Abstract
Dimensionality reduction approaches have been used to exploit the redundancy in a Cartesian coordinate representation of molecular motion by producing low-dimensional representations of molecular motion. This has been used to help visualize complex energy landscapes, to extend the time scales of simulation, and to improve the efficiency of optimization. Until recently, linear approaches for dimensionality reduction have been employed. Here, we investigate the efficacy of several automated algorithms for nonlinear dimensionality reduction for representation of trans, trans-1,2,4-trifluorocyclo-octane conformation—a molecule whose structure can be described on a 2-manifold in a Cartesian coordinate phase space. We describe an efficient approach for a deterministic enumeration of ring conformations. We demonstrate a drastic improvement in dimensionality reduction with the use of nonlinear methods. We discuss the use of dimensionality reduction algorithms for estimating intrinsic dimensionality and the relationship to the Whitney embedding theorem. Additionally, we investigate the influence of the choice of high-dimensional encoding on the reduction. We show for the case studied that, in terms of reconstruction error root mean square deviation, Cartesian coordinate representations and encodings based on interatom distances provide better performance than encodings based on a dihedral angle representation.
INTRODUCTION
Understanding the relationship between the dynamic nature of molecular structures and their properties and functions is of critical importance for many fields of research. However, obtaining an accurate representation of molecular motion remains a challenge. Experimental methods such as nuclear magnetic resonance (NMR) spectroscopy and x-ray crystallography are severely limited in their abilities to detect conformational motion at an atomic scale. Theoretical methods using force-field approximations of the potential energy surface are promising approaches for the simulation of changes in molecular conformation. Unfortunately, these approaches cannot be applied to many problems of interest due to the excessive calculation times required. For classical molecular mechanics approaches, calculation times are proportional to the number of degrees of freedom in the system, typically the 3N Cartesian coordinates in an N-atom system. Even when calculations can be performed in reasonable times, the volume of data generated makes interpretation and analysis difficult.
In most cases, a significant amount of redundancy exists due to the encoding of molecular structure in a 3N phase space and high similarities of the many conformations obtained during simulation. For the former, the variable interdependence is an obvious result of the constraints on atomic positions due to covalent bonds and energy barriers resulting from steric overlap. For larger molecules such as proteins, the number of constraints can increase due to the formation of stabilizing intramolecular hydrogen bonds and the formation of rigid secondary and supersecondary structural elements.1 These interdependencies between the degrees of freedom result in an inherently sparse phase space and suggest that a lower-dimensional representation of molecular structure can account for conformational changes. For example, a recent Lyapunov analysis of the folding of alanine peptides in aqueous solution resulted in effective dimensionalities of approximately three to five for peptides containing three to ten residues,2 much lower than the many thousands of dimensions required in a Cartesian representation. Interestingly, it was reported that the effective dimensionality decreases with peptide size despite the increase in the number of atoms. This effect was attributed to an increase in intramolecular interactions stabilizing the secondary structures of the larger peptides. It is noteworthy that this low dimensionality is observed not through the formation of stable hydrogen bonds, but hydrogen bonds that are formed and broken on a nanosecond time scale. Similar findings on the decrease in effective dimensionality with increasing system size have been obtained for other protein simulations.3
Investigation into the low intrinsic dimensionality of molecular dynamics (MD) may provide an answer to the Levinthal paradox for protein folding.4 Experimental investigations indicate that unfolded proteins do not exhibit purely random structure and may in fact retain structural properties present in the folded states.4, 5, 6 Computational evidence further suggests that excluded volume effects significantly constrain the unfolded ensemble.7 Lange and Grubmuller3 demonstrated that the effective variables from a 90% linear dimensionality reduction (i.e., retaining 10% of the number of original variables) for a 5 ns protein simulation explain most of the variance observed in a 200 ns long simulation. This result is significant because only one of the three conformational states found in the longer simulation was sampled during the short simulation. A similar finding was reported for equilibrium simulations of the reversible folding of a β-heptapeptide in a methanol solution.8 In this case, the effective variables explaining 69% of the collective atomic fluctuations were found to converge within 1 ns, despite the significantly longer time scales required for folding.
The inherent low dimensionality of molecular conformation suggests that methods can be developed that exploit redundancy for improving the efficiency of simulation, analysis, and optimization. For example, low-frequency concerted motions of atoms are known to occur in conformational rearrangements within proteins. These correlated motions are thought to be responsible for important biomolecular functions including protein folding, molecular recognition, induced fit and catalysis, allosteric signal transduction, and mechanical∕thermodynamic energy transport. However, analysis of these collective motions is very difficult to achieve with MD simulation. First, the timestep for numerical integration must be sufficiently small (femtoseconds) to account for high-frequency motions that influence the accuracy of the resulting trajectory and therefore limit the simulation time. Second, extracting the relevant low-frequency motions from the large amount of data generated is not trivial. These difficulties motivated the use of dimensionality reduction (in the form of quasiharmonic analysis or essential dynamics) as a successful approach for isolating low-frequency motions and obtaining improved sampling of the phase space for MD and Monte Carlo approaches.9, 10, 11, 12, 13, 14, 15, 16, 17 For example, by diagonalizing the covariance matrix of atomic displacements, it is often found that over 90% of the atomic motion in peptides or proteins is concentrated in correlated motions involving only 1%–5% of the degrees of freedom.3, 12, 18, 19 In addition to providing reduced complexity, an accurate representation for correlated molecular motions can aid in the interpretation of NMR and x-ray studies.20, 21, 22, 23, 24, 25, 26
It has been shown that dimensionality reduction can be used to extend the time scales of MD, and a theoretical framework for low-dimensional simulation with Langevin MD or metadynamics is a topic of current investigation.25, 27, 28 In addition to improving the efficiency of simulation, a logical extension is to utilize low-dimensional surrogate spaces for problems in optimization that occur in molecular recognition and self-assembly. Dimensionality reduction has already been used for efficient incorporation of protein flexibility into ligand docking studies, for example.29, 30 In another interesting approach, dimensionality reduction was utilized in comparative protein modeling to avoid false attractors in force-field based optimization by using evolutionary information to reduce the number of degrees of freedom for structure refinement.31
In addition to decreasing the complexity required for modeling flexibility in molecular structure, dimensionality reduction can be used to analyze the extensive data generated from simulation into intuitive results. By lowering the number of effective degrees of freedom, more meaningful visualizations might be obtained and undesirable effects from the so-called “curse of dimensionality” can be removed.32 In addition to filtering high-frequency motions, dimensionality reduction of molecular simulations can be utilized to identify discrete conformational substates24, 33, 34 and for understanding the extent of configuration space sampling and the topography of the system’s energy hypersurface.35 As reviewed by Altis et al.,36 dimensionality reduction can be utilized to obtain representations for reaction coordinates and free energy landscapes as well as the transition matrix between metastable conformational states.
While there is substantial evidence for correlated atomic motions within dynamic molecular structures, it is unclear which computational method is most appropriate for detecting these correlations. Historically, linear dimensionality reduction methods have been employed for the analysis of molecular structure. It seems, however, that this choice was made more for mathematical convenience rather than knowledge of the high-dimensional structure of a given phase space. Indeed, it is well known that if certain requirements of the input data are met, principal component analysis (PCA) results in a low-dimensional encoding with minimal residual variance and preservation of intersample distances, thereby providing a minimal reconstruction error. PCA is a robust, simple algorithm with no user-adjustable parameters. However, linear approaches tend to underestimate the proximity of points on a nonlinear manifold, leading to erroneous embeddings. Consequently, several authors have suggested that nonlinear dimensionality reduction methods may be more appropriate for molecular structure analysis.19, 36, 37 Lange and Grubmuller37 have used an information theoretic approach to show that covariance matrices cannot account for all correlations for the studied protein domains. In an investigation into the use of collective Langevin dynamics for the simulation of a peptide, the same authors observed that a nonlinear dimensionality reduction would require fewer effective degrees of freedom to obtain equally accurate embeddings and performed a manual construction of a nonlinear reduction from the linear one obtained with PCA.27
The design and analysis of nonlinear dimensionality reduction algorithms is an active area of research, and recent efforts have yielded new candidate methods for molecular structure analysis. Agrafiotis and Xu38 developed a nonlinear dimensionality reduction approach and applied it to conformational studies of methylpropylether. Nguyen39 used nonlinear PCA to analyze the free energy landscapes of peptides. Das et al. applied the Isomap40 algorithm to the analysis of folding simulation data for a coarse-grained bead model representative of a protein backbone, demonstrating that nonlinear dimensionality reduction provided a more accurate embedding of the reaction coordinates than linear techniques.41 However, despite their promise, it is difficult to accurately assess the efficacy of nonlinear dimensionality reduction algorithms for molecular structure analysis. The difficulty arises from two factors: (1) the intrinsic dimensionality for most problems is unknown, and (2) there is debate for some problems as to whether simulation approaches can provide sufficient sampling of the phase space to facilitate an accurate analysis of dimensionality reduction.3, 13, 24, 25, 27, 42
In this paper, we investigate the ability of well-known nonlinear dimensionality reduction algorithms to identify accurate, low-dimensional substructures in the conformation space for an eight-membered ring. We chose this particular molecule for several reasons. First, the ring closure problem provides an interesting dimensionality reduction benchmark where mathematical insight into the underlying manifold can be obtained. Second, although the Cartesian representation of an eight-membered ring involves 72 dimensions, there are effectively only two degrees of freedom, and therefore a dense sampling of all ring conformations can be obtained. Finally, the dynamic structure of eight-membered rings has been extensively studied and low-energy conformations have previously been identified.
