Abstract
Purpose
Studies comparing two or more vaccine platforms have historically evaluated each platform based on its ability to induce an immune response and may conclude that one vaccine is more efficacious than the other(s), leading to a recommendation for development of the more effective vaccine for clinical studies. Alternatively, these studies have documented the advantages of a diversified prime and boost regimen due to amplification of the antigen-specific T-cell population. We hypothesize here that two vaccine platforms targeting the same antigen might induce shared and distinct antigen-specific T-cell populations, and examined the possibility that two distinct vaccines could be used concomitantly.
Experimental design
Using recombinant poxvirus and yeast vaccines, we compared the T-cell populations induced by these two platforms in terms of serum cytokine response, T-cell gene expression, T-cell receptor phenotype, antigen-specific cytokine expression, T-cell avidity, and T-cell antigen-specific tumor cell lysis.
Results
These studies demonstrate for the first time that vaccination with a recombinant poxvirus platform (rV/F-CEA/TRICOM) or a heat-killed yeast vaccine platform (yeast-CEA) elicits T-cell populations with both shared and unique phenotypic and functional characteristics. Furthermore, both the antigen and the vector play a role in the induction of distinct T-cell populations.
Conclusions
In this study, we demonstrate that concurrent administration of two vaccines targeting the same antigen induces a more diverse T-cell population that leads to enhanced antitumor efficacy. These studies provide the rationale for future clinical studies investigating concurrent administration of vaccine platforms targeting a single antigen to enhance the antigen-specific immune response.
Keywords: Vaccinia, Saccharomyces cerevisiae, Carcinoembryonic antigen (CEA), T-cell populations, Antitumor immunity
Introduction
Numerous immunotherapy studies have been reported comparing different vaccine platforms that target the same antigen, in terms of their ability to induce immune cell activity and antitumor effects [1–9]. Such studies either choose the more efficacious vaccine for further study, or employ a diversified prime and boost strategy to amplify the T-cell response. We have previously reported the immune responses and antitumor efficacy of the diversified prime and boost vaccine regimen of recombinant vaccinia (rV) and recombinant fowlpox (rF) viruses containing murine B7-1, ICAM-1, and LFA-3 transgenes as well as the human carcinoembryonic antigen (CEA) transgene (rV/F-CEA/TRICOM) in preclinical models [6, 10–14]. Recently, we reported the immune responses and antitumor effects of a recombinant Saccharomyces cerevisiae (yeast-CEA) vaccine in preclinical models [15, 16]. The antitumor effects elicited by either vaccine are mainly attributed to the induction of a CEA-specific T-cell population. Here, we describe the first study to our knowledge to concurrently administer two different vaccine platforms targeting the same antigen in an antitumor model. The recombinant vaccinia and fowlpox vectors infect APCs and express transgenes via poxvirus promoters. The yeast vaccine is developed by transfection of Saccharomyces cerevisiae with a recombinant yeast plasmid, which then expresses the recombinant protein (e.g. CEA) internally. The heat-killed yeast when employed as a vaccine is then internalized by DCs at which time the protein is released in the APC and processed. Thus the CEA antigen delivered by either vaccine may be processed and presented differently by APCs, possibly leading to the induction of distinct T-cell populations. Previous in vitro studies have also shown that an array of cytokines is released by DCs upon exposure to the yeast vaccine [15]. We hypothesized that due to both vaccine platform- and antigen-specific effects, the rV/F-CEA/TRICOM and yeast-CEA vaccines would induce distinct T-cell populations, and that concurrent administration of the two vaccines would thus result in a more diverse T-cell population.
This study demonstrates for the first time that both the vaccine platform and the antigen can have a role in the induction of T-cell populations with both shared and unique cytokine responses, gene expression profiles, and T-cell receptor phenotypes. Furthermore, T-cell lines developed from vaccinated CEA-transgenic (CEA-Tg) mice have different avidity and cytolytic activity in vitro. These studies indicate that phenotypically and functionally distinct T-cell populations are induced by two distinct vaccine platforms targeting the same antigen. Because our study found that rV/F-CEA/TRICOM and yeast-CEA induced distinct T-cell populations, we hypothesized that concurrent administration of the vaccines may induce a more diverse T-cell population consisting of T cells generated from both vaccines. Finally, our study showed that the two vaccines may be combinatorially administered to improve antitumor efficacy. Our study differs from the typical diversified prime–boost studies in the literature. Here, we use two distinct vaccine platforms that when concurrently administered do not inhibit one another, and that induce a more diverse T-cell population upfront that is then boosted and expanded in magnitude with each subsequent vaccination. The concurrent administration of two vaccines targeting a single antigen could be advantageous for the treatment of cancer patients, in which a potent immune response may be created at the earliest stages of treatment. These results provide a rationale for the concurrent administration of two distinct vaccine platforms targeting a single antigen for the induction of a more diverse T-cell population directed against a tumor-associated antigen for the treatment of cancer.
