ1= −68.02, χ2=92.32, corresponding to one rotamer (χ1= −65, χ2=85) in the penultimate rotamer library. We therefore speculate that the conformation observed in the MD simulation may represent at least one of the ligand-free states of β2AR and the hydrophobic patch bridging ECL2 and TM7 may play a role in stabilizing this conformation. We referred this conformation to as a putative ligand-free conformation.
Figure 6.
left: variations of the F193 Cζ - K305 Cα distance (red line), the F193 O - K305 Nζ distance (blue line), and the K305 Nζ - D192 Cγ distance (black line) in one of the two carazolol-free standard MD simulations. right: snapshot structure of the β2AR receptor at 20 ns in one of the two carazolol-free standard MD simulations. F193 (labeled green stick) rotated toward TM7 and formed hydrophobic packing with the D192 (red stick) -K305 (blue stick) salt bridge as well as F194 (unlabeled green stick), Y308 and I309 (yellow stick). Carazolol (van der Waals spheres) is shown as a reference.
Exit from the putative ligand-free conformation
To obtain insights into how a ligand enters the binding pocket of the ligand-free conformation of the β2AR structure, we performed RAMD simulations by using the putative ligand-free conformation obtained in the standard MD simulation as input structure. The ligand, carazolol was taken from the crystal structure after superimposing the receptors. Because the ligand-binding pocket in the ligand-free conformation remained close to the crystal structure, which is consistent with the results of a microsecond-timescale simulation,22 in the modeled complex structure, carazolol was located at an almost identical position as in the crystal structure and also as in the crystal structure, its alkylamine and alcohol moieties were in the hydrogen distances with the side chains of D113 of TM3 and N312 of TM7.
100 RAMD simulations were performed with the same parameters used for the RAMD simulations of the carazolol-bound crystal structure. The statistic summary of the simulation results is listed in Table 2 and the egress trajectories are visualized in Figure 7.
Table 2.
Statistical summary of the RAMD simulations, in which the receptor conformation was the putative ligand-free conformation obtained from a standard MD simulation (see text for the detail). The results are separately listed from 60 simulations with 4 different accelerations and 40 simulations with the smallest acceleration. The latter results are in the gray-shaded rows.
| Pathway | Egress point | Number of trajectories |
Acceleration (kcal/Å·g) |
Trajectory length (ps) |
|---|---|---|---|---|
| A | Extracellular opening | 29 (9a, 20b) | 0.20, 0.23, 0.25, 0.30 | 42–702 |
| 29 (8a, 21b) | 0.20 | 48–970 | ||
| B | H4–H5 cleft | 9 | 0.20, 0.23, 0.25, 0.30 | 34–722 |
| 5 | 0.20 | 272–634 | ||
| C | H5–H6 cleft | 2 | 0.20, 0.25 | 86, 644 |
| 0 | 0.20 | |||
| F a | H6–H7 cleft | 3 | 0.20, 0.30,0.30 | 452, 86, 106 |
| 0 | 0.20 | |||
| G | H2–H7 cleft | 2 | 0.23, 0.30 | 108, 42 |
| 0 | 0.20 | |||
| H | H2–H3 cleft | 3 | 0.20, 0.20, 0.30 | 160, 744, 204 |
| 0 | 0.20 | |||
| I | Behind ECL2 | 7 | 0.20, 0.23, 0.30 | 38–868 |
| 3 | 0.20 | 220, 436, 716 | ||
| no egress | 5 | 0.20 | 1000 | |
| 2 | 0.20 | 1000 |
carazolol passed out of the receptor from the “left” side (sub-pathway A1) and the “right” side (sub-pathway A2) of the D192-K305 salt bridge, respectively.
pathways C and D in Table 1 were not observed here.
Figure 7.
Paths of the center of mass of carazolol in the RAMD trajectories starting from the putative ligand-free conformation. left: the first set of 60 simulations with four different acceleration magnitudes; right: the second set of 40 simulations with the smallest acceleration used in the first set. The color scale is from yellow (start) to blue (end). The starting conformation of the receptor is shown in cartoon. For clarity, the second extracellular loop of residues 173 to 196 and the solvated lipid bilayer environment are not shown. The distribution of the paths is similar to in Figure 2, but sub-pathway A2 showed significantly higher frequency than sub-pathway A1 (see text for the detail).
