Abstract
There are several medical application areas that require the segmentation and separation of the component bones of joints in a sequence of images of the joint acquired under various loading conditions, our own target area being joint motion analysis. This is a challenging problem due to the proximity of bones at the joint, partial volume effects, and other imaging modality-specific factors that confound boundary contrast. In this article, a two-step model-based segmentation strategy is proposed that utilizes the unique context of the current application wherein the shape of each individual bone is preserved in all scans of a particular joint while the spatial arrangement of the bones alters significantly among bones and scans. In the first step, a rigid deterministic model of the bone is generated from a segmentation of the bone in the image corresponding to one position of the joint by using the live wire method. Subsequently, in other images of the same joint, this model is used to search for the same bone by minimizing an energy function that utilizes both boundary- and region-based information. An evaluation of the method by utilizing a total of 60 data sets on MR and CT images of the ankle complex and cervical spine indicates that the segmentations agree very closely with the live wire segmentations, yielding true positive and false positive volume fractions in the range 89%–97% and 0.2%–0.7%. The method requires 1–2 minutes of operator time and 6–7 min of computer time per data set, which makes it significantly more efficient than live wire—the method currently available for the task that can be used routinely.
Keywords: MRI, image segmentation, live wire, model-based segmentation, kinematics
INTRODUCTION
Motivation
There are several medical research applications which require the segmentation of the bones of a human skeletal joint in medical images. We cite here two areas—joint motion analysis, and image-guided surgery. In motion analysis, the idea is to acquire 3D tomographic images of the joint in several positions for each type of motion that is being investigated (for example, flexion-extension, inversion-eversion, internal-external rotation), and then by appropriately analyzing the images to determine how the 3D geometric architecture of the bones changes from position to position. The main goal for motion analysis is to understand normal motion, to characterize deviations caused by various anomalies that may affect joint function, and to discern how effective are procedures to treat these conditions. Although many techniques based on 2D imaging (radiography, fluoroscopy, slice-only) and external tracking of markers attached to skin have been investigated (see Ref. 1 for a review), 3D imaging approaches are vital to capture full 3D motion information. Several such efforts are currently under way that employ MRI and CT (examples2, 3, 4, 5). In image-guided orthopedic surgery (examples6, 7, 8), bony structures need to be identified in preoperative images, not only for optimal preoperative planning but also for intraoperative guidance by registering in the same space all intraoperative information (images, internal organs of the patient, instruments, and tools) with the preoperative images. Our own motivation for the work presented in this article comes from the first application of joint motion analysis.
Previous work
Although the literature on image segmentation is vast,9 there is little research dedicated to the problem of segmenting the bones at a joint in medical imagery. In our application context, this problem poses some special challenges (discussed below). And consequently, well-known segmentation frameworks such as deformable boundary,10 level-set11 water-shed,12 graph-cut,13 clustering,14 and fuzzy connectedness15 approaches, we believe, will not yield a level of precision, accuracy, and efficiency that is required by the application when directly employed, or will require considerable research and development to bring them to a form with an acceptable level of performance in routine use. Therefore, our review scope will be confined to those methods which are designed specifically for segmenting the bones at joints in MR and CT imagery.
The segmentation and separation of the bones at a joint in MR and CT images pose the following five main challenges: (Ch1) Bones are situated very close to each other and the partial volume effect exacerbates their segmentation and separation. (Ch2) The extent of this effect depends often on the orientation of the slice planes with respect to the articulating bone surfaces. Since there are multiple bones at a joint and their surfaces are usually curved, it is impossible to select a slice plane (for MRI) that is optimal in orientation for reducing this effect. (Ch3) In the case of MRI, bone does not give much signal, and connective tissues, such as ligaments and tendons that attach to bone, behave like bone in this respect. This makes the demarcation of boundaries of bones that come close together with the presence of connective tissues even more challenging. (Ch4) In the case of CT, the thinner aspects of cortical bones pose a challenge similar to the one described in (Ch3) for MRI, namely that thin bone regions resemble soft tissue and skin regions. When these entities come close together at a joint, the absence of boundary specific information causes difficulties. (Ch5) Although each of the bones at a joint undergoes a rigid transformation in moving from one joint position to another, because of their relatively independent motions, the collection of bones and the soft tissue structures produces a strange admixture of nonlinear rigid and elastic transformations.
The papers that dealt with the problem of segmenting bone in MR images, and particularly focusing on separating the bones at a joint in MR and CT images, are sparse. Dogdas et al.16 and Hoehne et al.17 used mathematical morphology to segment the skull in 3D human MR images. Heinonen et al.18 used thresholding and region growing to segment bone in MR image volumes. These studies did not consider the problem of separating the bones at an articulating joint. Lorigo et al.19 applied a texture-based geodesic active contour method to segment bones in knee MR images. Sebastian et al.20 combined active contour, region growing, and region competition for segmentation of the carpal bones of wrist in 3D CT images. Reyes-Aldasoro et al.21 applied a subband filtering based K-means method to segment bones, tissues, and muscles in knee MR images. Grau et al. applied an improved watershed transform for knee cartilage segmentation.22 Hoad et al.6 combined threshold region growing with morphological filtering to segment lumbar spine in MR images for computer-assisted surgery purposes. Udupa et al.15 used the live-wire method23 to segment bones of the ankle complex in MR images. The studies in Refs. 16, 17, 18, 22 did not demonstrate how effective these general methods of segmenting bones in MR images would be in separating the bones at a joint. Particularly, there is no strategy in these methods that addresses the challenges denoted in (Ch1)–(Ch3) above. The methods in Refs. 19, 20, 23 employ slice-by-slice strategies, as such they demand a significant amount of user time, even if it is just for ascertaining that the segmented results are acceptable in every slice. Reyes-Aldasoro et al. in Ref. 21 did not address the problem of separating the bones of a joint. Further, the bones of the ankle joint complex and spine are more challenging to segment than those of the knee owing to the reasons identified in (Ch1)–(Ch3) above, and due to the fact that the bones of the former are smaller and more compactly packed. Similar to Ref. 21, Hoad and Martel in Ref. 6 did not address the problem of separating the component bones (vertebrae) of the spine, which, in view of (Ch1) and (Ch4) above, is a more challenging problem than just delineating all bone only.
Purpose, rationale, outline of paper
In our ongoing studies of the kinematics of the ankle complex,26 the shoulder joint,27 and of the spinal vertebrae28 involving cadaveric specimens as well as human subjects, we acquire multiple 3D (volume) images for each subject corresponding to multiple situations. These situations include various combinations of the following cases: Multiple subjects∕specimens; multiple positions of the joints; under different load conditions; preinjury conditions; postsurgery conditions; different types of surgical reconstructions; and different longitudinal time points. The number of 3D images generated for the same subject∕specimen in these tasks ranges from 10 to 30. In these images, it is fair to assume that each of the component bones of the joint complex has not changed its shape but the spatial arrangements of these bones forming the joint assembly are different in different images. This leads to a unique situation from the segmentation perspective as discussed further in the next paragraph. The study of the kinematics of the joint complex under the various conditions delineated above requires the segmentation of each component bone of the joint complex in each 3D image of the set of (10–30) 3D images generated as above for each subject∕specimen. Owing to the difficulties (Ch1)–(Ch5) mentioned above, and because of the lack of availability of a more efficient method, we have been using the live-wire method for segmenting bones. Live wire is a slice-by-slice method wherein the user steers the segmentation process by offering recognition help and a computer algorithm performs delineation. Because of user help, (Ch1)–(Ch5) are overcome, but the process becomes very demanding in terms of user time, although it is more reproducible and efficient than manual boundary tracing and it never calls for post hoc correction. Although we have coped with the live-wire method so far with the number of subjects being 20–50 and the number of 3D images being a couple of hundreds, live wire is becoming impractical for greater number of subjects.