Materials and methods
Mice and tumor cell lines
For in vitro stimulation of lymphocytes, female C57BL/6 (H-2b) mice were obtained from the National Cancer Institute, Frederick Cancer Research and Development Facility (Frederick, MD). A breeding pair of C57BL/6 mice homozygous for expression of the human CEA gene (CEA-Tg) was generously provided by Dr. John Shively (City of Hope, Duarte, CA). Homozygosity for CEA expression was confirmed by PCR analysis of mouse-tail DNA [13]. Six- to eight-week-old female mice were used for all experiments, and were housed in micro-isolator cages under pathogen-free conditions in accordance with AAALAC guidelines. Experimental studies were carried out under approval of the NIH Intramural Animal Care and Use Committee. The target tumor cell line EL-4 (H-2b, thymoma) was obtained from American Type Culture Collection (Manassas, VA). LL/2 murine lung adenocarcinoma tumor cells were gifted by Dr. Chandan Guha (Albert Einstein College of Medicine, New York, NY). LL/2 murine lung carcinoma cells expressing human CEA (LL2-CEA) were generated by retroviral transduction with CEA cDNA, as previously described [17]. Cells were maintained in complete medium (RPMI or DMEM supplemented with 10% fetal bovine serum, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin).
Vaccine constructs
Recombinant vaccinia (rV) and recombinant fowlpox (rF) viruses containing murine B7-1, ICAM-1, and LFA-3 genes as well as the human CEA gene (rV/F-CEA/TRICOM) have been previously described [10, 12]. The murine GM-CSF-expressing rF virus (rF-GM-CSF) has been previously described [18]. A recombinant S. cerevisiae construct expressing the full-length CEA protein (yeast-CEA) has been previously described [15]. Yeast-CEA was produced and heat-killed for these studies as previously described [19].
Vaccination schedules
For serum cytokine analysis, CEA-Tg mice (n = 2) were vaccinated with 1 × 108 pfu rV-CEA/TRICOM or 4 YU/animal (1 YU = 107 yeast particles) of yeast-CEA as previously described [16]. In all other studies for the rV/F-CEA/TRICOM vaccine group, CEA-Tg mice were primed with 1 × 108 pfu rV-CEA/TRICOM admixed with 1 × 107 pfu rF-GM-CSF on day 0, and boosted every 7 days with 1 × 108 pfu rF-CEA/TRICOM admixed with 1 × 107 pfu rF-GM-CSF. For the rest of this manuscript, this vaccine will be designated rV/F-CEA/TRICOM. In the yeast-CEA vaccine group, CEA-Tg mice were vaccinated every 7 days with yeast-CEA (4 YU/mouse). Mice receiving the combination of rV/F-CEA/TRICOM and yeast-CEA vaccines were primed with 1 × 108 pfu rV-CEA/TRICOM administered subcutaneously on the dorsal right flank, and with 4 YU/mouse of yeast-CEA delivered subcutaneously on the inner legs and shoulder blades. The separation of the yeast-CEA dose over multiple sites has previously been described [16], and has been employed here to separate not only the yeast-CEA vaccine, but also the rV/F-CEA/TRICOM to target multiple draining lymph nodes in the mouse. Mice in the combination group were boosted at 1-week intervals for the remainder of the study with 1 × 108 pfu rF-CEA/TRICOM and yeast-CEA (4 YU/mouse).