Sub-pathway A2 gained popularity
Comparing Table 1 and Table 2, we can see that the extracellular opening was still the predominant egress pathway, which was observed in 29 of the 55 egress trajectories in the first set of 60 simulations and 29 of 38 egress trajectories in the second set of 40 simulations. However, in contrast to in the RAMD simulations with the ligand-bound crystal structure, where the “left” cleft (between TM5, TM6 and TM7) and the “right” (between TM2, TM3 and TM7) of the extracellular opening showed an equal frequency of being used as the points of egress, here the “right” cleft (sub-pathway A2) was used in 20 of the 29 trajectories along pathway A in the first simulation set and 21 of 29 in the second simulation set, respectively. The increased popularity of sub-pathway A2 was probably due to the shift of the salt bridge towards TM4 and the “left” area became smaller after the formation of the hydrophobic patch, partially covering the extracellular surface opening. Therefore, the results may suggest that the “right” cleft has a higher possibility to serve as a more specific ligand-entry site on the extracellular surface in the presence of the hydrophobic bridge. Of the 41 trajectories along sub-pathway A2, in 25 trajectories, the ligand pushed F193 rotated out of the path, and in 16 other trajectories, the ligand passed underneath F193 by bending its heterocycle ring toward the tail so that forming transient face-to-face π-packing with the phenyl ring of F193, which then caused ca. 90 degree rotation of F193 after the passage. Figure 8 shows the interactions between carazolol and the ECL2-TM7 bridge in two of such egress trajectories. However, in the 17 trajectories along sub-pathway A1 (the “left” cleft of the extracellular opening), the ligand showed minor interactions with F193, mainly by a sweeping motion of its chain tail.
Figure 8.
Snapshots showing interactions between carazolol and the ECL2-TM7 hydrophobic bridge in two egress trajectories, in which the putative ligand-free conformation of the β2AR receptor was used. Left: carazolol interrupted the ECL2- TM7 hydrophobic cluster by pushing F193 rotated away; right: carazolol passed underneath F193 and formed face-to-face packing with the phenyl ring of F193. Two movies showing the egress trajectories are available in Supporting Information (Movies 3–4).
Notably, the D192-K305 salt bridge was much better preserved in the simulations, remained intact in 44 of the 58 trajectories along the extracellular opening pathway, compared with the exclusive breakage in the egress simulations starting from the ligand-bound crystal structure. This indicated that the salt bridge was stabilized by the formation of the ECL2-TM7 hydrophobic bridge, in consistence with the standard MD simulation results as shown in Figure 6.
In addition to the common pathways (pathway A, B, C and F) observed in both the ligand-bound crystal structure and the putative ligand-free conformation, three new pathways mainly involving TM2 and ECL2 (pathways G, H, and I, see Table 2) were observed in the putative ligand-free conformation while pathways D and E mainly involving TM1 were absent. This may reflect the subtle differences in the inter-helical packing between the two receptor conformations although the backbone RMSD is only 1.8Å and the extracellular opening was the only opening in both conformations
Ligand exit was retarded
Figure 9 shows the spatial occupancy maps computed from the 100 RAMD simulations starting with the putative ligand-free conformation. Overall, the distribution of the occupancy clouds is similar to in Figure 5. The major difference observed is that the maximum area in the map at d12 = 11 Å became smaller and two comparable maximum areas appeared in the maps at d12 = 13 and 15Å. Although high occupancy can be resulted from a larger number of snapshots staying in similar positions in a single trajectory and/or focused multiple trajectories, the widely spread clouds in those maps indicate that the former situation was more likely. The high occupancy areas appeared in the maps at d12 = 13 and 15Å therefore indicate that ligand exit became slower in the putative ligand-free conformation than in the ligand-bound crystal structure. In fact, the average egress trajectory length was 311.28 ps in the putative ligand-free conformation compared to 243.35 ps in the ligand-bound crystal structure and 5 more simulations did not achieve ligand exit in the putative ligand-free conformation (see Table 1 and Table 2). The ECL2-TM7 hydrophobic bridge in the putative ligand-free conformation may count for the slower exit as it partially covers the extracellular opening and makes more difficult for the ligand to move further away from the initial position to completely escape the binding pocket. The blocking effect of the ECL2-TM7 hydrophobic bridge can be further noticed in the maps at d12 = 17 and 19Å, where the central areas corresponding to the hydrophobic patch region were blank, which means that the hydrophobic patch was not used as an egress point along pathway A and thus the D192-K305 salt bridge was remained, in consistence with the structural observation of the trajectories as shown in Figure 8. In addition, the smaller maximum area in the map at d12 = 11Å and lack of minor maximum area in the map at d12 = 9 may indicate that it was easier for the ligand to move away from the initial position in the putative ligand-free conformation than that in the crystal structure and moving up was easier than moving down, which was most likely due to the loser ligand-receptor contacts in the modeled complex structure
Figure 9.