In this paper, we describe an efficient method for enumeration of the conformations of eight-membered rings. Because the choice of representation for high-dimensional encoding [for example, dihedral angles (DHAs) versus Cartesian coordinates] has been a topic of recent debate,36 we use the ring enumeration to generate four different high-dimensional encodings for comparison. We provide an empirical verification of the manifold dimensionality for ring atoms and substituents. We compare the efficacy of several canonical dimensionality reduction algorithms for finding low-dimensional representations of molecular structure. Finally, we compare results from enumeration to those obtained using samples from room temperature MD.
Eight-membered rings
Saturated cyclic compounds have been studied extensively since the 19th century.43 Of these, eight-membered rings have been the most popular subject due to the existence of multiple conformers of similar energy, a complicated potential energy surface resulting from the ring closure constraint, and significant steric influence from hydrogen atoms on preferred molecular conformations.43, 44, 45, 46 For these same reasons, eight-membered rings pose an interesting challenge for dimensionality reduction algorithms. In Cartesian space, a saturated 8-ring requires 72 dimensions to represent a conformation. Taking changes in bond lengths and angles as negligible, a conformation can also be represented by eight variable DHAs. We can intuitively reduce this number to five DHAs by forcing the first three atoms to lie in the xy-plane. The placing of the remaining atoms 4,…,8 of the ring, with fixed bond angles and bond lengths, is accomplished by choosing the values of the five dihedrals, t2,…,t6 where we use the convention that the ith dihedral ti is formed by the atoms i−1,i,i+1,i+2 and we identify atom i+8 with atom i. This appears to require five dihedrals in order to construct the ring. The same result of five DHAs is achieved using the Cremer–Pople47 puckering coordinates.
Interestingly, it can be shown that there are only two independent variables due to the ring closure constraint.48 This is a consequence of the fact that the bond angles at atoms 8 and 1 as well as the bond length between these two atoms have not been used in the construction, and thus fixing these three degrees of freedom to prescribed values introduces three constraints among the five torsions, reducing the number of independent variables among them to 2. This result allows for an excellent benchmark for dimensionality reduction algorithms. First, we can perform a dense sampling of the two independent variables to obtain all relevant conformations of eight-membered rings. Second, we can expect, under the assumption of fixed bond lengths and angles, the phase space of 8-rings to lie on a 2-manifold in the higher dimensional spaces. From the Whitney embedding theorem,32 we can expect a successful dimensionality reduction to smoothly embed the samples in a minimum of two dimensions and a maximum of five dimensions.
Although cyclo-octane is the most commonly studied 8-ring, complications arise due to symmetry. Therefore, we consider a substituted cyclo-octane, trans, trans-1,2,4-trifluorocyclo-octane for all studies in this paper to remove any symmetry issues.
High-dimensional encodings
The most common encoding of a molecular conformation under classical force fields is given by the 3N Cartesian coordinates for an N-atom system. This encoding is intuitive in that the distance between conformations is related to the root mean squared deviation (RMSD) of the Euclidean distances between each distinct pair of atoms. Cartesian coordinates are not the only choice, however. For example, high strain energies for bond stretching and angle bending typically result in relatively small deviations relative to movements due to changes in bond torsions. Consequently, a high-dimensional encoding utilizing only DHAs with fixed bond lengths and angles reasonably approximates both the low-energy conformations of molecules and their corresponding energies.18, 48 The dihedral encoding offers two advantages over a Cartesian encoding. First, it provides a natural separation between internal coordinates and overall translation and rotation of a given molecule. The use of Cartesian coordinates, on the other hand, requires RMSD fitting to ensure that the Eckart conditions are satisfied.49 Second, the encoding is a dimensionality reduction in and of itself. In proteins, for example, the ratio of the number of DHAs to Cartesian coordinates is about 1:8.18
DHA encodings do introduce some complications, however. DHAs are periodic variables and therefore a Euclidean distance metric might not be appropriate for dimensionality reduction. To address this issue, a circular variable transformation (CVT) has been proposed to transform the input data from dihedral space to a linear metric coordinate space using trigonometric functions or a complex representation of angles.36 A related but unaddressed issue results from the complicated relationship between the magnitude of change in DHAs and the resulting change in molecular conformation. For example, a change of a few degrees in a DHA located near the middle of a long chain molecule causes a drastic change in the overall conformation of the molecule, while a change in the DHAs of terminal atoms results in no change in conformation.48 While Cartesian RMSD represents an intuitive metric for quantifying conformational differences, one can imagine circumstances where a large change in DHAs ultimately results in very similar conformations. Some authors have reported preliminary results showing that an angle-based analysis of conformational data is very sensitive to noise and prone to error.50 Further, allowing fluctuations in bond lengths and angles might have the effect of making the DHAs more flexible,18 as evidenced by differences obtained from normal mode analysis when using DHAs rather than Cartesian coordinates.
Interatom distances represent a fourth alternative encoding of molecular structures, and the set of related techniques, known as distance geometry, has been extensively developed in the context of molecular structure studies due in part to its application to NMR structure determination.51 However in the present context it presents two immediate disadvantages. First, for larger systems, the dimensionality is actually increased from 3N to N(N−1)∕2. Second, in order to recover structure, an additional embedding is required to calculate the atomic positions from the interatom distances. Nonetheless, it has been reported that PCA performed on interatom distances is more powerful for the purposes of topographical discrimination than is PCA using either Cartesian coordinates or DHAs.52, 53 Additionally, this approach also provides a natural separation between internal motion and overall rotations and translations. In this paper, we compare the effectiveness of dimensionality reduction using all four encoding schemes: Cartesian coordinates, DHAs, DHAs with CVT, and interatom distances.
METHODS
Dimensionality reduction
In general, dimensionality reduction algorithms provide a method for taking a set of samples {x1,…,xn}⊂RD and calculating a corresponding low-dimensional representation {y1,…,yn}⊂Rd. Because dimensionality reduction is often used for visualization, some algorithms do not generate an explicit map from the high-dimensional coordinates to the low-dimensional representation. For many applications, however, it is desirable to have an explicit forward map, Φ(x):RD→Rd, that gives the low-dimensional representation of an arbitrary point x and an explicit reverse map ϕ(y):Rd→RD that gives the high-dimensional representation of an arbitrary point y. This allows for mapping new samples that were not available at the time of the initial reduction and also provides a common metric for comparison of algorithms. Therefore, for the purposes of this work, we consider dimensionality reduction as the problem of generating Φ and ϕ from a training set of n samples, XD×n=(x1,…,xn). Because some methods do not generate explicit maps, we describe an approach for generating maps from a dimensionality reduction below.
We evaluate the performance of each algorithm using the reconstruction error. Ideally, a forward map of an arbitrary point followed by a reverse map will give the same point back. Typically, the reconstruction error is given by ∥x−ϕ(Φ(x))∥. Here, however, we must compare between different high-dimensional encodings. Additionally, since we are using molecular conformations as samples, RMSD between a molecular structure and the reconstructed structure offers a more intuitive approach. Therefore, if the high-dimensional encoding is a Cartesian coordinate, we use the reconstruction RMSD (ϵ) as a metric,
| (1) |
where a=D∕3 gives the number of atoms in the molecule x. If the high-dimensional encoding is not Cartesian, the Cartesian representations of x and ϕ(Φ(x)) are used to calculate the RMSD. Throughout the paper we use the term “molecular RMSD” in order to distinguish from a statistical definition that is not normalized by the number of atoms. For each reduction method, we evaluate the algorithm performance using the mean molecular RMSD, ∑ϵ∕m, for a test set of m molecules not present in the training set.