Cytokine expression profiles
For serum cytokine analysis, vaccinated mice (see vaccination schedule above) were bled on days 0, 2, and 4 post-vaccination and serum was isolated. Cytokine expression was analyzed using a Th1/Th2 and proinflammatory cytokine panel by Linco Diagnostic Services (St. Charles, MO). To measure cytokines secreted by CD8+ T cells from mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA (n = 5), CD8+ T cells were bulk cultured and restimulated in the presence of CEA-572-579 peptide (GIQNSVSA, designated CEA-572) or CEA-526-533 peptide (EAQNTTYL, designated CEA-526) (10 μg/ml) as previously described [16]. Cytokine levels were measured using the mouse Inflammatory Cytokine Cytometric Bead Array Kit and the mouse Th1/Th2 Cytokine Cytometric Bead Array kit (BD Biosciences, San Jose, CA) according to the manufacturer’s instructions.
T-cell receptor (TCR) profiles
RNA was isolated using the RNeasy Mini Kit (Qiagen, Inc., Valencia, CA) according to the manufacturer’s instructions. RNA was then used in RT-PCR reactions using the Invitrogen SuperScript® First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. Vα and Vβ genes were amplified using primers and conditions previously described for 19 Vα and 24 Vβ genes [20, 21]. PCR products were analyzed using the Agilent 2100 Bioanalyzer and Agilent DNA 1000 Reagent Kit (Agilent Technologies, Santa Clara, CA) by on-chip electrophoresis according to the manufacturer’s instructions. Agilent 2100 Expert Software (version B.02.06SI418 [Patch 01]) was used to identify PCR products by size (bp) and quantity (nmol/L). For each sample, quantities of each gene present were summed, and for each gene, a percent of the total TCR Vα or Vβ repertoire was calculated.
A mouse Vβ TCR screening panel (BD Pharmingen, San Jose, CA) consisting of monoclonal antibodies specific for mouse TCR Vβ 2, 3, 4, 5.1 and 5.2, 6, 7, 8.1 and 8.2, 8.3, 9, 10, 11, 12, 13, 14, and 17 was used to identify TCR Vβ expression at the protein level by flow cytometry using a FACScan cytometer (Becton Dickinson).
cDNA oligoarray
CEA-Tg mice were either untreated or vaccinated 3 times at 1-week intervals with rV/F-CEA/TRICOM or yeast-CEA. On day 33, splenocytes were harvested and RNA was isolated. T- and B-cell activation, chemokines and chemokine receptors, and common cytokines cDNA oligoarrays (SABiosciences, Frederick, MD) were used to investigate changes in gene expression. Genes were considered up-regulated or down-regulated if their normalized intensity ratio was ≥2 or ≤0.5 (a 2-fold cutoff), respectively, according to manufacturer’s recommendations.
CEA-specific CTL cell lines and in vitro assays
CEA-526-specific and CEA-572-specific T-cell lines generated from mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA were maintained in culture with CEA-526 or CEA-572 peptide (1 μg/ml) and IL-2 (10 U/ml) with fresh irradiated APCs. To measure the ability of the T-cell lines to lyse 111In-labeled targets, various ratios of T cells were incubated with labeled targets in triplicate at 37°C and 5% CO2 in 96-well U-bottom plates. In certain studies, anti-MHC class I blocking antibody (H2Db, BD Pharmingen) was used to distinguish between TCR-mediated and NK-like cytotoxicities. Radioactivity in supernatants was measured using a gamma-counter (Corba Autogamma, Packard Instruments, Downers Grove, IL). Percentage of tumor lysis was calculated as follows: % tumor lysis = [(experimental cpm – spontaneous cpm)/(maximum cpm – spontaneous cpm)] × 100. To evaluate the avidity of CEA-specific CTL lines, tumor-killing activity was tested as previously described [22]. Data were averaged and graphed as ∆% specific lysis. To normalize groups within each experiment, data were also expressed as percentage of maximum lysis versus peptide concentration. Finally, the natural logarithm of the normalized data was plotted against peptide concentration. The avidity of each T-cell population was defined as the negative log of the peptide concentration that resulted in 50% maximal target lysis [22, 23] and was expressed in nM. The HIV-gag-390-398 peptide (SQVTNPANI, designated HIV-gag peptide) was used as a negative control in this experiment. MHC class I-peptide tetramers specific for CEA-526 and CEA-572 were obtained from Beckman Coulter (Fullerton, CA). Where indicated, CTL activity was converted to lytic units (LU), as described by Wunderlich et al. [24].
Tumor therapy studies
For therapy studies involving LL2-CEA tumors, 6- to 8-week-old female CEA-Tg mice were injected i.v. in the tail with 3 × 105 LL2-CEA cells in a volume of 100 μl. Four days post-tumor implantation, mice were primed and then boosted as described above. To enumerate lung metastases, lungs from the mice killed were inflated, stained with India ink, and fixed in Fekete’s solution [25].