The computed spatial occupancy maps of the ligand from 100 RAMD trajectories as a function of receptor-ligand center-to-center distance d12=9, 11, 13, 15, 17, and 19Å. The starting conformation of the receptor is the putative ligand-free conformation with a hydrophobic patch bridging ECL2-TM7. Red means high occupancy and blue low occupancy. Note: each map has its own color scale.
Summary and Implication
In this work, ligand exit pathways in the β2AR receptor were investigated by applying the random acceleration molecular dynamics (RAMD) method. In the RAMD simulations using the carazolol-β2AR bound crystal structure as input structure, the ligand was observed to exit the receptor predominantly through the extracellular surface opening, which was referred to as pathway A and a few other pathways through inter-helical clefts were observed with significantly lower frequencies. Ligand exit along pathway A involved the breakage of the D192-K305 salt bridge that linked the ECL2 to the extracellular top of the TM7. The hydrogen bonds formed between D113 (TM3) and N312 (TM7) and the alkylamine-alcohol tail of carazolol were the major resistance encountered by the ligand during exiting from the receptor via diverse routes and no second barrier was notable.
Standard molecular dynamics (MD) simulations were used to investigate the dynamics of the receptor structure and a putative ligand-free conformation of the receptor was obtained in a carazolol-absent MD simulation. In this conformation, the ECL2-TM7 salt bridge was expanded to a hydrophobic patch due to the rotation of F193 of ECL2 toward TM7, which represented an energetically more favorable conformation than the ligand-bound conformation. We therefore infer this new conformation to be the ligand-free conformation of the β2AR receptor. By using this new conformation in the RAMD simulations, we found that the extracellular opening (pathway A), in particular the cleft formed by TM2, TM3 and TM7 (sub-pathway A2) was the predominant egress pathway, where carazolol showed significant interactions with the ECL2-TM7 hydrophobic bridge especially F193. These results allowed us to propose an inference about the ligand entry pathway, which is that the ligand, very likely with its ring head, first dives into the cleft between TM2, TM3 and TM7 at the extracellular opening, then passes through the ECL2-TM7 junction by interacting with F193 and reaches the other end of the binding pocket (the cleft between TM4, TM5, and TM6), and finally this orientation is stabilized by polar interactions between its polar tail with receptor residues such as D113, Y316 and N312. This binding process could result in the inward conformation of F193 as observed in the ligand-bound conformation.