We may solve this problem in two stages. In the first stage, addressed in this article, all component bones of a given joint will be segmented by using live wire in one 3D image. This segmented result will then be used as a rigid model to delineate automatically these same bones in all the rest of the 3D images pertaining to the same joint. In the second stage, not addressed in this article, to segment the first 3D image, our aim is to replace live wire by a more efficient strategy that will afford a level of precision and accuracy which is comparable to that of live wire. It is tempting to suggest if a model-based approach, such as deformable boundary,10 active shape model,29 or active appearance model,30 may offer a viable solution to the whole problem. Our rationale for opting for the two stage approach is as follows. First, deformable model-based approaches10, 29, 30 will introduce untrue variations in the shape of a specific bone among different images of a given joint and will fail to guarantee that the two segmentations of the same bone in two different positions corresponding to one of the situations mentioned above represent a rigid geometric transformation of the same bone model. Second, in our preliminary efforts, we found that the final position of the model produced by these methods does not often quite agree with the boundary perceivable in the image slices because of Ch1–Ch5. Correcting for these errors so that the results are comparable with the live-wire method in boundary placement would call for either considerable user help or a significant effort to modify the methods. Although these shortcomings may not matter in certain types of analysis, in our application, wherein accurate architectural description of the bones and how this architecture changes from one condition to another are crucial, this matters. If a straightforward rigid registration method is employed among the images, it fails to locate the bones correctly as illustrated in Fig. 1. These reasons and the mixed rigid and elastic nature of the transformations undergone by the images directed us toward the two-stage approach mentioned above.
Figure 1.
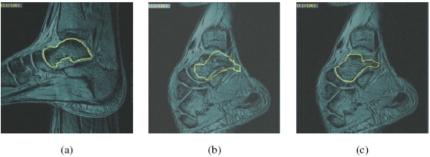
(a) A typical image slice from a 3D MR data set of the left ankle joint of a healthy volunteer at one position. Boundary of the talus delineated by using the live-wire is illustrated on the image. (b) An image slice of the same ankle joint scanned at another position. The 3D data set of (a) was registered with the 3D data set of (b) by using Mutual Information, and the intersection of the talus, delineated from the 3D data set of (a), with the current image slice of (b), is depicted after registration. The mismatch in identifying the talus is clear. (c) Location of the talus bone in the 3D data set of (b) by using the proposed rigid-model based approach.
The complete methodology of our approach including how the rigid model is first created from the first 3D image and how this is subsequently used to segment the same bone in all other 3D images is described in Sec. 2. In Sec. 3, we carry out an evaluation of this method in terms of its precision, accuracy, and efficiency by considering two joint systems—the ankle complex imaged via MRI and the spine imaged via CT. In Sec. 4, we summarize our conclusions and give some pointers on the second stage of this work that is currently under way. A preliminary version of this paper appeared in the conference proceedings of the SPIE 2005 medical imaging symposium.31
The problem addressed in this paper can also be considered from the viewpoint of image registration for devising a solution. The transformations undergone by images in dealing with the registration problem in general range from linear rigid and affine and nonlinear elastic transformations to complex mixtures of rigid and affine, rigid and elastic, and affine and elastic transformations. The situation dealt with in this article corresponds to a mixture of rigid and elastic transformations. It is perhaps more challenging than pure elastic transformation for devising a registration solution. Instead of taking a fundamentally registration approach, we have pursued a predominantly segmentation strategy in this article for several reasons. First, we needed an immediate solution to delineate bones’ boundaries in our application. Second, although a registration approach would perhaps lead to a fundamental advance, the solution would be more general and complicated than the one proposed here. Finally, even after a sophisticated registration operation, we believe, from our simple experiments with registrations, that the delineation will need to be adjusted which is what led to our current segmentation propagation approach, although the registration transformation we have employed is simple. Segmentation propagation has been used in the past in other applications, for example, in brain MRI for assessing change in brain volume over time.24, 25
METHODOLOGY
Notation, terminology, outline of method
We refer to a 3D volume image as a scene and represent it by a pair C=(C,f), where C, called the scene domain, is a 3D rectangular array of voxels, and f is the scene intensity function which assigns to every voxel v∊C an integer called the intensity of v in C. Let C1,C2,…,Cn be the scenes acquired for the same joint complex (such as the ankle) for the same subject under different conditions (exemplified earlier) wherein we may assume that the component bones of the joint complex have not changed shape. Our aim is to segment a given target bone B in all these scenes such that the segmentations differ only by a rigid transformation (and by any digitization artifacts caused by the different positions and orientations) of B in the different scenes. Since the solution methodology is the same for any n⩾2, we assume from now on that we are given two scenes C1=(C1,f1) and C2=(C2,f2) corresponding to, what we shall from now on refer to generically as, two different positions (Position 1 and Position 2) of the joint complex. For any scene C=(C,f), we denote its segmentation that captures bone B, by a binary scene Cb=(Cb,fb) where fb is such that, for any voxel c∊C, fb(c)=1, if c is determined to be in the bone, and fb(c)=0, otherwise. Our goal is to obtain binary scenes and representing segmentation of B in C1 and C2.
Overall, the segmentation methodology consists of the following steps:
S1: Segmentation of the bone in scene C1 corresponding to Position 1.
S2: VOI (volume of interest) selection corresponding to the bone in C1 and C2.
S3: Registration of the VOI scenes of C1 and C2.
S4: Matching the segmentation corresponding to the VOI scene of C1 with the VOI scene of C2.
S4.1 Describing the relative intensity pattern in the vicinity of the bone boundary.
S4.2 Formulating an energy functional for the match.
S4.3 Optimization of energy functional.
S4.4 Computation of the segmented bone for C2.
These steps are described in more detail in the following sections.