Statistical analysis
GraphPad Prism version 4.0a for Macintosh (GraphPad Software, San Diego, CA) was used to perform statistical analyses on in vivo data. A 2-tailed, nonparametric Mann–Whitney test was performed for the average number of tumors per mouse at day 45. A log-rank (Mantel-Cox) test was performed for mice bearing >9 pulmonary tumor nodules at day 45 which were deemed to have ≤1 week to live. All values were calculated at a 95% confidence interval and a p value ≤0.05 was considered significant.
Results
Vaccine induction of cytokines and chemokines
We first investigated the role of the vaccine in inducing cytokine and chemokine host innate immune responses in vivo that may subsequently influence CEA-specific T-cell responses. CEA-Tg mice were vaccinated with rV-CEA/TRICOM or yeast-CEA. Serum was collected and analyzed for a panel of cytokines using a Th1/Th2 and pro-inflammatory cytokine panel. As shown in Fig. 1, rV-CEA/TRICOM (closed squares) induces a predominantly Th1-type cytokine profile, where MIP1α, RANTES, GM-CSF, and IL-12p70 levels are high and IL-5 levels are low (Fig. 1a, b, c, and i, respectively). In contrast, yeast-CEA vaccination (open circles) induces a mixed Th1/Th2 cytokine profile with increased levels of IL-6, IL-1α, and IL1β (Fig. 1d, g, and h, respectively), low levels of MIP1α, RANTES, IL-13, and IL-5 (Fig. 1a, b, f, and i, respectively). These data indicate that vaccination with rV-CEA/TRICOM versus yeast-CEA induces expression of different cytokines, suggesting that different T-cell populations could be induced by each of the vaccine platforms.
Fig. 1.
Vaccination with rV-CEA/TRICOM or yeast-CEA induces differential serum cytokine profiles. CEA-Tg mice (n = 2) were vaccinated with rV-CEA/TRICOM (filled squares) or yeast-CEA (open circles). Serum was collected at 0, 2, and 4 days, pooled and analyzed for a panel of cytokines including a MIP1α, b RANTES, c GM-CSF, d IL-6, e IL-12p70, f IL-13, g IL-1α, h IL-1β, and i IL-5. Data were presented as pg/ml of cytokine on each day
Vaccination with rV/F-CEA/TRICOM versus yeast-CEA induces distinct TCR repertoires
We next sought to determine if vaccination with either platform induces CD8+ T-cell populations with distinct TCR repertoires. CEA-Tg mice were vaccinated with either rV/F-CEA/TRICOM or yeast-CEA as described in the “Materials and methods” section. Untreated mice served as a negative control (Fig. 2a and d). Spleens from vaccinated mice were harvested 14 days post-vaccination and pooled. RT-PCR reactions were performed using 19 Vα- and 24 Vβ-specific primers. PCR products were then analyzed and the percentage of the total TCR repertoire was calculated for each gene (Fig. 2). The TCR Vα profiles of splenocytes from untreated mice and mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA indicate that each group has a distinct TCR Vα expression profile (Fig. 2b and c). The expression of 12 of the 19 Vα genes was similar between T cells induced by both vaccines, while 7 Vα genes (Vα5, Vα6, Vα7, Vα8, Vα13.1, Vα34S-281, and Vα5Τ) are unique to T-cell populations from one vaccine compared to the other (Fig. 2b and c). Comparison of the Vβ repertoires from these same animals indicated that, with a few exceptions, the Vβ profiles also differ among the two groups of mice (Fig. 2e and f). The expression of 14 out of the 24 Vβ genes was similar between T cells induced by both vaccines. Yet the vaccines induce unique Vβ genes as well. As shown in Fig. 2e and f, 10 Vβ genes were uniquely expressed by T cells from either rV/F-CEA/TRICOM or yeast-CEA-vaccinated CEA-Tg mice (Vβ1, Vβ4, Vβ5.1, Vβ5.2, Vβ5.3, Vβ8.1, Vβ8.3, Vβ9, Vβ10, and Vβ20). These data indicate that the Vα and Vβ TCR repertoires of T cells from mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA have both shared and unique patterns of TCR gene expression. It is unknown, however, if these differences are due to different processing and presentation of the CEA antigen by the different vector-infected cells, or are due to the vaccine platforms themselves. The TCR repertoires of T-cell lines specific for two different CEA epitopes created from CEA-Tg mice vaccinated with the two vaccine platforms are described below.