However, for the β1 and the β3 subtypes of the βAR receptors, the mechanisms of ligand entry and exit along the extracellular opening can be different because these is no salt bridge linking ECL2 to the extracellular top of TM7. The residues corresponding to the D192-K305 salt bridge are two aspartic acids in β1, and alanine and glycine in β3. Lack of the salt bridge in β1 and β3 would result in no tight packing junction between ECL2 and TM7 even in the presence of the conserved phenylanaline at the position of F193 as in β2, therefore, ligands may not experience the same steric obstacle as when entering the binding pocket of β2, on the other hand, ECL2 may be more flexible and less stable, which could lead to a lower stability of inactive states of the β1 and the β3 subtypes. In addition, the two negatively charged aspartic acids in β1 may act as an electrostatic sensor, attracting the positively charged catecholamines down into the binding pocket directly, and in this case the alkylamine-alcohol tail may enter first. Also, the electrostatic interactions can prevent the bound ligand from escape from the binding-pocket. (see Supporting Information Figure S3 for a homology model of the β1 structure that was built based on the β2 structure by using the MOE software (Chemical Computing Group, Inc.)). We hypothesize that the amino acid differences in the entrance to or exit from the ligand-binding pocket may contribute to the receptor subtype-selectivity of ligand-binding in the βAR subfamily. Unfortunately, there have no experimental data available for the critical residues like D192, K305 or F193. A important reason could be that these residues are not located in the transmembrane binding pocket and the mutagenesis analysis reported to date was mainly guided by the homology models of β2AR that were constructed based on the bovine rhodopsin structure and in these models the ECL2 residues had to resemble a beta-stranded structure as in rhodopsin, thus the formation of the D192-K305 salt bridge or the hydrophobic patch involving F193 was not possible. However, this hypothesis could be tested by site-directed mutagenesis or chimeric-receptor experiments focusing on the ECL2 loop and the TM7 top residues, in particular the sites corresponding to residues D192, K305 and F193 in β2.
Although it is not clear at present to what extent the differences in the mechanisms of ligand entry and exit as well as receptor stability contribute the subtype-specificity of ligand binding, this work for the first time identified the role of a unique ECL2-TM7 junction in ligand entry and exit in β2AR, which provides important information for understanding ligand-binding specificity in the βAR subfamily.
Methods
Modeling of β2AR with phospholipid bilayer and water
The modeling of β2AR was based on the crystal structure in which a partial inverse agonist carazolol is bound (resolution 2.2 Å, PDB entry: 2RH1). The T4 lysozyme molecule that was used to aid crystallization by replacing the intracellular loop 3 of β2AR was removed and the intracellular loop 3 was not modeled back in. Thus, there were 282 residues in the β2AR structure, including residues from D29 to L230 and from K263 to L342. In addition, nine buried crystal water molecules as well as a palmitic acid that was covalently bonded to C341 were kept. The hydrogen atoms were added by using the REDUCE program.23
To mimic the membrane environment, the modeled β2AR was inserted into a patch of phospholipid bilayer generated by the Membrane module in the VMD1.8.6 program,24 which consisted of 114 palmitoyloleoyl-phosphatidylcholine (POPC) lipid molecules and had a dimension of ca. 75 Å × 75Å. The POPC lipid was used because it is the main constituent of lipid bilayers. The embedded β2AR together with the membrane was then solvated in a TIP3P water box and charge neutralized by using the tLeap module in the AMBER8 program.25 This added 22480 solvent water molecules and 3 chloride ions and resulted in 87,435 atoms in total. The system without the ligand was modeled similarly except that carazolol was deleted from the crystal structure, and 22481 solvent water molecules and 2 chloride ions were added and the total number of the atoms was 87,392.
The parameters of the POPC lipid molecules, the palmitic acid and the carazolol ligand were derived by using the Antechamber module in the AMBER8 program and the GAFF force field,26 in which the POPC parameters have been previously used in the MD simulations of the bovine rhodopsin18 and were also evaluated by Jojart and Martinek.27 Three disulfide bonds were added for C106 – C191, C184 –C190 and C341-palmitic acid, respectively. The parameters of protein residues were assigned based on the AMBER ff03 force field.28 The parameter files (POPC.prep, CAU.prep and PLM.prep) of the POPC lipid molecule, the carazolol molecule and the palmitic acid are available in Supporting Information.
MD and RAMD simulations
The MD simulations were carried out by using the AMBER8 program. The modeled system was first subject to a two-stage energy minimization. In the first stage of 2000 steps, the receptor was restrained to its crystallographic positions by a harmonic potential with a force constant of 32 kcal/mol/Å2 while the others were unrestrained. In the second stage of 2000 steps, no restraint was applied. After energy minimization, the whole system was subjected to a gradual heating from 10K to 300K in 50 ps and with constant volume while the receptor and the head groups of the lipid molecules were restrained by a force constant of 32 kcal/mol/Å2. The restraints on the head-groups were used to prevent the tails from interdigitating, which would otherwise result in the reduction of the bilayer thickness. The system was then shifted to constant temperature of 300K and pressure of 1.0 atm. The restraints were removed from the lipid molecules after 500ps and from the receptor after another 500 ps. The simulations continued for production runs without any restraint. The bonds involving hydrogen atoms were constrained by using the SHAKE algorithm. The time step was 1 fs, and the non-bonded interactions were updated every 10 time steps.