Detailed description of the segmentation method
S1: Segmentation of the bone in C1
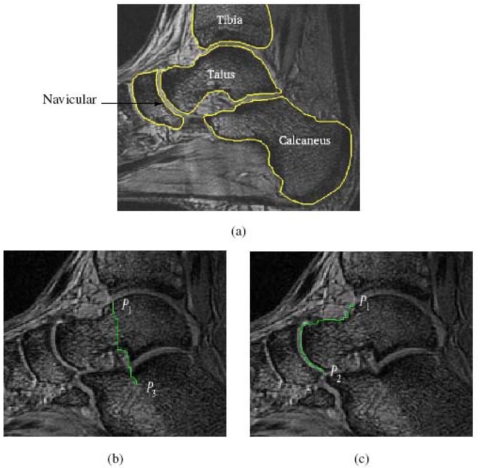
Our current approach for this step utilizes user help in a slice-by-slice manner, but somewhat differently for MRI and CT scenes as described below. For MRI scenes, to segment B in C1, we employ the live wire method.23 Live wire is a user-steered slice-by-slice segmentation method in which the user begins by positioning the cursor on the boundary in a slice display of the scene and selects a point. For any subsequent position of the cursor, a live-wire segment is displayed in realtime which represents an optimum path from the initial point to any current position of the cursor. In particular, if the cursor is positioned near the boundary, the live wire snaps onto the boundary. The cursor is placed as far away from the first position as possible with the live wire snapped onto the boundary, and a new point is set by depositing the cursor. Typically, 3–7 points deposited on the boundary in this fashion are adequate to delineate a bone in each slice of a given scene. In live wire, pixel vertices are considered to be the nodes of a directed graph, and each pixel edge is considered to be oriented and represents two directed arcs. That is, if a and b are the end points (vertices) of a pixel edge, then the two directed arcs are (a,b) and (b,a). Each directed arc is assigned a cost, and the live wire method finds an optimum path between any two points P1 and P2 specified in the scene as a sequence of directed arcs (oriented pixel edges) such that the total cost along the path is the smallest. This formulation and the facility to assign costs based on training (see Ref. 23 for details) allow overcoming some of the challenges (Ch1)–(Ch4) mentioned earlier. One aspect of this is illustrated in Fig. 2 which shows one slice of a MRI scene of the ankle joint complex and a live wire segment (oriented optimal path) defined between two specified points. The orientedness of pixel edges imposes an orientation on the path, which allows the live wire method to resolve between two boundary segments that come very close (in the figure, the boundaries of the navicular and the talus, the talus and tibia, the talus and calcaneus) which are otherwise very similar in their intensity properties. In the figure, the live wire segment from P1 to P2 on the boundary of the talus has an orientation opposite to the orientation of the boundary segment of the navicular that comes very close to this live wire segment, and they have very different cost structures. This is why live wire can resolve closely situated boundaries and boundaries of very thin objects.
Figure 2.
Slice of an MRI scene of the ankle joint and the live wire segment displayed between two pairs of points (b) P1 to P3, and (c) P1 to P2. The four bones that come close together and their boundaries are shown in (a) for reference.
A three-dimensional version of live wire, called iterative live wire,42 is also available in our software system40 which is in general more efficient than but offers precision and accuracy comparable to the 2D version. It operates as follows. The user performs 2D live wire in one slice of the 3D scene, preferably in the slice passing through the middle of the object. The user then selects the next slice. The algorithm then projects the points P1,…,Pn selected by the user on the previous slice to the current slice and performs live wire between successive projected points in the current slice. Since the projected points are likely to be near the actual boundary, owing to the orientedness and snapping property of live wire, the middle portions of the live wire segment between each pair Pi, Pi+1 of points will lie on the actual boundary in the current slice. The algorithm then selects midpoints of these segments and performs live wire between successive midpoints. The entire process is repeated iteratively until convergence. From the user’s perspective, as soon as he selects the next slice, this entire process is executed instantaneously and the resulting contour is displayed. If the result is acceptable, the user selects the next slice again. Otherwise, he will perform the usual live wire on this slice. Typically the user can skip 4–5 slices in this automatic mode before having to steer live wire manually. The user directs the iterative live wire process on both sides of the starting (middle) slice until all slices are covered.
There are essential differences between live wire and active contour methods. These are described and illustrated in papers published on live wire. Here we briefly summarize them. First, live wire methods are formulated as a graph problem and globally optimum contours (although in a piecewise manner) are found via dynamic programming unlike most active contours which are affected by local minima. Second, live wire is by design user steered, calling for no post hoc correction, whereas active contours are designed to be “hands-off” methods, requiring correction if the method goes wrong. Third, because of the use of pixel edges as boundary elements and consideration of orientedness, live wire can negotiate boundaries that are situated even one pixel apart and boundaries of very thin (one pixel thick) objects. In most existing active contour formulations these are difficult to resolve. Finally, live wire and active contours both have noise resistance properties, the former due to the shortness of optimum paths and the latter due to contour smoothness incorporated into the energy function. However, their relative behavior depends much on the cost function used for the former and the energy functional employed for the latter.
For CT scenes, our approach to segment B in C1 is to draw a curve on the slice such that the curve encloses only B and no other bones. This masked region is subsequently thresholded to segment B. We note that precise drawing of the curve on the boundary is needed only in those parts where B comes in contact with other bones. In other parts, we have to draw the curve roughly just to enclose B.
The output of Step S1 is a binary scene representing a segmentation of B in C1.
S2: VOI (Volume Of Interest) selection in C1 and C2
The purpose behind Steps S2 and S3 is twofold. First, when the motion of B from C1 to C2 is substantial, if Step S4 to search for the bone in C2 utilizing its segmentation is carried out on the entire scene C2, any search strategy may be misled by local optima of the objective function utilized for search. Second, such a search would be computationally expensive, too. If a VOI enclosing B in C1 is determined roughly and if the matching VOI for B in C2 is also specified roughly, then both of these are overcome effectively. Since we have the exact segmentation of B in C1 from Step S1, a subscene of C1 corresponding to a VOI of B can be determined automatically. This is done by determining a rectangular box whose faces are parallel to the coordinate planes of the scene coordinate system of C1 and which encloses B with a gap of a few (5–10) voxels all around. A VOI of the same size is then specified in C2 whose location is adjustable manually (via slice display on one appropriately selected slice of C2). The orientation of this VOI is also such that the faces of the VOI box are parallel to the coordinate planes of C2. In this step, thus, the following entities are output: C1s, C2s—the VOI subscenes of C1 and C2; —the VOI subscene of . This step is akin to manual initialization done in many registration and segmentation methods and specifies the rough location of B in C2.
S3: Registration of the two VOI scenes
Registration is commonly used for initialization in model-based segmentation methods. We employ registration in the same spirit—to facilitate the model-based search process and not for achieving perfect matching. However, the registration in the case of joints is very difficult and more challenging in the sense that it can be considered to be neither purely rigid nor purely nonrigid (Ch5). The rationale for selecting a VOI in Step S2 is to make it possible for us to focus mostly on bone, and particularly on the bones of interest, and hence mostly on the rigid component of motion. The aim of Step S3 is therefore to register roughly C1s with C2s.
We employ different methods of registration while dealing with MRI and CT scenes. For MRI scenes, C1s is registered with C2s by maximizing the mutual information32 between C1s and C2s. For CT scenes, we use a method based on landmarks.33 The rationale for this choice is that, in CT scenes, the entire bony structure can be segmented (but without separating the component bones) very easily by thresholding the scene, toward the goal of creating a 3D rendering of this structure for Position 1 and Position 2 and identifying the corresponding landmarks in these renderings. (In MRI scenes, such an approach is infeasible.) Since thresholding, isosurface creation,34 rendering,35 and landmark selection, and registration can all be accomplished at interactive speeds on modern PCs even for extremely large data sets, there is no need to create VOI scenes in Step S2 when segmenting CT scenes. (Therefore, for CT scenes, we can assume that C1s=C1 and C2s=C2.) For bringing the bone model of B represented by to the close proximity of the boundary of B in C2, however, landmarks must be selected on the surface of B. The approach, therefore, involves thresholding C1 and C2 to create isosurfaces, rendering these isosurfaces, selecting the corresponding landmarks on the surface of B in the two renditions, and subsequently approximately registering the two isosurfaces by using these landmarks. The main reason for taking the landmark-based approach for CT scenes is that, since the domains of C1s and C2s are much larger for CT scenes than for MRI scenes, the mutual information method would take a substantially longer time for registration.