Fig. 2.
Distinct TCR repertoires are induced from vaccination with rV/F-CEA/TRICOM or yeast-CEA. CEA-Tg mice (n = 5) were primed with rV-CEA/TRICOM on day 0 and boosted on days 7 and 14 with rF-CEA/TRICOM. CEA-Tg mice (n = 5) were primed on day 0 and boosted on days 7 and 14 with yeast-CEA. On day 35, mice were killed, spleens were harvested, and splenocytes were pooled. RNA was extracted from splenocytes and underwent RT-PCR using primers specific for Vα (a–c) or Vβ (d–f) genes. Vα profiles for a no treatment, b rV/F-CEA/TRICOM, and c yeast-CEA are shown. Vβ profiles for d no treatment, e rV/F-CEA/TRICOM, and f yeast-CEA are shown. Each gene was quantified by calculating % of total TCR repertoire. Asterisks indicate genes that are uniquely expressed in T cells from mice vaccinated with one vaccine compared to the other
Vaccination with rV/F-CEA/TRICOM or yeast-CEA induces both shared and unique gene expression in response to vaccine platform and antigen
To investigate the effects of both vaccine platform and antigen on the gene expression of splenocytes, cDNA oligoarrays were used to analyze expression of 252 genes involved in T- and B-cell activation, chemokines, chemokine receptors, and cytokines in splenocytes of CEA-Tg mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA. Table 1 shows that for each array, both rV/F-CEA/TRICOM and yeast-CEA induce changes in expression of the same genes, including up-regulation of 26 genes by at least 2-fold, the majority of which are involved in cytokine signaling. In addition, both vaccines up-regulated Ltb4r2, a leukotriene receptor involved in chemotaxis of immune cells, genes involved in T-cell proliferation, such as secreted phosphoprotein-1 (Spp1, or osteopontin), and the tumor suppressor Inha. At the same time, each vaccine platform induces unique changes in expression of several genes (Table 1, bold). Yeast-CEA down-regulates genes involved in chemotaxis of immune cells, such as Ccl12, Cxcl9, Ccr9, while rV/F-CEA/TRICOM does not alter the expression of any of these genes. The results from this experiment indicate that the two vaccine platforms induce changes in gene expression that are both shared and unique.
Table 1.
Vaccination with rV/F-CEA/TRICOM or yeast-CEA induces expression of both shared and unique genes involved in T- and B-cell activation, chemokines and chemokine receptors, and cytokines
| Vaccination | Genes involved in T-cell and B-cell activation | Chemokines and chemokine receptors | Cytokines |
|---|---|---|---|
| rV/F-CEA/TRICOM /No treatment | |||
| Genes up-regulated ≥2-fold | H60, lgbp1b, Il11, Il4 | Inha (3.03), Ltb4r2, Bmp10, Bmp5 | Il17c, Il17f, Inhba, Fgf10, Gdf2, Gdf5, Gdf8, Ifna2, Ifna4, Ifnbl, Il13, Il17b, Il25, Il19, Ilf10, Ilf5, Ilf6, Ilf8, Il20, Il3, Il9 |
| Genes down-regulated ≥2-fold | Ms4al, Sppl (4.00) | Il1rn (4.29) | |
| Yeast-CEA/no treatment | |||