Standard MD simulations were carried out for two systems of with and without the carazolol ligand. For each system, two different simulations were conducted by varying the speed of heating the system at the beginning of the simulations. The production runs lasted for 28.4 ns and 28.2 ns, respectively for the simulations with the ligand, and 35.1 ns and 33.4 ns, respectively for those without the ligand.
Ligand egress was simulated by using the Random Acceleration Molecular Dynamics (RAMD) method implemented in the AMBER8 program, which was kindly provided by Dr. Rebecca Wade at the EML Research. In the RAMD simulations, a small, artificial, randomly oriented force was applied to the center of mass of carazolol. The direction of the force was kept for 40 time steps of 2 fs. If, during this time, carazolol moved more than 0.01 Å, the direction of the force was maintained, otherwise a new direction was chosen randomly. A simulation is terminated either when the mass center distance between the ligand and the receptor has reached 30Å or the simulation time has reached 1ns. Due to the expulsion effect of the extra force, the dissociation process can be dramatically accelerated and thus, the ligand egress time is not comparable to the experimental timescale. The experimental dissociation rate constant of the carazolol-β2AR complex was measured to be 2.5×10−3 min−1.29 The starting structure of the RAMD simulations was the last snapshot after the restraint on the lipid molecules was removed (the restraint on the protein was still on) in one of the two standard MD simulations for the carazolol bound system. Therefore, the carazolol-β2AR structure at the beginning of the RAMD simulations was almost identical to the crystal structure but equilibrated within the solvated lipid bilayer environment.
Four different magnitudes of the acceleration were used, 0.20, 0.23, 0.25 and 0.30 kcal/Å·g along with different random number generation seeds. This range of the acceleration magnitudes has been used in several other RAMD applications,15,16,18 in which the ligands have similar masses to carazolol (299.39 g/mol), such as rhodopsin’s chromophore retinal with a mass of 320.86 g/mol18 and cytochrome P450s’ substrate progesterone with a mass of 314.46 g/mol16. RAMD simulation trajectories vary with different random number generation seeds and acceleration magnitudes and a smaller acceleration magnitude often leads to slower ligand exit. To investigate the statistical effects of these parameters, we performed two sets of RAMD simulations for each system, in which the first set of 60 simulations were carried out with four different acceleration magnitudes as mentioned above, and the second set of 40 simulations were with the smallest acceleration used in the first set.
In the RAMD simulations, the ligand is allowed to explore the weakness of the receptor structure and thus “find” the possible paths to exit the binding pocket in a feasible computational time. The most likely egress pathway and the exit mechanisms can be suggested by statistical analysis of the simulation trajectories.