In both approaches, the rigid transformation will be denoted, for future reference, by τ0. This transformation is derived from the process of registration applied to to produce the (approximately) registered binary scene . To make sure that is as close a representation of as possible, for the interpolation operation involved in converting into , we use a procedure similar to shape-basedinterpolation:36 A distance transform is first applied to to convert it into a gray scene with the convention that the distance from boundary is positive for 1-valued voxels in and negative for 0-valued voxels. This scene is then interpolated (tri-linearly) and the resulting gray scene is converted into binary scene by thresholding the interpolated scene at 0.
Figures 34 illustrate the operations underlying Step S3 for an MRI and a CT scene pertaining to the ankle complex and the cervical spine, respectively.
Figure 3.
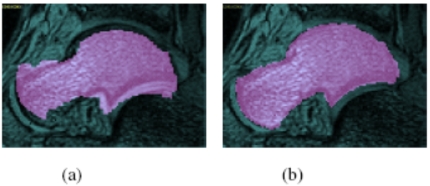
(a) One slice of an MRI scene C2 of an ankle in Position 2 with the corresponding slice of a segmented binary scene , obtained from C1 of the same ankle in Position 1 per Step S2, and overlaid on C2. (b) The same slice of C2 as in (a) with the corresponding slice of , obtained after registration as per Step S3, and overlaid on C2. The bone of interest B is the talus.
Figure 4.
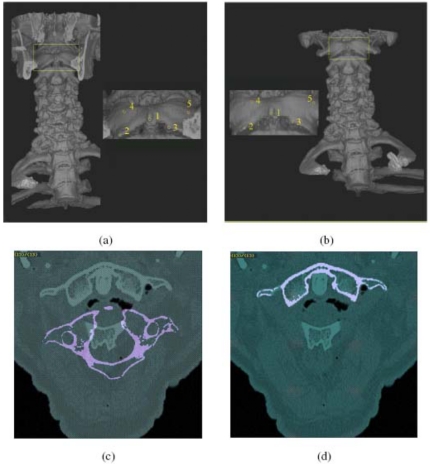
A 3D rendition of a cervical (and part of thoracic) spine in (a) Position 1 (neutral) and (b) Position 2 (45° of head-neck flexion) obtained from CT scenes C1 and C2. The insets show closeup views with landmarks indicated on the first cervical vertebra. (c) and (d) are identical to those in Figs. 3a, 3b but correspond to the CT example of (a) and (b).
S4: Matching the segmentation in Position 1 to the scene C2 in Position 2
While produced in Step S3 is usually in the close proximity of the boundary of B in scene C2, it by itself is usually not adequate as a segmentation of B in C2. The purpose of Step S4 is to use as a model of B and search in C2 for a position and orientation of this model that best matches the boundary and the intensity pattern in C1 in the vicinity of the model boundary with those of C2, and to subsequently obtain the final segmentation of B in C2. This is accomplished by using the following four substeps described in S4.1–S4.4 below.
S4.1: Describing the regional relative intensity pattern in the vicinity of the boundary of B in C1.
For any set S of voxels, we use the notation R(S) to denote the set of all points in R3 within (and on the boundary of) all cuboids represented by the voxels in S. For any voxel, c, a digital ball of radius ρ centered at c is the set of voxels,
| (1) |
where ‖d−c‖ denotes the distance between c and d. For any X⊂R3, we use the notation τ(X) to denote the set of points resulting from applying the same rigid transformation τ to each point in X. The regional relative intensity patternPτ,ρ,C(c) at a voxel c∊C in a scene C=(C,f) under a rigid transformation τ is a set of ordered pairs of points p and normalized intensities μτ,ρ,C(p)
| (2) |
where
| (3) |
|D| is the cardinality of D, and f′ is an appropriate interpolant of f. In our method, tri-linear interpolation is used to determine f′ from f. In other words, when τ is an identity transformation (I), D represents the digital ball of radius ρ centered at c that is within the domain of C, Pτ,ρ,C(c) represents the intensity pattern in C, within D, normalized by the mean intensity within D [the denominator in Eq. 3]. When τ is not an identity, D denotes the set of points within R(C) that represent a rigid transformation (by τ) of the centers of voxels within Bρ(c). The purpose of such a τ is to take a ball defined in C1 over to the domain of C2 for ascertaining the regional relative intensity pattern in C2, as will become clear in the next section.
The relative intensity pattern at a voxel within a segmented bone is intended to capture the local gestalts formed by the spatial distribution of intensities inside the bone and in other neighboring tissue regions such as muscles, ligaments, and cartilages. Since the relative intensity patterns are normalized by the respective mean values, they are not likely to be affected by background inhomogeneity. Thus, it is a fair assumption that, given an imaging protocol, a subject, and a bone, the variations among relative intensity pattern values are mostly due to noise.
S4.2: Formulating an energy function for the match.
The matching measure as an energy function defines a function of six independent variables corresponding to the 6 degrees of freedom of the rigid transformation. Let V0 denote the set of all voxels in with an intensity value of 1. The energy function is intended to express the total disagreement of regional relative intensity patterns between voxels in V0, computed from C1, and points in τ(V0) projected onto C2, computed from C2. Let V1={c∊V0|τ(c)∊R(C2)}. The energy function is then defined by
| (4) |
In the above equation, the situation corresponding to the voxels in V0 being mapped outside the domain of the second scene C2 (i.e., |V1|=0) will not happen because of the registration-based initialization in Step S3. Also, note that, p′ is a point in R(C2) obtained by transforming the point p by τ. The numerator represents the sum of squares of the difference in relative intensity pattern expressed in Eq. 3. The denominator is just a normalizing factor which represents the total number of voxels for which the difference in relative intensity pattern is computed. Following the fact that the energy function captures total disagreement, the optimization technique described in the next step seeks that transformation τ which minimizes this energy. The starting point for this search is τ0 which gave rise to .
S4.3: Optimization of the energy function.
For our application, the optimum of the energy function is assumed to correspond to the transformation that correctly matches the segmentation with the scene C2 at Position 2. Powell’s method (UOBYQA–unconstrained optimization by quadratic approximation)37 is used to find the minimum of the energy function and to get the corresponding rigid transformation parameters. Optimization is often performed in a multiresolution manner, as this is expected to decrease the sensitivity of the method to local minima. In our implementation, the multiresolution technique is utilized in the sense that the scenes are down sampled (repeatedly by a factor of 2) to three resolution levels; optimization is started in the lowest resolution, and the results are passed on to the higher level sequentially.
S4.4: Computation of the segmented bone for C2.
Once an optimum transformation τO is determined, is computed from by using the shape-based interpolation strategy described earlier in Step S3.
RESULTS AND EVALUATION
In this section, we demonstrate both qualitatively, through 2D and 3D image display, and quantitatively, through evaluation experiments, the extent of effectiveness of the new 3D model-based segmentation strategy. Two different application areas—motion analysis of the ankle complex and of the cervical spine by using MRI and CT, respectively—are considered. Image data in both these applications have been previously obtained independent of the research described in this article. Since the method currently available (that we are aware of) for the routine practical segmentation and separation of bones of joints is live wire, our method of evaluation, based on the framework suggested in Ref. 38, will focus on the analysis of precision, accuracy, and efficiency of the proposed method as compared to those of live wire. Since live wire is a slice-by-slice user-steered method, wherein the boundary delineation done by a piecewise optimal strategy is approved (and corrected if necessary) by the user, we will consider the segmentation performed by live wire to constitute a surrogate of true segmentation for assessing the accuracy of the new method.