Many algorithms and variants have been proposed for the problem of nonlinear dimensionality reduction including independent component analysis,54, 55 kernel PCA,56 self-organizing maps,57 neural network autoencoders,58 locally linear embedding (LLE),59 Isomap,40 and others.60 Here, we compare three canonical nonlinear algorithms to PCA: Isomap, LLE, and a neural network autoencoder.
PCA
PCA is a linear dimensionality reduction approach that has been widely applied to problems in almost every field of experimental science. The goal of PCA is to find a coordinate representation for data where the most variance is captured in the least number of coordinates. This representation can be found by performing an eigenvalue decomposition (or singular value decomposition) such that the resulting eigenvectors∕singular vectors provide an orthonormal basis for the data while the eigenvalues∕singular values provide information on the importance of each basis vector. Given the training set X, a row-centered matrix is calculated as , where and mD×1 gives the row means. Eigendecomposition of the training set covariance matrix, , is performed to give UPUT. The forward map is then given by , where is the matrix composed of the first d columns of U corresponding to the eigenvectors with the largest eigenvalues. The reverse map is calculated as . The reconstruction error for PCA will be zero for d>=D−z, where z is the number of nonzero eigenvalues in P. A review of PCA, its history, examples, and applications can be found in Refs. 32, 60, 61.
LLE
LLE is a nonlinear dimensionality reduction method. LLE is performed by first solving for the location of each sample xi in terms of its neighbors. For each sample, the neighbors are determined as all samples within a ball of specified radius centered on the sample or as the k nearest neighbors. A weight matrix W is obtained by determining the weights in a linear combination of neighbors that best reconstruct each sample,
| (2) |
where W=(wij). This problem has a closed form solution and assures not only that each approximation xi≈∑jwijxj lies in the subspace spanned by the k neighbors of xi but also that the solution W is invariant to translation, rotation, and rescaling. These properties allow, by design, calculation of a linear mapping that is also invariant to translation, rotation, and rescaling. This mapping from the n data samples xi to the low-dimensional embedding yi is performed by minimizing the embedding cost function
| (3) |
In this case, the weights wij are fixed and the low-dimensional coordinates are optimized. This is a quadratic minimization problem with a unique global minimum. It can be solved as a sparse n×n eigenvalue problem where the bottom d nonzero eigenvectors provide the embedding (the bottom eigenvalue is zero). From Eq. 2, it can be seen that LLE assumes that a sample and its neighbors can be treated in a linear fashion. Global structure is maintained due to the overlap of neighbors in each local patch in the embedding cost function. A detailed description of LLE can be found in Refs. 59, 62.
Because the low-dimensional representation is optimized directly in Eq. 3, no explicit maps are generated. Here, we use ΦNRM and ϕNRM to perform mapping in terms of the initial LLE reduction as described below.
Isomap
Isomap is an alternative nonlinear dimensionality reduction algorithm, first introduced in Ref. 40. The first step in the Isomap algorithm is to impose a graph structure G(V,E,W) on the input data set X. Each sample xi∊X is represented by a node vi∊V, and two nodes vi and vj are connected by an edge (vi,vj)∊E with weight wij∊W if xi is a neighbor of xj. Neighbors are calculated in the same manner as performed in LLE. The weight of wij is given by the Euclidean distance between xi and xj. The second step in Isomap involves computation of the shortest paths between all nodes in G. These distances are stored pairwise in a matrix DG. The distance matrix DG is intended to represent the distances between all samples on the manifold—the geodesic distances. Because these distances are Euclidean for each sample and its neighbors, Isomap makes the same assumption of local linearity as LLE. Unlike LLE, global distances between all neighbors are explicitly calculated with the graph approximation to geodesic distances.
Because all pairwise distances are available, multidimensional scaling (MDS) can be applied to DG to perform a low-dimensional embedding. MDS is a variant of PCA that starts with a distance matrix DG, converts the distance matrix to an inner product matrix, and calculates the eigenvalue decomposition of the resulting matrix (see, e.g., Havel et al.63). For the case presented here, this is performed by squaring each element in the distance matrix DG, double centering the resulting matrix, and performing the eigenvalue decomposition to give UPUT. The low-dimensional embedding is then given by , where is the matrix comprised by the first d columns of U corresponding to the eigenvectors with largest eigenvalues, and is the diagonal matrix containing the square roots of the largest d eigenvalues.
Like LLE, Isomap does not calculate explicit maps in order to perform an embedding. Here, we use ΦNRM and ϕNRM to perform mapping in terms of the initial Isomap reduction as described below.
Autoencoder neural networks
An autoencoder performs dimensionality reduction via a bottleneck architecture neural network. Autoencoders were originally introduced sometime in the early 1990s,32 but they have not been widely applied due to the extreme difficulty of the optimization problem associated with training the resulting network. However, a method was recently proposed for pretraining an autoencoder neural network using a restricted Boltzmann machine (RBM) in order to accelerate the optimization process.58 This method was used to obtain impressive results on a very large benchmark data set of handwritten digits.
The autoencoder introduced in Ref. 58 consists of weighted sums and compositions of the well-known function σ(x)=1∕(1+exp(x)). These functions are separated into distinct layers, with interconnections between functions in adjacent layers defining the network structure. At each layer in the network, inputs into the next layer consist of terms of the form σ(bj+∑iviwi), where bj represents a bias, wi represents a weight, and vi represents an input from the previous network layer. The inputs to the first layer are taken to be the components of the original vectors in our data set X={x1,…,xn}. The weights and biases are then optimized such that the mean reconstruction error 1∕n∑ixi−ϕAE(ΦAE(xi)) is minimized (where ΦAE is the forward map and ϕAE is the reverse map given by the network).
To provide an illustrative example, suppose we have a data set X with native dimension 784, for which we want to construct a two-dimensional (2D) embedding. We first define a network structure such as 784–1000–500–250–2, where the integers in the sequence represent the number of σ functions in each layer. When appropriately trained, this structure will perform a reduction of 784-dimensional data to a two-dimensional embedding. The justification for the initial increase in dimension to 1000 is that because the σ functions are inherently binary, we may experience a loss of information when going from normalized data in [0,1] to values in 0,1; the possible loss of information resulting from this process is potentially counterbalanced by an initial increase in dimensionality. The encoding structure is then mirrored to form a 2–250–500–1000–784 decoding network structure. The encoder and decoder networks are then joined and training is performed on the aggregate network.
As mentioned above, the optimization procedure for obtaining the autoencoder weights proceeds in two steps. In the first step, a RBM is trained. This training is performed for a user specified number of iterations. In the second step, the autoencoder weights are fine tuned using back propagation (BP). This step is also performed for a user specified number of iterations. In both cases a training set is used for the optimization and a test set is used to avoid overtraining. The training set is also split into batches to avoid overtraining, as well as to improve algorithm speed. During each iteration all of the batches are used in sequence.
The layers of the neural network and corresponding weights yield an analytic expression for both the forward (ΦAE) and reverse (ϕAE) maps that are optimized during training. This allows for future mapping of arbitrary points.
Neighbor reconstruction mapping
LLE and Isomap produce a low-dimensional embedding Yd×n={y1,…,yn}⊂Rd from the samples in X without generating an explicit map. Here, we have considered dimensionality reduction as a problem of finding the maps Φ and ϕ from training data. For LLE and Isomap, we accomplish this with the maps ΦNRM(X,Y,x) and ϕNRM(X,Y,y) that allow for dimensionality reduction to be performed on future samples based on the initial embedding of training data. A natural choice for these maps is some method that retains the positioning of a sample relative to its neighbors in the training set. Because LLE and Isomap assume that a sample and its neighbors are locally linear, we can perform the mapping using a linear combination of a sample’s k neighbors,
| (4) |
and
| (5) |
That is, the training set neighbors for an arbitrary point x or y can be identified in the input dimensionality and used to determine the sample mapping based on their positions (xi or yi) in the desired dimensionality. The question is how to choose the weights wi. The equations bear a strong resemblance to the reconstruction approach used in LLE [Eq. 2], and it has been suggested that this same approach can be used to map new samples.62 In this case, wi are determined in a least-squares optimization with a closed form solution. There are issues in implementing this approach, however. For the case when the number of neighbors k is greater than the intrinsic dimensionality of the manifold, the solution for wi is not unique. Because it can be desirable that k is variable and because the intrinsic dimensionality is not necessarily known a priori, it is not straightforward to decide when the problem must be conditioned to provide a unique solution. While this is worth investigating, for this work we have chosen wi to be the inverse Euclidean distance between the sample and the neighbor i. This approach allows for an arbitrarily high number of neighbors, however, will clearly fail in the case when a sample is outside the convex hull of its neighbors (due to the constraint that wi is positive).