Computation of ligand spatial occupancy maps
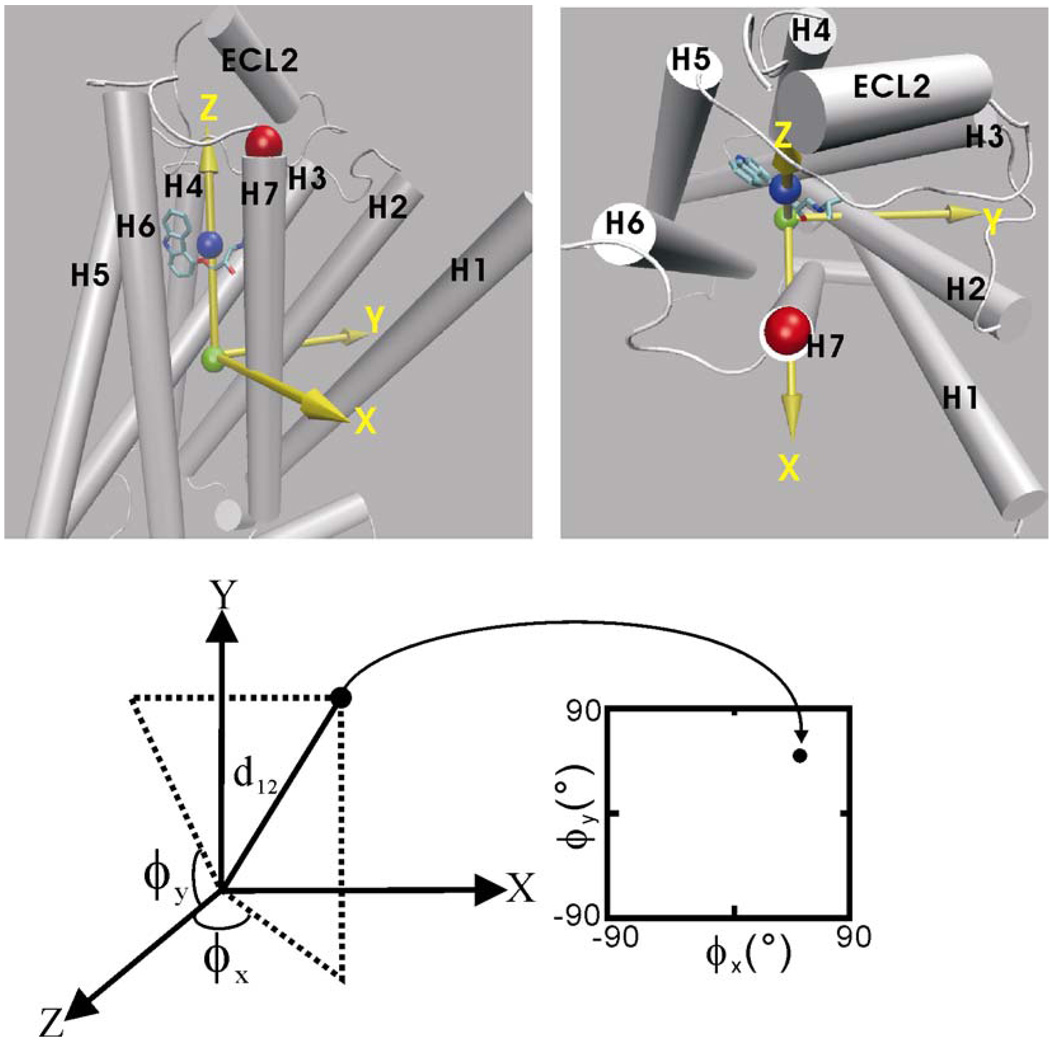
The RAMD simulation trajectories were also used to create the spatial occupancy maps of the ligand. The space was defined by constructing a reference coordinate system based on the initial receptor-ligand complex structure. The origin of the reference coordinate system was positioned at the mass center of the receptor. The z-axis direction points to the mass center of the ligand, see Figure 10. The y-axis was defined (orthogonal to the z-axis) by the vector from the origin to the K305:CA atom. The x-axis was defined orthogonal to the y- and z-axes. As a result, the positive and negative directions of the x-axis point to TM7 and TM4, respectively. The positive and negative directions of the y-axis point to TM2 and TM5–TM6 cleft, respectively. The coordinates from the RAMD trajectories were then transformed to new coordinates under the reference coordinate system. To present the spatial position of the ligand on a 2-dimensional map, two azimuth angles, ϕx and ϕy were defined as used by Spaar and Helms30in their work of analyzing Brownian dynamics simulation trajectories: ϕx is the angle from the positive z-axis to the orthogonal projection of the point in the zx- plane and ϕy is the angle from the positive z-axis to the orthogonal projection of the point in the zy- plane, see Figure 10. Therefore, the positions of the ligand at a certain distance from its mass center to the receptor mass center (d12, and d12=11.6Å in the carazolol-bound complex structure) can be projected onto an ϕx ϕy -plane. The ranges of the ϕx, ϕy angles are between −90 and 90 degrees and the signs correspond to those of the x, y coordinates of the position. Therefore, based on the ϕx, ϕy angles, one can easily identify how far the vector from the origin (the receptor mass center) to the current ligand mass center deviates from the z-axis (the initial direction of the vector), and furthermore, based on the directions of the x,y-axes relative to the receptor structure, one can locate the relative position of the ligand in the receptor structure. For example, a position with d12 = 17Å, ϕx = 20°, ϕy = 20° indicates that the ligand is located in the cleft formed by the z-axis, TM2 and TM7 and close to the extracellular surface opening. An occupancy map of the ligand is a two-dimensional population histogram defined by the ϕx, ϕy angles at a certain center-to-center distance d12. In the computation of occupancy maps, the bin sizes of the ϕx, ϕy angles were 2°.