Scene data
Ankle complex imaged via MRI
To investigate the robustness of the proposed method, two kinds of ankle image data are utilized—the first involving large rotations of the component bones and the second involving small rotations. For the large rotation case, the data analyzed in this study were acquired on a commercial 1.5T GE MRI machine, by using a coil specially designed for the study. During each acquisition, the foot of the subject was locked in a nonmagnetic device.39 This allows the control of the orientation and the motion of the foot by regular increments of 10° from neutral position (0° of pronation, 0° of supination) to the extreme positions of an open kinematic chain pronation to supination motion. The acquisition of one scene takes 5 min for each position; eight positions were acquired from 20° of pronation to 50° of supination. Each volume image is made up of sixty slices of 1.5 mm thickness. The imaging protocol used a 3D steady-state gradient echo sequence with a TR∕TE∕Flip angle=25 ms∕10 ms∕25°. Each slice is 256×256 pixels and the pixels are of size 0.55 mm×0.55 mm. The slice orientation was roughly sagittal. Ten pairs of scenes acquired for two extreme positions—20° of pronation and 50° of supination—corresponding to ten different subjects were used to test our method.
For the small rotation case, the following MR imaging protocol was used for scanning each foot. Each foot was held in an ankle loading device26 which allowed the ankle to be stressed in a controlled manner into different positions in nonstressed neutral and stressed anterior drawer and inversion positions. Each ankle was imaged in a 1.5 T GE MRI scanner by using a 3D fast gradient echo pulse sequence with TR∕TE∕flip angle=11.5 ms∕2.4 ms∕60°. The field of view was 18 cm with a 512×256 matrix size and a slice thickness of 2.1 mm. The slice orientation was roughly sagittal. Ten pairs of small rotation scene data acquired in neutral position and in stressed inversion, corresponding to ten different subjects, are used to test our method.
Cervical spine imaged via CT
The CT images analyzed in this article have been acquired for ten male unembalmed cadavers on a Siemens Volume Zooming Multislice CT scanner. For all acquisitions, the cervical spine underwent rotation at each of 0°, 10°, 20°, 30°, and 45° in flexion, and 10°, 20°, and 30° in extension. Images were obtained by using a slice thickness of 1.0 mm, a slice spacing of 1.5 mm, and pixel size of 0.23–0.35 mm.
Qualitative analysis
Several forms of 2D and 3D displays are presented in this section under the two application areas to demonstrate the quality of segmentation results. All results are obtained by using the 3DVIEWNIX software system.40
2D Display of the segmentation results
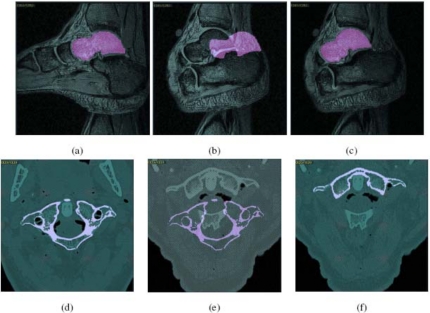
Figure 5a shows one slice of an input scene C1 pertaining to the foot of a normal subject and the same slice of obtained by live wire for the talus bone. Figure 5b shows this slice of of talus superimposed on the same numbered slice of C2. A large displacement of the talus in Position 2 can be readily seen although this displacement due to motion is in three dimensions. Figure 5c demonstrates this slice of C2 with the corresponding slice of obtained by using the model-based method. The segmentation agrees well as ascertained visually, with the underlying gray level image. An analogous example appears in Figs. 5d, 5e, 5f for the cervical spine application.
Figure 5.
(a) Talus in Position 1, segmented by using the live wire method, overlaid on the corresponding slice of an input scene C1. (b) Segmented talus from Position 1 overlaid on the same numbered slice of the scene C2 in Position 2. (c) Talus in Position 2, segmented by using the model-based method, overlaid on the corresponding slice of C2. (d)–(f) Similar to (a)–(c) but for a CT scene and for the first cervical vertebra.
3D Display examples
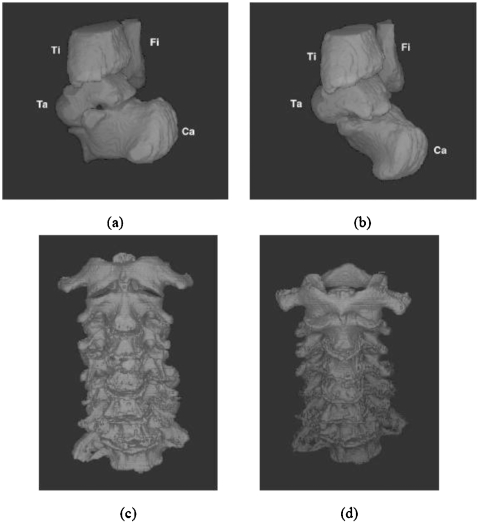
Figures 6a, 6b display 3D surface renditions of an ankle complex corresponding to two positions wherein the bones of interest are talus, calcaneus, tibia, and fibula. In Position 1 [(a)], the bones were segmented by using live wire, and in Position 2 [(b)], they were segmented by using the model-based method. Figures 6c, 6d similarly show 3D renditions of the cervical vertebrae in two positions, where vertebrae C1–C7 in Position 1 were segmented by manual masking and thresholding as described previously, and in Position 2 they were segmented by using the model-based method.
Figure 6.
(a) Three-dimensional display of talus, calcaneus, tibia, and fibula which were segmented by using live wire from a scene corresponding to Position 1. (b) The same bones segmented by using the model-based method from a scene corresponding to Position 2 of the same foot. (c), (d) Cervical vertebrae C1–C7 segmented from scene in Position 1 by manual interaction (c) and in Position 2 by model-based method (d).
3D Display of the segmentation results and principal axes of bones
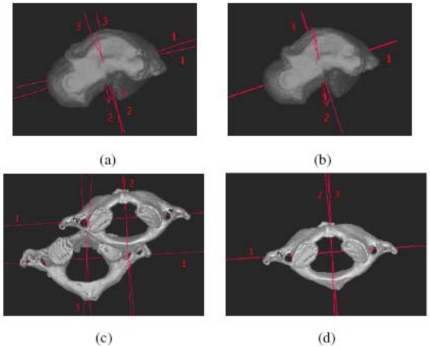
Figure 7 demonstrates the effectiveness of the proposed method by displaying the bones, segmented by using different methods, and their principal (inertial) axes.39 Figure 7a shows the talus surface segmented by using the live wire method with its principal axes in Position 1 overlaid on the same talus surface and also the principal axes of the surface segmented by using live wire in Position 2. The angle between the major axes shows the movement of the talus in the two positions. Figure 7b shows the talus surface segmented by using the model-based method with its principal axes in Position 2 overlaid on the principal axes of its surface obtained by using live wire in the same position. The angle between the major axes is now reduced to about 1°. Figures 7c, 7d show analogous results for the first cervical vertebra. The angle between the major axes and the translation between the centroids demonstrate a large displacement of the vertebra in the two positions in (c). This is reduced to about 1° in the angle between the major axes and to near zero displacement of the centroids in (d).
Figure 7.
(a) Talus surface segmented by using the live wire method with its principal axes in Position 1 overlaid on the talus surface in Position 2, both segmented by using the live wire method. (b) Talus surface segmented by using the model-based method with its principal axes for Position 2 and the principal axes for the surface in the same position obtained by using the live wire method. (c), (d) Similar to (a) and (b) but for the first cervical vertebra of a cadaveric body. Note that in (b) and (d) only the segmented surface is displayed.