| Genes up-regulated ≥2-fold | Rag1 (3.48), H60 (3.25), Igbp1b (3.25), Il11 (3.25), Il4 | Bdnf, Ccl20, Cmtm2a, Cmtm5, Cxcl15, Gdf5, Ccl17, Inha (4.92), Ltb4r2 (4.92), Bmp10, Bmp5 | Gdfl, Fgf10, Gdf2, Gdf5, Gdf8, Ifna2, Ifna4, Ifnbl, Il13, Il17b, Il25, Il19, Ilf10, Ilf5, Ilf6, Ilf8, Il20, Il3, Il9 |
| Genes down-regulated ≥2-fold | Tnfrsf13c (6.06), Ms4al (3.25), Spp1 | Ccl12 (3.03), Ccl8 (3.73) Ccr9, Ccrl2, Csf2, Cx3cr1, Cxcl10, Cxcl13, Hif1a, Inhbb, Lif (3.48), Gusb, Cxcl9 (4.92) | Gdf3(4.0), Il10, Csf-2, Fasl, Tnf, Tnfrsf11b, Tnfsf15 |
Genes in bold are up-regulated/down-regulated specifically by splenocytes from mice vaccinated with either rV/F-CEA/TRICOM or yeast-CEA, but not by both. Fold changes, >3-fold are noted in parentheses
rV/F-CEA/TRICOM and yeast-CEA induce functionally distinct T-cell populations
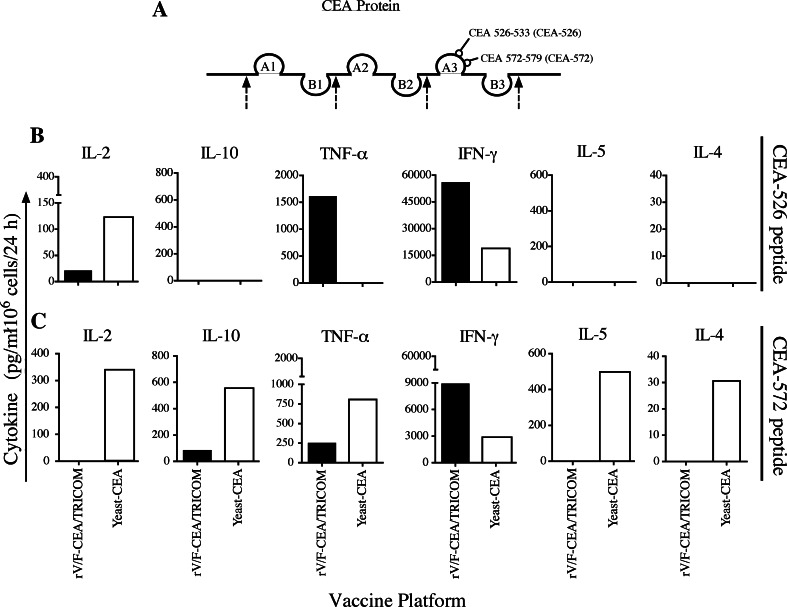
To determine the antigen-specific response of T-cell populations induced by the vaccines, we investigated the cytokines produced by T cells from vaccinated animals after in vitro stimulation with either of two discrete CEA epitopes (CEA-572 and CEA-526) (Fig. 3a).
Fig. 3.
Vaccination with rV/F-CEA/TRICOM or yeast-CEA induces distinct cytokine profiles in response to in vitro stimulation with two discrete CEA-specific epitopes. a CEA protein showing the discrete, non-overlapping CEA-526 and CEA-572 epitopes on the A3 loop of domain III. CEA-Tg mice (n = 5) were primed with rV-CEA/TRICOM (solid bars) on day 0 and boosted on days 7 and 14 with rF-CEA/TRICOM; CEA-Tg mice (n = 5) were primed on day 0 and boosted on days 7 and 14 with yeast-CEA (open bars). On day 33, mice were killed and spleens pooled and put into bulk cultures with either b CEA-526 or c CEA-572 peptide for 7 days. IL-2, IL-10, TNF-α, IFN-γ, IL-5, and IL-4 were measured by cytokine bead array (pg/ml/1 × 106 cells) after lymphocytes were restimulated for 24 h with CEA-specific peptide or VSVN peptide control. All data have been normalized to the VSVN peptide control. Please note the differences in the scales of each graph
We also observed that two different CEA epitopes induce different levels of cytokine production from T cells from vaccinated animals. Higher levels of TNF-α are secreted in response to CEA-526 after rV/F-CEA/TRICOM vaccination compared to yeast-CEA (Fig. 3b, closed bar), yet yeast-CEA vaccination produces significantly higher levels of TNF-α when T cells are stimulated with CEA-572 peptide (Fig. 3c, open bar). Also, T cells from yeast-CEA vaccination induce higher levels of IL-2 compared to T cells from rV/F-CEA/TRICOM vaccination, when stimulated with the CEA-526 and CEA-572 peptides (Fig. 3b and 3c, open bars). Similarly, after vaccination with rV/F-CEA/TRICOM, T cells induce higher levels of IFN-γ compared to yeast-CEA vaccination in response to the CEA-526 and CEA-572 peptides (Fig. 3b and c, closed bars).