Figure 10.
Definition of the reference coordinate system used in the computation of the ligand spatial occupancy maps. Upper: the green and blue balls indicate the mass centers of the receptor and the ligand, respectively. The red ball indicates the position of K305:CA used for the definition of the y-axis. The x, y, z-axes are represented by yellow arrows. The positive and negative directions of the x-axis point to TM7 and TM4, respectively. The positive and negative directions of the y-axis point to TM2 and the TM5–TM6 cleft, respectively. Lower: projection of a ligand position onto the ϕx ϕy-plane at a receptor-ligand center-to-center distance d12 (see text for the definition of the ϕx and ϕy angles). Note: for visualization, the xy-plane is placed on the page.
Acknowledgment
We thank Dr. Rebecca Wade for providing the codes of the RAMD method, Dr. R. Gabdoulline for helpful discussions, and UC Davis Genome Center for computer support. This work was supported in part by research grants from NIH (GM64458 and GM67168 to YD). Usage of VMD and AMBER is gratefully acknowledged.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supporting information Figures S1–S3, Movies 1–4 and the parameter files of the POPC lipid molecule, the carazolol molecule and the palmitic acid molecule.
References
- 1.Taylor MR. Pharmacogenetics of the human beta-adrenergic receptors. Pharmacogenomics J. 2007;7:29–37. doi: 10.1038/sj.tpj.6500393. [DOI] [PubMed] [Google Scholar]
- 2.Weinberger M. Long-acting beta-agonists and exercise. J. Allergy Clin. Immunol. 2008;122:251–253. doi: 10.1016/j.jaci.2008.05.030. [DOI] [PubMed] [Google Scholar]
- 3.Giembycz MA, Kaur M, Leigh R, Newton R. A Holy Grail of asthma management: toward understanding how long-acting beta(2)-adrenoceptor agonists enhance the clinical efficacy of inhaled corticosteroids. Br. J. Pharmacol. 2008;153:1090–1104. doi: 10.1038/sj.bjp.0707627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Yao XJ, Weis WI, Stevens RC, Kobilka BK. GPCR Engineering Yields High-Resolution Structural Insights into {beta}2 Adrenergic Receptor Function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 5.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta(2) adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 6.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, Stevens RC. High-Resolution Crystal Structure of an Engineered Human {beta}2-Adrenergic G Protein Coupled Receptor. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hanson MA, Cherezov V, Griffith MT, Roth CB, Jaakola VP, Chien EY, Velasquez J, Kuhn P, Stevens RC. A specific cholesterol binding site is established by the 2.8 A structure of the human beta2-adrenergic receptor. Structure. 2008;16:897–905. doi: 10.1016/j.str.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaakola V-P, Griffith MT, Hanson MA, Cherezov V, Chien EYT, Lane JR, Ijzerman AP, Stevens RC. The 2.6 Angstrom Crystal Structure of a Human A2A Adenosine Receptor Bound to an Antagonist. Science. 2008;322:1211–1217. doi: 10.1126/science.1164772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Le Trong I, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 10.Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 A crystal structure. J. Mol. Biol. 2004;342:571–583. doi: 10.1016/j.jmb.2004.07.044. [DOI] [PubMed] [Google Scholar]
- 11.Avlani VA, Gregory KJ, Morton CJ, Parker MW, Sexton PM, Christopoulos A. Critical role for the second extracellular loop in the binding of both orthosteric and allosteric G protein-coupled receptor ligands. J. Biol. Chem. 2007;282:25677–25686. doi: 10.1074/jbc.M702311200. [DOI] [PubMed] [Google Scholar]