Quantitative analysis
We use the framework described in Ref. 38 for evaluating the proposed segmentation method. In this framework, a method’s performance is assessed by three sets of measures—precision, accuracy, and efficiency. Precision here refers to the reproducibility of the segmentation results after taking into account all subjective actions that enter into the segmentation process. Accuracy relates to how well the segmentation results agree with the true delineation of the objects, and efficiency indicates the practical viability of the method, which is determined by the amount of time required for performing computations and providing any user help needed in segmentation. The measures that are used under each of these groups and their definitions are given under the following sections that describe each of these factors. The objects of interest for our two applications are: Talus, calcaneus, tibia, fibula, navicular, cuboid in foot MRI, and vertebrae C1–C7 in cervical spine CT. Since similar performance has been observed for all objects, we present here the measure for two representative objects in each area—talus and calcaneus in the foot MRI and C1 and C3 in the spine CT.
Precision
For segmentation in the foot application, precision is affected mainly by the way an operator selects the VOI (Step S2). To evaluate this effect, one operator repeated the segmentation experiment two times with 20 pairs of MR scenes, considering ten pairs each of small and large rotation scenes. Here, “small rotation” means the translation and rotation between the bones in the two different positions are low. For example, the data sets with the angle between the major principal axes of a bone in the two positions of the order of 5° or less will be considered as small rotation scenes. For large rotation scenes, the angle between the major axes of the bone is greater than 20°. Let V1 and V2 be the set of voxels constituting the segmentation of the same object region in two repeated trials. For precision, we use two measures. The first measure is (|V1∩V2|∕|V1∪V2|)×100 to estimate the overlap agreement, where |X| denotes the cardinality of set X. The second measure is simply the similarity of the volume enclosed by the surface of the bones, defined by (1−(‖V1|−|V2|∕|V1∪V2‖))×100. The reason for considering this second measure is as follows. Even when the actual mismatch between segmentations obtained in two repeated trials is small, the extent of overlap can be large because of interpolation and digitization effects, especially coming from the slices at the ends of the scene. This effect is minimized if we consider similarity of volumes, although just the similarity of volumes alone does not indicate repeatability of segmentation. However, given acceptable overlap, volume similarity indicates repeatability.
Table 1 lists these measures for the proposed method for segmenting the talus and calcaneus. Mean and standard deviation over the scene population are listed. For segmentation of spine in CT scenes, precision is affected mainly by the way an operator selects the landmarks (Step S3). As above, one operator repeated the segmentation experiment two times to evaluate this effect with ten pairs of CT scenes. Table 1 lists the precision of the method for segmenting vertebrae C1 and C3. Both measures are very high for the talus and calcaneus indicating the high level of precision of the method. The overlap measures are lower for the vertebrae due to their complex shape and substantially higher end-slice effect than in the foot bones. However, the volume similarity is high for the vertebrae; therefore, we may conclude that the method achieves high precision for the second application also. This is also confirmed by our qualitative examination of all results on slice display which consistently indicate excellent agreement with the scene.
Table 1.
The mean and standard deviation of precision measures estimated from 20 scenes for two bones of the foot, and for two vertebrae of the spine.
| Small rotation | Large rotation | Vertebra C1 | Vertebra C3 | |||
|---|---|---|---|---|---|---|
| Talus | Calcaneus | Talus | Calcaneus | |||
| Overlap | 99.56±0.25 | 99.64±0.21 | 98.70±0.43 | 98.79±0.42 | 85.52±1.25 | 86.64±1.21 |
| Volume | 96.50±0.22 | 99.55±0.14 | 96.34±0.23 | 99.40±0.21 | 96.42±0.31 | 96.76±0.24 |
Accuracy
Of the three factors used to describe the effectiveness of a segmentation method, accuracy is the most difficult to assess. This is due mainly to the difficulty in establishing the true delineation of the object of interest in the scene. Consequently, an appropriate surrogate of truth is needed in place of true delineation.
For the foot MR images, we have taken the segmentation resulting from the live wire method in Position 2 as this surrogate of truth. We utilized the same data sets as those employed in the assessment of precision for both applications. In both applications, as mentioned previously, all data sets have been previously segmented by experts in the domain by using live wire in the foot MRI application and via the interactive method described under Step S1 in the spine application. These segmentations for the second scene in each pair were used as true segmentations and those for the first scene were used to provide the model. For any scene, let be the segmentation result (binary scene) output by our method for which the true delineation result is . Let C=(C,f) be any given scene. The following measures, called true-positive volume fraction (TPVF) and false-positive volume fraction (FPVF) are used to assess the accuracy of the proposed method. Here |X| denotes the cardinality of set X and the operations on binary scenes have the obvious interpretations akin to those on sets. TPVF indicates the fraction of the total amount of tissue in the true delineation. FPVF denotes the amount of tissue falsely identified. Cd is a binary scene representation of a reference superset of voxels that is used to express the two measures as a fraction. In our case, we took the entire scene domain to represent this superset. We may define FNVF and TNVF in an analogous manner.38 Since these measures are derivable from Eqs. 5, 6, TPVF and FPVF are sufficient to describe the accuracy of the method (see Ref. 38 for details).
| (5) |
| (6) |
Table 2 lists the mean and standard deviation values of FPVF and TPVF achieved for the two bones in the two application areas by the model-based method over the two sets of data. As alternative measures, the table also lists the mean and standard deviation of the distance between the centroids and the angle between the major principal axes of the bones depicted in and .
Table 2.
Mean and standard deviation of FPVF, TPVF, distance between geometric centroids, and the angle between the major principal axes for talus, calcaneus, C1 and C3 achieved by the model-based method.
| Measure | Small rotation | Large rotation | Vertebra C1 | Vertebra C3 | ||
|---|---|---|---|---|---|---|
| Talus | Calcaneus | Talus | Calcaneus | |||
| FPVF | 0.51±0.05 | 0.20±0.04 | 0.67±0.02 | 0.20±0.06 | 0.23±0.02 | 0.33±0.04 |
| TPVF | 96.97±1.04 | 97.00±1.10 | 96.40±1.20 | 96.78±1.22 | 88.62±1.33 | 89.69±1.53 |
| Distance (mm) | 0.30±0.02 | 0.29±0.04 | 0.34±0.03 | 0.31±0.03 | 0.19±0.01 | 0.17±0.01 |
| Angle (degree) | 1.05±0.45 | 1.02±0.40 | 1.27±0.20 | 1.20±0.29 | 1.01±0.20 | 1.03±0.22 |
We note that in the expression of FPVF and TPVF, there is a phenomenon similar to the one alluded to under the description of precision which arises from end slices. Its effect can be seen for the more complex shaped vertebrae in terms of their lower TPVF than those for the talus and calcaneus, although the segmentations appear to be visually highly acceptable. This is why we added the measures of distance and angle listed in Table 2. The proposed method produces highly accurate segmentations overall.
Efficiency
The method is implemented in C language within the 3DVIEWNIX software system and is executed on an Intel Pentium IV PC with a 1.7 GHZ CPU under the Red Hat Linux OS version 7. In determining the efficiency of a segmentation method, two aspects should be considered—the computation time (Tc) and the human operator time (To). The mean Tc and To per data set estimated over ten data sets for each application area for each bone are listed in Table 3. To measured here does not include the operator time required in segmenting the bones in Position 1. In the first application area, To includes the specification of VOI and the subsequent verification to ensure that the VOI is adequate over all slices. In the second application area, To involves the time taken to specify the landmarks for registration. We note that the operator time required in both applications is not unreasonable (being 1–2 min), and neither is the computational time (6–7 min).