Estimating intrinsic dimensionality
We have described methods for obtaining a map Φ(x):RD→Rd for dimensionality reduction. How do we determine d? One obvious choice is to determine some metric for quantifying the success of dimensionality reduction and evaluate the reduction performance at different embedding dimensionalities. For PCA and MDS, this metric can be the residual variance. The eigenvalues obtained in these approaches give the variance in each dimension, and therefore the sum of the d+1 to D eigenvalues is a measure of the variance that is not accounted for in the reduction. When this value is near zero, little is gained from adding a dimension. Although LLE also solves an eigenproblem, the eigenvalues obtained have been shown to be unreliable in determining d.62
An alternative metric utilized in Isomap40 is a geodesic distance correlation residual given by 1−R2(DG,DY), where R2(DG,DY) is the correlation coefficient between geodesic distances DG and distances in the low-dimensional space DY. This metric requires knowledge of the geodesic distances, however. For linear subspaces, the geodesic distances are given by the Euclidean distances. Otherwise, a method for estimating the geodesic distances, such as the one provided in Isomap, must be utilized. As discussed earlier, a more general method that allows comparison between different algorithms is the reconstruction error.58, 64
The approaches listed above are often cited as methods for estimating the intrinsic dimensionality of a manifold. However, they all rely on dimensionality reduction methods that attempt an embedding of sample data in a space with lower dimensionality. Therefore, these approaches are really only suitable for estimating the smooth embedding dimensionality. This subtlety is important because the Whitney embedding theorem32 dictates that a smooth embedding of a d-manifold may require as many as 2d+1 dimensions. Knowledge of the smooth embedding dimensionality is desirable for performing dimensionality reduction. For determining the intrinsic dimensionality, however, methods such as local-PCA (Ref. 65) might be more accurate for manifolds with complex structure. This is because they do not rely on a single-coordinate embedding of the entire manifold.
Algorithm implementations
We have implemented each of the four dimensionality reduction algorithms in a high-speed, multithreaded C++ library for computing intrinsic dimensionality estimates, low-dimensional embeddings, and forward and reverse mappings.66 Multithreading was performed using OPENMP and through an interface to multithreaded BLAS and LAPACK routines available in the Intel Math Kernel Library (MKL). For PCA and Isomap, we performed eigendecomposition using the relatively robust representation algorithm.67 For LLE, the divide-and-conquer algorithm was used.68 As discussed above, our extended implementations of LLE and Isomap support computation of forward and reverse maps via reconstruction from neighboring points. When the geodesic distance graph in Isomap is disconnected, each connected component is embedded separately using MDS, yielding nonoverlapping regions in the low-dimensional embedding space. This allows for forward and reverse mappings in the case of disconnected graphs, but will produce erroneous results for extrapolation outside of any individual component.
Ring enumeration
For this work, we have utilized two approaches for sampling trans, trans-1,2,4-trifluorocyclo-octane conformations—a deterministic enumeration and MD simulation. For the former, we assume that bond lengths and angles are invariant, and therefore we only require an enumeration of carbon atoms in each conformation (hydrogen and fluorine atoms are at default positions). As will be described, this treatment is advantageous in that it allows for a dense sampling of all ring conformations regardless of energy. Additionally, the conformational space is known to be 2D. Because bond angles and lengths will exhibit small changes, we also perform analysis of samples obtained from MD simulation.
For ring enumeration, we set all eight bond lengths to a constant, canonical value, and all bond angles to the same value of 115°. There are eight free torsions, from which two are used as control parameters and sampled to a prescribed resolution. The remaining six torsions are adjustor variables, set by the requirement of loop closure. As is well known,69 the loop closure problem can be reduced to the solution of a generalized eigenvalue problem of degree 16, and thus there can be at most 16 solutions, corresponding to the real generalized eigenvalues. Our numerical results indicate that the solution manifold is smooth, in agreement with a recent study of the same problem by Porta et al.70 An earlier study of the problem given by Manocha and Xu71 was not carried out at a sufficient resolution to allow for a comparison.
We employed two distinct algorithms for this calculation in order to guard against algorithm related degeneracies: (1) our own implementation of Lee and Liang’s72 algorithm (LL for the solution of the 7R problem of inverse kinematics) and (2) the triaxial loop closure algorithm of Coutsias et al.,69, 73 (TLC). Both algorithms result in an optimal formulation as a generalized eigenproblem of degree 16, whose formulation and solution requires less than 10 ms on a 2 GHz Pentium processor. Our results for the two algorithms were in close agreement for the case where torsions separated by two degrees of freedom, e.g., t5 and t8, were used as control parameters. Employing different combinations of control dihedrals produced alternative representations for the conformation space, which were shown by detailed pointwise comparison to be equivalent.
Some of the advantages of using this particular problem and data set are as follows. (1) The underlying manifold is smooth. (2) Its dimensionality is known, but its structure is nontrivial, although completely known in principle. (3) It is compact and can be completely bounded in torsion space: for any set of solutions, each of the torsions is bounded in a closed subinterval of [−t*,t*], where t*<180°. (4) However, the manifold does appear to have nontrivial topology (not proven), involving cycles in terms of generalized coordinates or pseudorotations.74 (5) Molecular dynamics studies may be employed to obtain local coverings of subsets of the manifold. (6) The data set is 2D to very high accuracy and provides a sufficiently fine cover of the manifold since the underlying eigenproblem is mostly very well conditioned.75 (7) Although the actual minimal dimensionality of a smooth embedding is not known, it has a strict upper bound of five. (8) The structure of this manifold is of somewhat generic character, which may be expected to be found in any situation where localized torsional motions of constrained flexible molecular chains may occur.
Test data sets
The enumerated conformations were used to generate four sets of input data for use in testing the various dimensionality reduction algorithms. The DHA data set consists of samples encoding the conformations using ring DHAs, yielding a native dimensionality of 8. The CVT data set consists of samples encoding the conformations using the circular variable transform,36 in which each DHA is represented by a pair specifying the angle sine and cosine; the CVT data set has native dimensionality 16. Samples in the XYZ data set encode conformations using the Cartesian coordinates of the ring, hydrogen, and fluorine atoms, yielding native dimensionality 72. All atoms were initialized in default positions and then minimized in the MM3 force field76 to a RMS gradient of 0.01 kcal∕mole Å with the ring atoms frozen. Minimization was performed using the program TINKER. Finally, samples in the IPD data set encode conformations using the interparticle distances between each distinct pair of atoms, yielding a native dimensionality of 276. For each conformation, we used the corresponding Cartesian encoding in the XYZ data set to calculate the MM3 intramolecular potential energy for each sample, again using TINKER.
RESULTS
Ring enumeration and clustering
The actual data set used for the present study employs torsions t6 and t8, a combination that was seen to give the simplest representation. This combination necessitated the use of the LL algorithm, as the TLC algorithm requires that the adjustor torsions form three coterminal pairs. The sampling of the control torsions was carried out at 0.5° increments, and the values of the adjustor and control torsions were tabulated for all alternative conformations. This particular representation was found to produce 0, 2, 4, or 6 solutions as the control torsions ranged in the intervalst6∊[−179.75°,180.25°] and t8∊[180°,180°], where solutions were found for t6∊[−131.25°,131.25°] andt8∊[−131.5°,131.5°]. The domain of values that give a nonzero number of solutions was seen to be connected. Preliminary analysis of the conditioning of the eigenvalue problem indicates that the resolution was adequate to capture all bifurcations of real eigenvalues. Certain interesting confluences of eigenvalues and degeneracies were observed, independent of the algorithm employed, but were found to occur away from actual real eigenvalue bifurcations. A careful analysis of these data will be presented elsewhere.75 The calculation resulted in 1 031 644 conformations. By comparison, the detailed study reported by Porta et al.70 employed distance geometry methods, requiring roughly 4 months of computer time.
Uniform conformational sampling in the space given by two independent torsion variables does not yield uniform sampling in either Cartesian or dihedral space. To correct for this situation, we performed clustering on each of the four data sets generated from the enumeration, further subdividing the resulting samples into distinct training and test sets. Clustering was performed by randomly choosing a sample and adding it into the training or test set only if the Euclidean distance (with respect to a given encoding) to all other points in the selected set was greater than some threshold dt; we repeated the process until all samples were evaluated. Because the training and test sets were generated independently, samples appearing in both sets were identified and removed from the test set. We used dt=0.12 to construct the XYZ data set, resulting in 8375 and 8229 samples for the training and test sets, respectively. For the DHA data set, we used dt=0.04, respectively. yielding 7830 and 7692 samples for the training and test sets. For the CVT data set, we used dt=0.01, resulting in 7790 training set samples and 7678 test set samples. For the IPD data set, we simply calculated all of the interparticle distances from the XYZ data set to give training and test sets of 8375 and 8229, respectively.