- 12.Jager D, Schmalenbach C, Prilla S, Schrobang J, Kebig A, Sennwitz M, Heller E, Trankle C, Holzgrabe U, Holtje HD, Mohr K. Allosteric small molecules unveil a role of an extracellular E2/transmembrane helix 7 junction for G protein-coupled receptor activation. J. Biol. Chem. 2007;282:34968–34976. doi: 10.1074/jbc.M705563200. [DOI] [PubMed] [Google Scholar]
- 13.Klco JM, Wiegand CB, Narzinski K, Baranski TJ. Essential role for the second extracellular loop in C5a receptor activation. Nat. Struct. Mol. Biol. 2005;12:320–326. doi: 10.1038/nsmb913. [DOI] [PubMed] [Google Scholar]
- 14.Luedemann SK, Lounnas V, Wade RC. How do substrates enter and products exit the buried active site of cytochrome P450cam? 1. Random expulsion molecular dynamics investigation of ligand access channels and mechanisms. J. Mol. Biol. 2000;303:797–811. doi: 10.1006/jmbi.2000.4154. [DOI] [PubMed] [Google Scholar]
- 15.Winn PJ, Ludemann SK, Gauges R, Lounnas V, Wade RC. Comparison of the dynamics of substrate access channels in three cytochrome P450s reveals different opening mechanisms and a novel functional role for a buried arginine. Proc. Natl. Acad. Sci. U S A. 2002;99:5361–5366. doi: 10.1073/pnas.082522999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schleinkofer K, Sudarko, Winn PJ, Luedemann SK, Wade RC. Do mammalian cytochrome P450s show multiple ligand access pathways and ligand channelling? EMBO Rep. 2005;6:584–589. doi: 10.1038/sj.embor.7400420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Carlsson P, Burendahl S, Nilsson L. Unbinding of retinoic acid from the retinoic acid receptor by random expulsion molecular dynamics. Biophys J. 2006;91:3151–3161. doi: 10.1529/biophysj.106.082917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang T, Duan Y. Chromophore channeling in the G-protein coupled receptor rhodopsin. J. Am. Chem. Soc. 2007;129:6970–6971. doi: 10.1021/ja0691977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perakyla M. Ligand unbinding pathways from the vitamin D receptor studied by molecular dynamics simulations. Eur. Biophys. J. 2009;38:185–198. doi: 10.1007/s00249-008-0369-x. [DOI] [PubMed] [Google Scholar]
- 20.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr. Opin. Struct. Biol. 2001;11:224–230. doi: 10.1016/s0959-440x(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 21.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 22.Dror RO, Arlow DH, Borhani DW, Jensen M, Piana S, Shaw DE. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc. Natl. Acad. Sci. U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Word JM, Lovell SC, Richardson JS, Richardson DC. Asparagine and glutamine: using hydrogen atom contacts in the choice of side-chain amide orientation. J. Mol. Biol. 1999;285:1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 24.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 25.Case DA, Cheatham TE, 3rd, Darden T, Gohlke H, Luo R, Merz KM, Jr, Onufriev A, Simmerling C, Wang B, Woods RJ. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 27.Jojart B, Martinek TA. Performance of the general amber force field in modeling aqueous POPC membrane bilayers. J. Comput. Chem. 2007;28:2051–2058. doi: 10.1002/jcc.20748. [DOI] [PubMed] [Google Scholar]
- 28.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, Caldwell J, Wang J, Kollman P. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J. Comput. Chem. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 29.Burgisser E, Lefkowitz RJ, Delean A. Alternative Explanation for the Apparent "Two-Step" Binding Kinetics of High-Affinity Racemic Antagonist Radioligands. Mol. Pharmacol. 1981;19:509–512. [PubMed] [Google Scholar]
- 30.Spaar A, Helms V. Free Energy Landscape of Protein-Protein Encounter Resulting from Brownian Dynamics Simulations of Barnase:Barstar. J. Chem. Theo. Comput. 2005;1:723–736. doi: 10.1021/ct050036n. [DOI] [PubMed] [Google Scholar]