Table 3.
Mean operator time To and computational time Tc (in minutes) for the talus, calcaneus, C1, and C3 in the two application areas.
| Time | Talus | Calcaneus | C1 | C3 |
|---|---|---|---|---|
| To | 1 | 1 | 2 | 2 |
| Tc | 6 | 6 | 7 | 7 |
CONCLUDING REMARKS
Computer assisted biomechanical analysis, biomechanical modeling, surgery planning, and image-guided surgery of joints require the segmentation and separation of bones at the joints. This article offers a practical solution via a rigid model-based strategy that can be readily used. The bones segmented in the scene corresponding to the first position are used as a model to seek, under rigid transformations, a segmentation of the same bones in scenes corresponding to subsequent positions of the joint. The only parameter in the proposed method is the size ρ of the ball that is used in defining regional relative intensity pattern. After some initial experimentation, we have used ρ=5 in all our experiments. Our evaluation suggests that, for bones of the foot in MR images, the method achieves a segmentation [TPVF,FPVF] of about [97%, 0.4%] and [translation, rotation] of [0.3 mm, 1.1°] with the live wire method used as a reference. For the cervical vertebrae in CT images, these segmentation accuracy measures are [89%, 0.3%] and [0.18 mm, 1.02°]. These errors are well within the extent of displacement and rotation that we seek to measure by using such image-based methods.
Since this is a case of intra subject and intra modality registration, one may surmise if Step S3 with mutual information, cross correlation, or sum of squared differences as a criterion would solve the problem and if Step S4 is really needed. When both the rotation and translation of bone B are small from Position 1 to Position 2, this strategy produces acceptable results, although S4 always improves the result. However, when rotation or∕and translation is∕are large, S3 along (together with S2) produces unacceptable results. Step S3 in this manner can be thought of as solving the object recognition problem in segmentation. S4 then completes the finer object delineation step aided by recognition. Since it is difficult to predict the nature of bone movement for the particular situation at hand, inclusion of Step S4 is always to be preferred.
One may also surmise if an atlas based registration methodology would form a solution to the entire problem. This is certainly feasible and may constitute a powerful and very general solution. The solution methodology, however, is likely to be more complicated. A proper atlas will have to be constructed first. Because of (Ch5) and large rotations and∕or translations, a single atlas, corresponding to, say, the neutral joint position, may not be adequate. Or it may have to be a four-dimensional atlas. Our proposed solution is certainly a first cut simple approach to address this challenging segmentation∕registration problem.
A problem that was not addressed in this paper is the segmentation of the bones in the images corresponding to the first position with a degree of automation higher than that afforded by live wire or iterative live wire. We are currently investigating a family of methods based on live wire, active shape, and appearance models, and their combined hybrid strategies for this purpose. The idea in these strategies is to build methods of high degree of automation that are tightly integrated into the regimen of user steering in the spirit of live wire, so that, they take just as much help as is needed by the operator without requiring post hoc correction. Another issue that needs attention in the future is isoshaping.41 Long bones, such as the tibia and the fibula, which are covered to different extents in the images in the two positions owing to a limited field of view, do not really posses a boundary in the vicinity of their shafts where they are cut off by the limited field of view. The ideas of “shape centers” utilized in isoshaping of these bones (a process of trimming the shafts of these bones automatically so that the bone assumes the same shape in segmentations coming from all positions) can also be employed to recognize such aspects of the bone and to handle these in a manner different from the manner in which the real aspects of the boundary are handled.
ACKNOWLEDGMENTS
The research reported here is supported by DHHS Grant Nos. AR46902, NS37172, and EB004395. The authors are grateful to Dr. Stacie Ringleb, Dr. Carl Imhouser, and the Catharine Sharpe Foundation for help with the acquiring of the MR and CT image data. The authors would also like to thank Synthes Spine for partly funding the project related to motion analysis of the spine.
References
- Hirsch B. E., Udupa J. K., and Stindel E., Tarsal joint kinematics via 3D imaging, in 3D Imaging in Medicine, 2nd ed. (CRC, Boca Raton, FL, 2000), Chap. 9. [PubMed] [Google Scholar]
- Udupa J. K., Hirsch B. E., Samarasekera S., and Goncalves R., “Joint kinematics via 3-D MR imaging,” in Visualization in Biomedical Computing, Proc. SPIE 10.1117/12.131118 1808, 664–670 (1992). [DOI] [Google Scholar]
- Sint Jan S. V., Giurintano D. J., Thompson D. E., and Rooze M., “Joint kinematics simulation from medical imaging data,” IEEE Trans. Biomed. Eng. 10.1109/10.649989 44, 1175–1184 (1997). [DOI] [PubMed] [Google Scholar]
- Fiepel R. M., “Three-dimensional motion patterns of the carpal bones: An in-vivo study using three-dimensional computed tomography and clinical applications,” Surg. Radiol. Anat. 21(2), 165–170 (1999). [DOI] [PubMed] [Google Scholar]
- You B. M., Siy P., Anderst W., and Tashman S., “In vivo measurement of 3-D skeletal kinematics from sequences of biplane radiographs: Application to knee kinematics,” IEEE Trans. Med. Imaging 10.1109/42.929617 20(6), 514–525 (2001). [DOI] [PubMed] [Google Scholar]
- Hoad C. L. and Martel A. L., “Segmentation of MR images for computer assisted surgery of the lumbar spine,” Phys. Med. Biol. 10.1088/0031-9155/47/19/305 47, 3503–3517 (2002). [DOI] [PubMed] [Google Scholar]
- Fuente M. D., Ohnosorge J. A., Schkommodau E., Jetzki S., Wirtz D. C., and Radermacher K., “Fluoroscopy-based 3-D reconstruction of femoral bone cement: A new approach for revision total hip replacement,” IEEE Trans. Biomed. Eng. 10.1109/TBME.2005.844032 52(4), 664–675 (2005). [DOI] [PubMed] [Google Scholar]
- Chen J. X., Wechsler H., Mark Pullen J., Zhu Y., and Macmahon E. B., “Knee surgery assistance: Patient model construction, motion simulation, and biomechanical visualization,” IEEE Trans. Biomed. Eng. 10.1109/10.942595 48(9), 1042–1052 (2001). [DOI] [PubMed] [Google Scholar]
- Pal N. and Pal S., “A review on image segmentation techniques,” Pattern Recogn. 10.1016/0031-3203(93)90135-J 26, 1277–1294 (1993). [DOI] [Google Scholar]
- Kass M., Witkin A., and Terzopoulos D., “Snakes: Active contour models,” Int. J. Comput. Vis. 10.1007/BF00133570 1, 321–331 (1987). [DOI] [Google Scholar]
- Sethian J., Level Set Methods and Fast Marching Methods (Cambridge University Press, Cambridge, 1996). [Google Scholar]
- Vincent L. and Soille O., “Watersheds in digital spaces: An efficient algorithm based on immersion simulations,” IEEE Trans. Pattern Anal. Mach. Intell. 10.1109/34.87344 13(6), 583–598 (1992). [DOI] [Google Scholar]
- Boykov Y., Veksler O., and Zabih R., “Fast Approximate Energy Minimization via Graph Cuts,” International Conference on Computer Vision, Kerkyra, Corfu, Greece, pp. 377–384 (1999). [Google Scholar]
- Diday E. and Simon J. C., “Clustering analysis,” in Digital Pattern Recognition (Springer-Verlag, Berlin, 1980), pp. 47–94. [Google Scholar]
- Udupa J. K. and Samarasekera S., “Fuzzy connectedness and object definition: Theory, algorithms, and applications in image segmentation,” Graph. Models Image Process. 58(3), 246–261 (1996). [Google Scholar]
- Dogdas B., Shattuck D. W., and Leahy R. M., “Segmentation of the skull in 3D human MR images using mathematical morphology,” Proc. SPIE 4684, 1553–1562 (2002). [Google Scholar]
- Bomans M., Hoehne K. H., and Riemer M., “3D segmentation of MR images of the head for 3D display,” IEEE Trans. Med. Imaging 10.1109/42.56342 9(2), 177–183 (1990). [DOI] [PubMed] [Google Scholar]
- Heinonen T., Eskola H., Dastidar P., Laarne P., and Malmivuo J., “Segmentation of T1 MR scans for reconstruction of resistive head models,” Comput. Methods Programs Biomed. 54, 173–181 (1997). [DOI] [PubMed] [Google Scholar]
- Lorigo L., Faugeras O., Grimson E., and Keriven R., “Segmentation of bone in clinical knee MRI using texture-based geodesic active contours,” Medical Image Computation and Computer Assisted Interventions, Boston, October, 1998.