The data also show that T cells from vaccinated animals secrete different levels of various cytokines in response to a single CEA epitope. T cells from mice vaccinated with yeast-CEA secrete IL-2, IL-10, TNF-α, IFN-γ, IL-5, and IL-4 in response to the CEA-572 epitope (Fig. 3c, open bars); on the other hand, T cells from mice vaccinated with rV/F-CEA/TRICOM secrete significantly higher levels of IFN-γ compared to yeast-CEA in response to the CEA-572 peptide and lower levels of IL-10 and TNF-α in response to the CEA-572 epitope (Fig. 3c, closed bars). These results indicate that the T-cell populations induced by vaccination with rV/F-CEA/TRICOM or yeast-CEA are antigen-specific and functionally distinct.
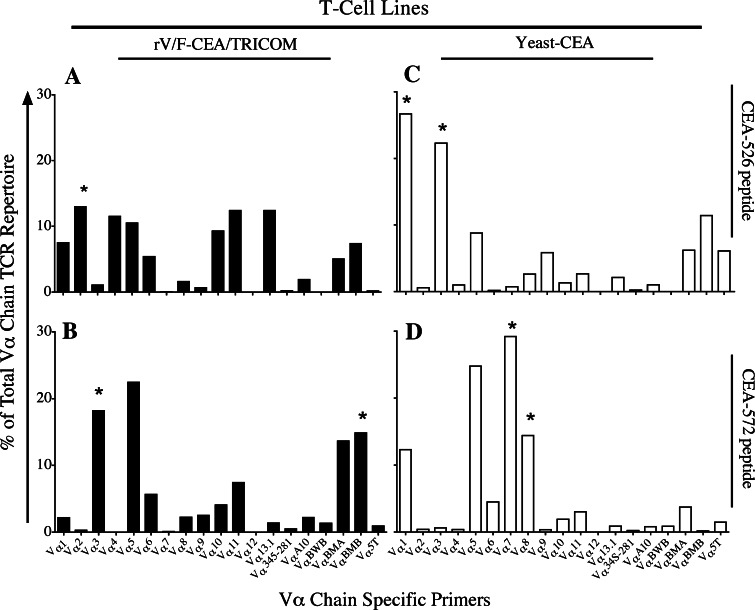
T-cell lines developed from mice vaccinated with rV/F-CEA/TRICOM versus yeast-CEA have distinct TCR repertoires and functional avidity
To further explore potential differences in the functionality of T cells from mice vaccinated with either rV/F-CEA/TRICOM or yeast-CEA, T-cell lines specific for either CEA-526 or CEA-572 peptide were created from vaccinated CEA-Tg mice as described in the “Materials and methods”. Vα TCR profiles from the 4 cell lines indicate that the T-cell populations have shared and distinct Vα TCR repertoires. In T cells from vaccinated mice stimulated with the CEA-526 epitope, the T cells have shared expression of 16 out of the 19 Vα genes and unique expression of 3 Vα genes (Fig. 4a and c). In T cells from vaccinated mice stimulated with the CEA-572 epitope, the T cells have shared expression of 15 out of the 19 Vα genes and unique expression of 4 Vα genes (Fig. 4b and d). Similar results were seen when Vβ TCR profiles were analyzed (data not shown). In addition, expression of selected Vβ TCR genes of the T-cell lines were confirmed by flow cytometry using commercially available monoclonal antibodies (data not shown). These data provide further evidence that the T-cell populations from mice vaccinated with either rV/F-CEA/TRICOM or yeast-CEA are both platform- and antigen-specific.
Fig. 4.