Empirical verification of manifold dimensionality
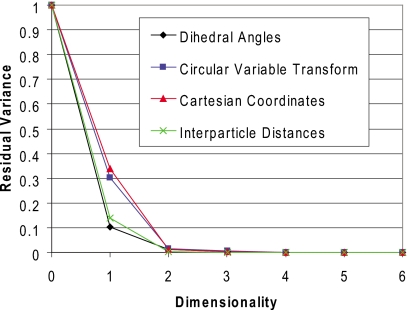
In order to verify that the manifold embedded in each native-dimensional space is indeed 2D, we performed a variant of local PCA,65 referred to here as point PCA, in order to estimate the intrinsic dimensionality. Taking the same approximations used in LLE and Isomap, we assume that a local region of a manifold given by a point and its k-nearest neighbors is approximately linear (local PCA differs from point PCA in that generalized clustering techniques such as vector quantization are used to determine locality). This assumption allows for estimation of intrinsic dimensionality by assessing the error in fitting each set of points to a lower-dimensional hyperplane. PCA can be utilized to perform this task; for a d-dimensional manifold, the residual variance should be near zero given an encoding with d principal components. For example, in the present case of a two-dimensional manifold, the neighborhood of each point should reside on a two-dimensional plane, and therefore the variance in the data should be explained entirely by the first two principal components. This is shown in Fig. 1 for each of the data sets using k=5 (Euclidean) neighbors per point, varying the number of principal components from 1 to 6. It is desirable to keep the number of neighbors small in this type of approach in order to reduce the fits to local regions of a manifold, reduce the computational time required, and reduce the total number of samples required. It is important to note, however, that the residual variance at k+1 dimensions with PCA should always be zero. For example, in the case presented here, the residual variance at six dimensions in Fig. 1 should always be zero regardless of the intrinsic dimensionality. Therefore the number of neighbors should always be adjusted to assure that it is higher than the intrinsic dimensionality.
Figure 1.
Empirical estimation of intrinsic dimensionality for the four data sets using point PCA. For DHAs and the circular variable transform, the bond torsions for the ring atoms are included. For the other data sets, hydrogen atoms are also included. For all data sets, an intrinsic dimensionality of 2 is obtained.
Algorithmic dimensionality reduction
We first evaluated each previously described dimensionality reduction algorithm considering only the ring (carbon) atoms in the various data sets. Our primary goal was to assess the ability of each algorithm to identify an embedded, low-dimensional manifold with low reconstruction error. For each combination of algorithm and data set, the training sets were used to compute forward and reverse maps for embedding dimensionalities ranging from 1 to 8. Each sample in the test set is mapped forward to the embedded manifold and subsequently reverse mapped to the native-dimensional space. Because it is difficult to directly compare the reconstruction error across disparate high-dimensional spaces, we utilize the mean molecular RMSD in Cartesian space as a comparative metric. Thus, each reverse-mapped native-dimensional encoding in the test set is (if necessary) converted to Cartesian coordinates, the resulting molecule is superimposed over the reference molecule, and the molecular RMSD between the two structures is calculated.
Several issues arise when converting samples from the DHA, CVT, and IPD data sets to Cartesian coordinates. For the CVT data set, DHAs are calculated from the mean of the angles obtained from the arcsine and arccosine of the transformed variables. Because reconstruction error can lead to transformed variables with values greater than 1, all such variables were truncated to 1 in the reconstructed CVT data set to avoid a complex result from the inverse transform. When translating IPD encodings to Cartesian space, a difficulty arises due to the fact that the distances do not necessarily preserve stereochemistry and therefore care must be taken in the embedding to prevent reflections that would alter the stereochemistry of the molecule through improper rotation (as given by sign changes in the eigenvectors used for embedding).
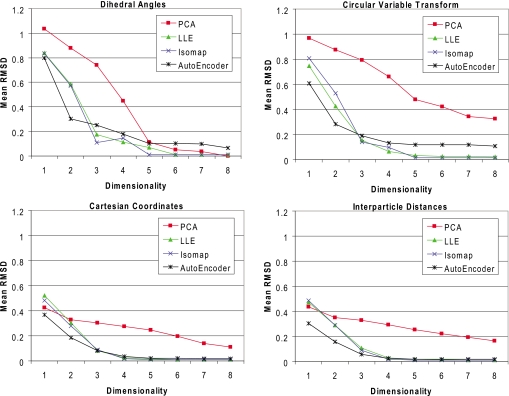
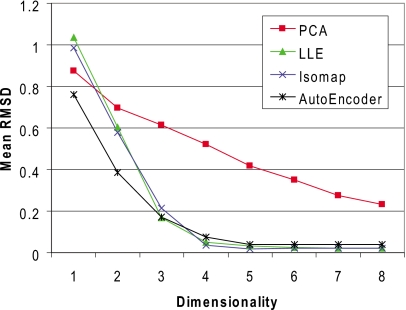
The RMSD reconstruction error for each algorithm on all four data sets, over the range of experimental embedding dimensionalities, is shown in Fig. 2. For Isomap, the results are relatively insensitive to the number of neighbors k chosen to form the adjacency graph; here, we use k=5. In contrast, the results for LLE were sensitive to the selection of k. We obtained the best results with k=10, which is the setting we used to obtain our experimental results. In the case of the autoencoder, a distinct test set is used during training to prevent overfitting. Here, approximately 20% of the samples in each baseline training set were used for this purpose. The RBM and BP batch sizes were, respectively, set to 200 and 100 in all runs. Further, 50 and 600 iterations were respectively used for RMB and BP training. The sizes of the network layers were set to {8 16 8 n} for the DHA data set, {16 32 16 8 n} for the CVT data set, {72 128 64 n} for the XYZ data set, and {276 384 64 n} for the IPD data set; n represents the target embedding dimensionality.
Figure 2.
Mean molecular RMSD obtained from reconstruction of the ring atoms for the four test sets using different dimensionality reduction algorithms. The error is obtained by mapping the samples in the test set to the low-dimensional embedding and subsequently employing the reverse map to reconstruct the molecule in the native-dimensional space.
The results from Fig. 2 demonstrate that the XYZ and IPD encodings produce the most accurate dimensionality reductions in terms of reconstruction error. The improved accuracy relative to the DHA and CVT encoding can be attributed to two causes. First, conversion of DHAs to Cartesian coordinates requires atoms to be placed relative to previously positioned atoms, which can result in an amplification of error. Second, the change in conformation associated with a change in a DHA is not independent of the other DHAs. While the CVT encoding performed slightly better than the DHA encoding in the case of nonlinear reduction algorithms, there is a drastic difference in the two encodings under PCA. This can be explained by the fact that the CVT is itself a nonlinear transformation. The results from the XYZ and IPD data sets are overall very similar with the IPD encoding performing slightly better at low embedding dimensionalities and the XYZ encoding performing slightly better at high embedding dimensionalities.
With the exception of results at embedding dimensionalities equal to 1 on the CRT and IPD data sets, the nonlinear dimensionality reduction techniques significantly outperform the baseline linear PCA algorithm. LLE and Isomap yield very similar results. In all cases, the autoencoder obtains lower RMSDs at embedding dimensionalities of 1 and 2 than Isomap and LLE, but with Isomap and LLE outperforming the autoencoder at larger embedding dimensionalities. However, autoencoder performance could potentially be improved by a better parametrization or longer optimization.