- Sebastian T. B., Tek H., Crisco J. J., and Kimia B. B., “Segmentation of carpal bones from CT images using skeletally coupled deformable models,” Med. Image Anal. 70(1), 21–45 (2003). [DOI] [PubMed] [Google Scholar]
- Reyes-Aldasoro C. C. and Bhalerao A., “Sub-band filtering for MR Texture Segmentation,” Medical Image Understanding and Analysis, Portsmouth, U.K., July, 2002, pp. 22–23.
- Grau V., Mewes A. U. M., Alcaniz M., Kikinis R., and Warfield S. K., “Improved watershed transform for medical image segmentation using prior information,” IEEE Trans. Med. Imaging 10.1109/TMI.2004.824224 23(4), 447–458 (2004). [DOI] [PubMed] [Google Scholar]
- Falcao A. X., Udupa J. K., Samarasekera S., and Sharma S., “User-steered image segmentation paradigms: Live wire and live lane,” Graph. Models Image Process. 60, 233–260 (1998). [Google Scholar]
- Holden M., Schnabel J. A., and Hill D. L., “Quantification of small cerebral ventricular volume changes in treated growth hormone patients using non-rigid registration,” IEEE Trans. Med. Imaging 10.1109/TMI.2002.806281 21(10), 1292–1301 (2002). [DOI] [PubMed] [Google Scholar]
- Calmon G. and Roberts N., “Automatic measurement of changes in brain volume on consecutive 3D MR images by segmentation propagation,” Magn. Reson. Imaging 18(4), 439–453 (2000). [DOI] [PubMed] [Google Scholar]
- Siegler S., Udupa J. K., Ringleb S. I., Imhauser C. W., Hirsch B. E., Odhner D., Saha P. K., Okereke E., and Roach N., “Mechanics of the ankle and subtalar joints revealed through a three dimensional stress MRI technique,” J. Biomech. 10.1016/j.jbiomech.2004.03.036 38(3), 567–578 (2005). [DOI] [PubMed] [Google Scholar]
- Rhoad R. C., Klimkiewicz J. J., Williams G. R., Kesmodel S. B., Udupa J. K., Kneeland J. B., and Iannotti J. P., “A new in vivo technique for 3D shoulder kinematics analysis,” Skeletal Radiol. 27, 92–97 (1998). [DOI] [PubMed] [Google Scholar]
- Simon S., Davis M., Odhner D., Udupa J., and Winkelstein B., “CT imaging techniques for describing motions of the cervicothoracic junction and cervical spine during flexion, extension, and cervical traction,” Spine (in press). [DOI] [PubMed]
- Cootes T. F., Taylor C. J., Cooper D. H., and Graham J., “Active shape models their training and application,” Comput. Vis. Image Underst. 10.1006/cviu.1995.1004 61, 38–59 (1995). [DOI] [Google Scholar]
- Cootes T. F., Edwards G. J., and Taylor C. J., “Active appearance models,” IEEE Trans. Pattern Anal. Mach. Intell. 10.1109/34.927467 23(6), 681–685 (2001). [DOI] [Google Scholar]
- Liu J., Udupa J. K., Saha P. K., Odhner D., Hirsch B. E., Siegler S., Simon S., and Winkelstein B. A., “Model-based 3D segmentation of the bones of joints in medical images,” Proc. SPIE (in press, 2005). [DOI] [PMC free article] [PubMed]
- Pluim J. P. W., Antoine Maintz J. B., and Viergever M. A., “Mutual information based registration of medical images: A survey,” IEEE Trans. Med. Imaging 10.1109/TMI.2003.815867 22(8), 986–1004 (2003). [DOI] [PubMed] [Google Scholar]
- Toennies K. D., Udupa J. K., Herman G. T., Wornom I. L., and Buchman S. R., “Registration of 3D objects and surfaces,” IEEE Comput. Graphics Appl. 10.1109/38.55153 10(3), 52–62 (1990). [DOI] [Google Scholar]
- Udupa J. K., “Multidimensional digital boundaries,” Graph. Models Image Process. 56(4), 311–323 (1994). [Google Scholar]
- Grevera G. J., Udupa J. K., and Odhner D., “An order of magnitude faster surface rendering in software on a PC than using dedicated rendering hardware,” IEEE Trans. Vis. Comput. Graph. 10.1109/2945.895878 6(4), 335–345 (2000). [DOI] [Google Scholar]
- Raya S. P. and Udupa J. K., “Shape-based interpolation of multidimensional objects,” IEEE Trans. Med. Imaging 10.1109/42.52980 9, 32–42 (1990). [DOI] [PubMed] [Google Scholar]
- Powell M. J. D., UOBYQA: Unconstrained optimization by quadratic approximation, Math. Program, DOI 10.1007/s101070100290.
- Udupa J. K., LeBlanc V. R., Zhuge Y., Imielinska C., Schmidt H., Hirsch B. E., and Woodburn J., “A framework for evaluating image segmentation algorithms,” Comput. Med. Imaging Graph. 10.1016/j.compmedimag.2005.12.001 30(2), 75–87 (2006). [DOI] [PubMed] [Google Scholar]
- Udupa J. K., Hirsch B. E., Hillstrom H. J., Bauer G. R., and Kneeland J. B., “Analysis of in vivo 3-D internal kinematics of the joints of the foot,” IEEE Trans. Biomed. Eng. 10.1109/10.725335 45, 1387–1396 (1998). [DOI] [PubMed] [Google Scholar]
- Udupa J. K., Odhner D., Samarasekera S., Goncalves R., Iyer K., Venugopal K., and Furuie S., “3DVIEWNIX: An open, transportable, multidimensional, multimodality, multiparametric imaging software system,” Proc. SPIE 10.1117/12.174042 2164, 58–73 (1994). [DOI] [Google Scholar]
- Saha P. K. and Udupa J. K., “Isoshaping rigid bodies for motion analysis,” Proc. SPIE 4684, 343–352 (2002). [Google Scholar]
- Souza A. and Udupa J. K., “Iterative live wire and live snake: New user-steered 3D image segmentation paradigms,” Proc. SPIE 6144, 1159–1165 (2006). [Google Scholar]