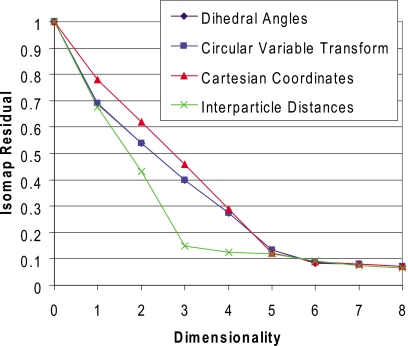
The results in Fig. 2 concern the error associated with reconstruction of molecular Cartesian coordinates from a low-dimensional embedding space, which indirectly captures the ability of dimensionality reduction algorithms to both identify embedded manifolds and the associated bidirectional mapping to∕from a native encoding. For certain applications, such as visualization of energy landscapes, this process—in particular the reverse map—is unnecessary and only a low-dimensional embedding of the training data is required. In this case, an alternative metric can be utilized: the geodesic distance correlation residual.40 This metric quantifies the correlation between (1) geodesic distances in the native high-dimensional space and (2) Euclidean distances in the embedded low-dimensional space. Geodesic distances are computed via the combination of adjacency graph construction and shortest-path computation that underpin the first phase of Isomap. Figure 3 illustrates the geodesic distance correlation residual (one minus the square of the standard correlation coefficient) for each of our four training sets; low-dimensional embeddings in all cases were obtained with Isomap. Under this metric, we find that the DHA and CVT encodings are slightly more effective at preserving the native geodesic distances in low-dimensional embeddings. Interestingly, the IPD encoding yields a dramatic improvement over alternative encodings for embedding dimensionalities of 3 and 4, potentially due to the fact that this encoding does not preserve stereochemistry.
Figure 3.
Correlation residual between Euclidean distances in the low-dimensional embedding space and geodesic distances in the native-dimensional space. Dimensionality reduction was performed using Isomap. Results for DHA and CVT data sets overlap.
Neither Fig. 2 nor Fig. 3 suggest an intrinsic dimensionality of 2 for any of the data sets, despite the fact that only two degrees of freedom were used in our data generation process. Although this would initially appear to be incorrect, we observe that the ring closure problem is multivalued in these two degrees of freedom, with multiple conformations corresponding to a single pair of torsion angles. Therefore, we cannot expect a successful embedding in only two dimensions. From the Whitney embedding theorem we can expect up to five dimensions to be required for a smooth embedding of the 2-manifold, and indeed, all nonlinear methods are able to embed each data set with minimal reconstruction error given four or five embedding dimensions. As discussed in Sec. 2, this result provides an excellent example of the differences in estimating intrinsic dimensionality and embedding dimensionality.
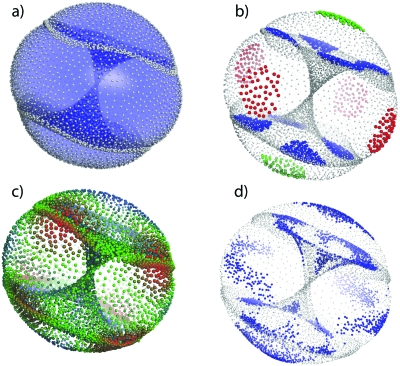
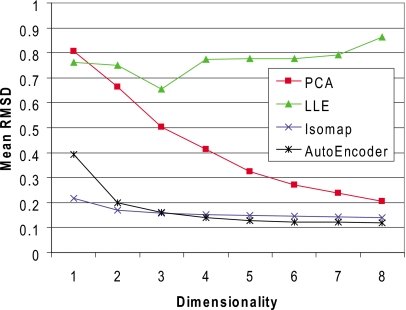
This issue is illustrated nicely by the three-dimensional Isomap embedding of the XYZ data set shown in Fig. 4a. The graphic shows a surface reconstruction of the embedding manifold in three dimensions; physical training samples are colored white. The manifold is essentially a two-dimensional hour-glass shaped surface that intersects a 2D ball shaped surface. Although the training samples are essentially embedded as a 2-manifold, singularities (at the surface intersections) result in a slightly elevated RMSD reconstruction error relative to other samples as shown for the XYZ data set in Fig. 2. While the error is relatively small (<0.1 Å), it is difficult to imagine a 2D embedding of this structure. The symmetrical structure of the manifold results from symmetry in the ring conformations (e.g., the relative position of fluorine atoms within a boat-chair conformation). Figure 4b shows, for example, all boat-chair conformations of the molecule on the inner surface of the hour-class substructure together with the crown and boat conformations on the outer surface of the ball-like substructure. The reference structures for Fig. 4b are taken from Evans and Boeyens,74 who present Cartesian coordinates of the canonical cyclo-octane conformations originally described by Hendrickson.46 When hydrogen and fluorine atoms are included in the dimensionality reduction, a very similar embedding is obtained [Fig. 4c]. While the reconstruction error is higher for this case at lower embedding dimensionalities, all nonlinear dimensionality reduction algorithms produce a low-error embedding in four to five dimensions (Fig. 5).
Figure 4.
3D embeddings of the XYZ data set using Isomap. (a) Surface reconstruction of the ring-atom manifold using training samples shown as white balls. (b) Color coding of ring conformations within 0.2 Å RMSD from the ideal boat-chair (blue), crown (green), and boat (red). (c) Embedding of XYZ data set using all atoms with training samples colored by log(MM3 energy), with red=high energy and blue=low energy. (d) Samples from MD simulation (blue) mapped onto the full XYZ manifold (light gray).
Figure 5.
Mean of the molecular RMSD obtained from reconstruction of all atoms for the XYZ test sets using different dimensionality reduction algorithms.
It is interesting to note that the singularities occur at relatively high energy conformations of the molecule [Fig. 4c]. For this reason, and also to evaluate the effects of sampling, we decided to perform tests on data generated from MD simulation rather than the enumerated conformations. In this case, bond angles and lengths are variable and high energy conformations are not sampled. Starting from 100 random conformations, MD simulations were performed in TINKER at 1.0 fs timesteps for 0.5 ns at 300 K with conformation samples taken every 0.5 ps. These results were then compiled and clustered as before using dt=0.7 to produce a training set with 5167 samples and a test set with 3610 samples. A plot of the conformations sampled (as mapped onto the Isomap manifold obtained from the XYZ data set) is shown in Fig. 4d. Boat-chair, crown, and boat conformations were all sampled; however, no interconversions between conformers were seen within any single 0.5 ns run.
The results from the dimensionality reduction on this data set are shown in Fig. 6 for Cartesian coordinates. Because the MD sampling occurred in distinct regions of the high-dimensional space, there was not a single connected graph, but rather multiple connected components. For Isomap, this can be handled with separate embeddings in nonoverlapping regions of the low-dimensional space. LLE, however, has no inherent method for dealing with multiple components. The resulting weight matrix constructed by LLE has no information on how to position components relative to one another and the algorithm performs very poorly as illustrated in Fig. 6. Isomap and the autoencoder perform well, however, and are able to obtain good results at a dimensionality of 2. For the MD sampling, we do not obtain near-zero errors for any of the first eight dimensionalities. Because there is no constraint on bond angles or bond lengths within the ring, hydrogen and fluorine atoms are able to deviate from their minimum energy orientations, and the sampling is random.
Figure 6.
Mean of the molecular RMSD obtained from reconstruction of samples obtained from MD simulation.
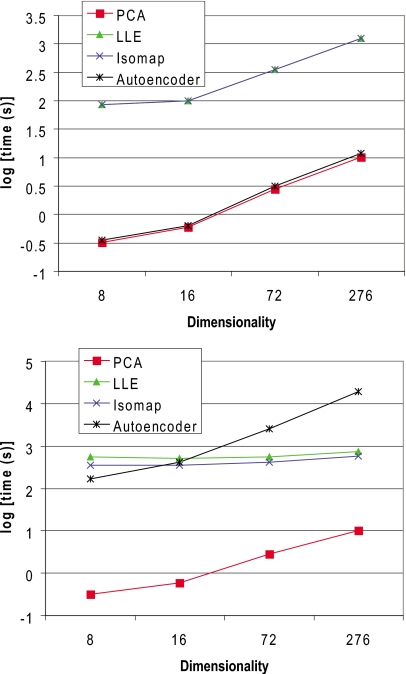
The run times for training and mapping on a single CPU core are shown in Figs. 78. Although the autoencoder has a linear time complexity in the number of samples (assuming constant optimization iterations), its run times far exceed those for the other methods for the sample sizes investigated. In terms of high dimensionality, Isomap and LLE scale much better than the autoencoder and require smaller run times. For mapping, PCA and the autoencoder are orders of magnitude faster. Since LLE and Isomap do not provide explicit maps, neighbors must be calculated for every sample to perform mapping.
Figure 7.
Run times on a single CPU core for each dimensionality reduction algorithm as a function of sample size for the XYZ data set.
Figure 8.
Run times on a single CPU core for each dimensionality reduction algorithm as a function of input dimensionality given by the four test sets. Top: Time required for obtaining the forward and reverse maps. Bottom: Time required for applying the forward map followed by the reverse map for 7500 test samples. PCA and autoencoder run times overlap as do LLE and Isomap times.
DISCUSSION AND CONCLUSIONS
We have used an eight-membered ring to demonstrate the importance of nonlinear correlations in molecular motion and also to demonstrate the efficacy of automated algorithms for nonlinear dimensionality reduction. For high-dimensional encodings ranging from 8 to 276 dimensions, these algorithms are able to provide low-error embeddings within the theoretical limit of 5 dimensions. We have chosen a relatively small molecule for performing these tests in order to avoid sampling issues and to allow for a benchmark against results that can be obtained analytically. However, it is important to note that many of these same constraints can be utilized for larger molecules such as proteins. For example, a similar loop closure constraint has been used for sampling flexible loops between rigid secondary structure elements in proteins.
DHAs have been proposed as a natural encoding for dimensionality reduction of molecular conformation due to the separation of internal degrees of freedom from overall motion. One issue with this approach is that sensitivity of DHAs to overall conformational changes within a molecule is not constant; relatively small changes in one DHA can cause drastic conformational changes while large changes in another can result in relatively little change. Here, an issue is that the conformational change resulting from deviation of one DHA is not independent of the other angles. A scheme for weighting DHAs based on conformational change is not trivial. For the case we studied, the use of DHAs (or the circular variable transform of these variables) resulted in poor reconstruction errors when a Cartesian RMSD is utilized as a metric. Interatomic distances provide another method of separating internal motion; however, they require an increase in the number of variables considered and an additional embedding must be performed to retrieve the Cartesian coordinates. Since it is relatively straightforward to remove any net rotation and translation of the entire molecule, we find a Cartesian coordinate representation to be the simplest approach for encoding molecular conformations.
We have evaluated three automated algorithms for nonlinear dimensionality reduction. In general, the performance of LLE and Isomap was very similar for the case presented. LLE is attractive from a theoretical standpoint in that only local Euclidean distances are considered. From a numerical standpoint, however, we have found the algorithm difficult to implement due to numerical issues in solving for the smallest eigenvalues and the problem of solving for reconstruction weights when the number of neighbors is larger than the intrinsic dimensionality of the manifold. The autoencoder performed best at low dimensionalities, generates fast explicit forward and reverse maps, and considers reconstruction error explicitly in the objective function. The main drawback for the autoencoder is the excessive run times required when the input dimensionality is high. While we have improved this to some extent with a multithreaded implementation, algorithmic changes or a distributed-memory parallel implementation may be necessary for efficiently investigating large problems.
Despite their simplicity, dimensionality reduction on eight-membered rings involves two complications that make for an interesting benchmark case. First, although the ring closure problem is 2D and possible conformations lie on a 2-manifold in the high-dimensional space, the problem is multivalued and the manifold cannot necessarily be embedded in two dimensions. However, for the applications presented in this paper, a strict preservation of topology is not necessarily required; rather, it is desirable to obtain a low-error representation in as few dimensions as possible. Therefore, algorithms that provide an implicit or explicit mechanism for “manifold tearing”65 might be desirable. For example, improved results at two dimensions were obtained when only the low-energy MD results were used. A second issue involves symmetry in the molecule. Here, we have used a trisubstituted ring in order to avoid any symmetry issues. In practice, however, algorithms capable of detecting or enforcing symmetry might be beneficial.77
Although we have focused dimensionality reduction approaches on the coordinated movements of molecules, dimensionality reduction in the general sense has been central to the field of molecular physics. Single particle representations for atoms, rigid-body approximations, and spherical78 and aspherical79 point-particle representations for groups of atoms have been utilized for some time to improve the efficiency of simulation and remain topics of current investigation.
ACKNOWLEDGMENTS
Sandia is a multipurpose laboratory operated by Sandia Corporation, a Lockheed-Martin Co., for the U.S. Department of Energy under Contract No. DE-AC04-94AL85000. E.A.C. acknowledges partial support from NIH-NIGMS Grant No. R01-GM081710. S.N.P. acknowledges support from the Boehringer fund for Biocomputing (Boehringer Ingelheim Pharmaceuticals, Ridgefield, CT).
References
- Dill K. A., Biochemistry 10.1021/bi00483a001 29, 7133 (1990). [DOI] [PubMed] [Google Scholar]
- Hegger R., Altis A., Nguyen P. H., and Stock G., Phys. Rev. Lett. 10.1103/PhysRevLett.98.028102 98, 028102 (2007). [DOI] [PubMed] [Google Scholar]
- Lange O. F. and Grubmuller H., J. Phys. Chem. B 10.1021/jp062548j 110, 22842 (2006). [DOI] [PubMed] [Google Scholar]
- Plaxco K. W. and Gross M., Nat. Struct. Biol. 10.1038/90349 8, 659 (2001). [DOI] [PubMed] [Google Scholar]
- Dinner A. R., Sali A., Smith L. J., Dobson C. M., and Karplus M., Trends Biochem. Sci. 10.1016/S0968-0004(00)01610-8 25, 331 (2000). [DOI] [PubMed] [Google Scholar]
- Onuchic J. N. and Wolynes P. G., Curr. Opin. Struct. Biol. 10.1016/j.sbi.2004.01.009 14, 70 (2004). [DOI] [PubMed] [Google Scholar]
- Pappu R. V., Srinivasan R., and Rose G. D., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.97.23.12565 97, 12565 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Groot B. L., Daura X., Mark A. E., and Grubmuller H., J. Mol. Biol. 10.1006/jmbi.2001.4655 309, 299 (2001). [DOI] [PubMed] [Google Scholar]
- Abseher R. and Nilges M., Proteins: Struct., Funct., Genet. 39, 82 (2000). [DOI] [PubMed] [Google Scholar]
- Amadei A., de Groot B. L., Ceruso M. A., Paci M., Di Nola A., and Berendsen H. J. C., Proteins: Struct., Funct., Genet. 35, 283 (1999). [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen A. B. M., and Berendsen H. J. C., Proteins: Struct., Funct., Genet. 10.1002/prot.340170408 17, 412 (1993). [DOI] [PubMed] [Google Scholar]
- Amadei A., Linssen A. B. M., deGroot B. L., vanAalten D. M. F., and Berendsen H. J. C., J. Biomol. Struct. Dyn. 13, 615 (1996). [DOI] [PubMed] [Google Scholar]
- deGroot B. L., Amadei A., Scheek R. M., vanNuland N. A. J., and Berendsen H. J. C., Proteins: Struct., Funct., Genet. 26, 314 (1996).8953652 [DOI] [Google Scholar]
- Noguti T. and Go N., Biopolymers 10.1002/bip.360240308 24, 527 (1985). [DOI] [PubMed] [Google Scholar]
- Brooks B. R., Janezic D., and Karplus M., J. Comput. Chem. 10.1002/jcc.540161209 16, 1522 (1995). [DOI] [Google Scholar]
- Janezic D. and Brooks B. R., J. Comput. Chem. 10.1002/jcc.540161210 16, 1543 (1995). [DOI] [Google Scholar]
- Janezic D., Venable R. M., and Brooks B. R., J. Comput. Chem. 10.1002/jcc.540161211 16, 1554 (1995). [DOI] [Google Scholar]
- Hayward S. and Go N., Annu. Rev. Phys. Chem. 10.1146/annurev.physchem.46.1.223 46, 223 (1995). [DOI] [PubMed] [Google Scholar]
- Teodoro M. L., Phillips G. N., and Kavraki L. E., J. Comput. Biol. 10.1089/10665270360688228 10, 617 (2003). [DOI] [PubMed] [Google Scholar]
- Bruschweiler R. and Case D. A., Phys. Rev. Lett. 10.1103/PhysRevLett.72.940 72, 940 (1994). [DOI] [PubMed] [Google Scholar]
- Case D. A., Acc. Chem. Res. 10.1021/ar010020l 35, 325 (2002). [DOI] [PubMed] [Google Scholar]
- de Groot B. L., Hayward S., van Aalten D. M. F., Amadei A., and Berendsen H. J. C., Proteins: Struct., Funct., Genet. 31, 116 (1998). [DOI] [PubMed] [Google Scholar]
- Meinhold L. and Smith J. C., Biophys. J. 10.1529/biophysj.104.056101 88, 2554 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romo T. D., Clarage J. B., Sorensen D. C., and Phillips G. N., Proteins: Struct., Funct., Genet. 10.1002/prot.340220403 22, 311 (1995). [DOI] [PubMed] [Google Scholar]
- Stepanova M., Phys. Rev. E 10.1103/PhysRevE.76.051918 76, 051918 (2007). [DOI] [PubMed] [Google Scholar]
- van Aalten D. M. F., Conn D. A., de Groot B. L., Berendsen H. J. C., Findlay J. B. C., and Amadei A., Biophys. J. 73, 2891 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange O. F. and Grubmuller H., J. Chem. Phys. 10.1063/1.2199530 124, 214903 (2006). [DOI] [PubMed] [Google Scholar]
- Spiwok V., Lipovova P., and Kralova B., J. Phys. Chem. B 10.1021/jp068587c 111, 3073 (2007). [DOI] [PubMed] [Google Scholar]
- Mustard D. and Ritchie D. W., Proteins: Struct., Funct., Genet. 10.1002/prot.20569 60, 269 (2005). [DOI] [PubMed] [Google Scholar]
- Zacharias M., Proteins: Struct., Funct., Bioinf. 54, 759 (2004). [DOI] [PubMed] [Google Scholar]
- Qian B., Ortiz A. R., and Baker D., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0404703101 101, 15346 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby M., Geometric Data Analysis (Wiley, New York, 2001). [Google Scholar]
- Caves L. S. D., Evanseck J. D., and Karplus M., Protein Sci. 7, 649 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitao A. and Go N., Curr. Opin. Struct. Biol. 10.1016/S0959-440X(99)80023-2 9, 164 (1999). [DOI] [PubMed] [Google Scholar]
- Clarage J. B., Romo T., Andrews B. K., Pettitt B. M., and Phillips G. N., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.92.8.3288 92, 3288 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altis A., Nguyen P. H., Hegger R., and Stock G., J. Chem. Phys. 10.1063/1.2746330 126, 244111 (2007). [DOI] [PubMed] [Google Scholar]
- Lange O. F. and Grubmuller H., Proteins: Struct., Funct., Bioinf. 62, 1053 (2006). [DOI] [PubMed] [Google Scholar]
- Agrafiotis D. K. and Xu H. F., Proc. Natl. Acad. Sci. U.S.A. 99, 15869 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen P. H., Proteins: Struct., Funct., Bioinf. 65, 898 (2006). [Google Scholar]
- Tenenbaum J. B., de Silva V., and Langford J. C., Science 10.1126/science.290.5500.2319 290, 2319 (2000). [DOI] [PubMed] [Google Scholar]
- Das P., Moll M., Stamati H., Kavraki L. E., and Clementi C., Proc. Natl. Acad. Sci. U.S.A. 10.1073/pnas.0603553103 103, 9885 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsera M. A., Wriggers W., Oono Y., and Schulten K., J. Phys. Chem. 10.1021/jp9536920 100, 2567 (1996). [DOI] [Google Scholar]
- Rocha W. R., Pliego J. R., Resende S. M., dos Santos H. F., de Oliveira M. A., and de Almeida W. B., J. Comput. Chem. 19, 524 (1998). [DOI] [Google Scholar]
- Bharadwaj R. K., Mol. Phys. 10.1080/002689700162630 98, 211 (2000). [DOI] [Google Scholar]
- Chen Z. and Escobedo F. A., J. Chem. Phys. 10.1063/1.1328069 113, 11382 (2000). [DOI] [Google Scholar]
- Hendrickson J., J. Am. Chem. Soc. 10.1021/ja01002a036 89, 7036 (1967). [DOI] [Google Scholar]
- Cremer D. and Pople J. A., J. Am. Chem. Soc. 10.1021/ja00839a011 97, 1354 (1975). [DOI] [Google Scholar]
- Go N. and Scheraga H., Macromolecules 10.1021/ma60014a012 3, 178 (1970). [DOI] [Google Scholar]
- Kudin K. N. and Dymarsky A. Y., J. Chem. Phys. 10.1063/1.1864872 122, 124103 (2005). [DOI] [PubMed] [Google Scholar]
- Teodoro M., G.Phillips, Jr., and Kavraki L., Annual Conference on Research in Computational Molecular Biology, Washington, DC, 2002. (unpublished).
- Crippen G. M. and Havel T. F., Distance Geometry and Molecular Conformation (Wiley, New York, 1988). [Google Scholar]
- Elmaci N. and Berry R. S., J. Chem. Phys. 10.1063/1.478992 110, 10606 (1999). [DOI] [Google Scholar]
- Pan P. W., Dickson R. J., Gordon H. L., Rothstein S. M., and Tanaka S., J. Chem. Phys. 10.1063/1.1830434 122, 034904 (2005). [DOI] [Google Scholar]
- Jutten C. and Herault J., Signal Process. 10.1016/0165-1684(91)90079-X 24, 1 (1991). [DOI] [Google Scholar]
- Hyvarinen A., Neural Computing Surveys 2, 94 (1999). [Google Scholar]
- Scholkopf B., Smola A., and Muller K. -R., Advances in Kernel Methods SV Learning (MIT, Cambridge, MA, 1999). [Google Scholar]
- Kohonen T., Self-Organizing Maps (Springer-Verlag, Berlin, 1995). [Google Scholar]
- Hinton G. and Salakhutdinov R., Science 10.1126/science.1127647 313, 504 (2006). [DOI] [PubMed] [Google Scholar]
- Roweis S. and Saul L., Science 10.1126/science.290.5500.2323 290, 2323 (2000). [DOI] [PubMed] [Google Scholar]
- Lee J. A. and Verleysen M., Nonlinear Dimensionality Reduction (Springer-Verlag, Berlin, 2007). [Google Scholar]
- Trefethen L. and Bau D., Numerical Linear Algebra (SIAM, Philadelphia, 1997). [Google Scholar]
- Saul L. and Roweis S., J. Mach. Learn. Res. 4, 119 (2004). [Google Scholar]
- Havel T., Kuntz I. D., and Crippen G. M., Bull. Math. Biol. 10.1016/S0092-8240(83)80020-2 45, 665 (1983). [DOI] [Google Scholar]
- Martin S. and Backer A., Proceedings of the ACM Symposium on Applied Computing (SAC), 2005. (unpublished), pp. 22–26.
- Lee J. A. and Verleysen M., Nonlinear Dimensionality Reduction (Springer, New York, 2007). [Google Scholar]
- The code is available for academic use by contacting one of the Sandia authors.
- Dhillon I. S. and Parlett B. N., Linear Algebr. Appl. 10.1016/j.laa.2003.12.028 387, 1 (2004). [DOI] [Google Scholar]
- Anderson E., Bai Z., Bischof C., Blackford S., Demmel J., Dongarra J., Du Croz J., Greenbaum A., Hammarling S., McKenney A., and Sorensen D., Lapack Users’ Guide 3rd ed. (Society for Industrial and Applied Mathematics, Philadelphia, 1999). [Google Scholar]
- Coutsias E., Seok C., Jacobson M., and Dill K., J. Comput. Chem. 10.1002/jcc.10416 25, 510 (2004). [DOI] [PubMed] [Google Scholar]
- Porta J., Ros L., Thomas F., Corcho F., Cant J., and Perez J., J. Comput. Chem. 10.1002/jcc.20733 28, 2170 (2007). [DOI] [PubMed] [Google Scholar]
- Manocha D. and Zhu Y., in ISMB, edited by Altman R. B., Brutlag D. L., Karp P. D., Lathrop R. H., and Searls D. B. (AAAI, Menlo Park, CA, 1994), pp. 285–293. [Google Scholar]
- Lee H. and Liang C., Mech. Mach. Theory 10.1016/0094-114X(88)90107-3 23, 219 (1988). [DOI] [Google Scholar]
- Coutsias E., Seok C., Wester M., and Dill K., Int. J. Quantum Chem. 10.1002/qua.20751 106, 176 (2006). [DOI] [Google Scholar]
- Evans D. G. and Boeyens J. C. A., Acta Crystallogr., Sect. B: Struct. Sci. 10.1107/S0108768188008730 44, 663 (1988). [DOI] [Google Scholar]
- Pollock S. and Coutsias E. (unpublished).
- Allinger N. L., Yuh Y. H., and Lii J. H., J. Am. Chem. Soc. 10.1021/ja00205a001 111, 8551 (1989). [DOI] [Google Scholar]
- Shah M. and Sorensen D. C., Proceedings of the 44th IEEE Conference on Decision and Control and European Control Conference, Seville, Spain, 12–15 December 2005. (unpublished), Vols. 1–8, pp. 2260–2264.
- Praprotnik M., Delle Site L., and Kremer K., Annu. Rev. Phys. Chem. 10.1146/annurev.physchem.59.032607.093707 59, 545 (2008). [DOI] [PubMed] [Google Scholar]
- Everaers R. and Ejtehadi M. R., Phys. Rev. E 10.1103/PhysRevE.67.041710 67, 041710 (2003). [DOI] [PubMed] [Google Scholar]