T-cell lines specific for the CEA-572 epitope from mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA have distinct TCR Vα profiles. CEA-Tg mice (n = 5) were primed with rV-CEA/TRICOM on day 0 and boosted on days 7 and 14 with rF-CEA/TRICOM. CEA-Tg mice (n = 5) were primed on day 0 and boosted on days 7 and 14 with yeast-CEA. Two weeks after the final vaccination, spleens were harvested and pooled, and splenocytes were bulk cultured with CEA-526 or CEA-572 peptide for 7 days. Lymphocytes were restimulated with fresh peptide, IL-2, and irradiated APCs every 7 days and kept in culture for in vitro experiments. TCR profile analysis was conducted after 18 stimulation cycles. Vα TCR repertoires of rV/F-CEA/TRICOM T-cell lines (black bars) maintained in the presence of a CEA-526 peptide and b CEA-572 peptide. Vα TCR repertoires of yeast-CEA T-cell lines (white bars) maintained in the presence of c CEA-526 peptide and d CEA-572 peptide. Results are expressed as % of total Vα chain TCR repertoire. Asterisks indicate genes that are uniquely expressed in T cells from mice vaccinated with one vaccine compared to the other
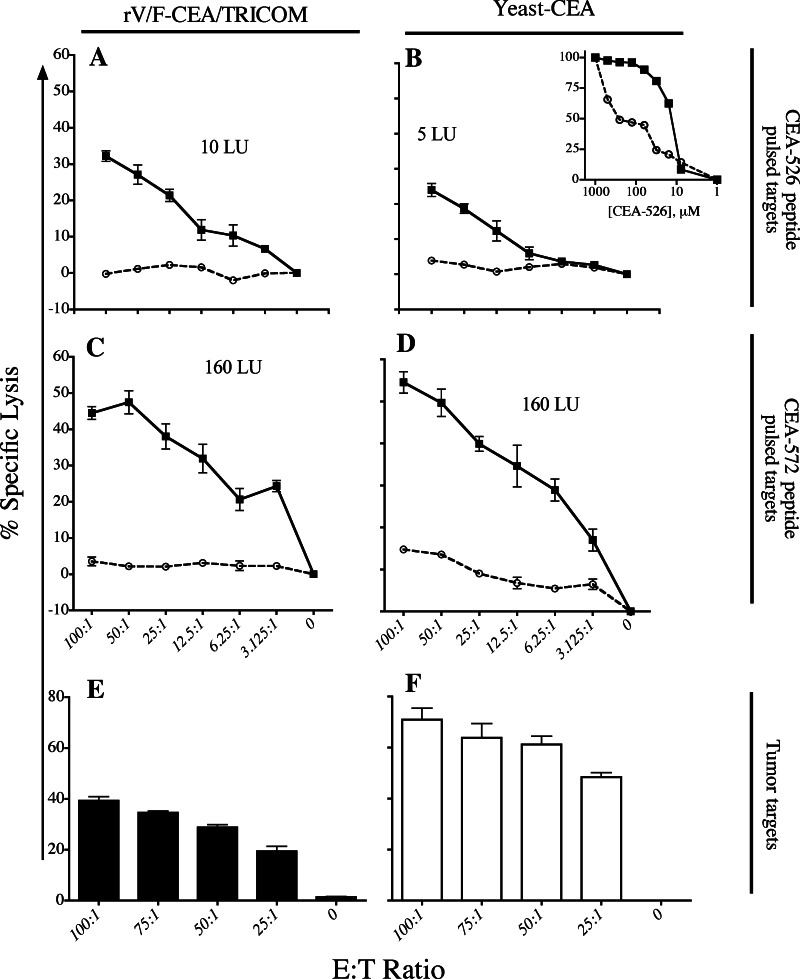
To characterize the functional differences between T cells from either vaccine, we compared the CEA-specific cytolytic activity of T cells generated from rV/F-CEA/TRICOM or yeast-CEA vaccination. The purity of the T-cell line cultures was confirmed via cell surface staining with monoclonal antibodies to identify CD8, CD4, and NK cells followed by flow cytometry (data not shown). In addition, tetramer staining using MHC class I-peptide tetramers specific for CEA-526 or CEA-572 confirmed peptide specificity for the T-cell lines (data not shown). Figure 5a and b shows that a CEA-526 peptide-specific T-cell line generated from rV/F-CEA/TRICOM has higher lytic activity compared to a T-cell line generated from yeast-CEA vaccination. T-cell lines specific for the CEA-572 epitope both demonstrated similar levels of cytolytic activity (Fig. 5c and d). Figure 5b inset shows that the CEA-526-specific T-cell line generated from rV/F-CEA/TRICOM vaccination had a 23.3-fold higher avidity than the CEA-526-specific T-cell line generated from yeast-CEA vaccination. The CEA-572-specific T-cell lines were also used in a CTL assay targeting 111In-labeled LL2-CEA cells. CEA-572-specific T cells from mice vaccinated with either rV/F-CEA/TRICOM or yeast-CEA were cultured for 20 weeks prior to this assay. Both T-cell lines specific for the CEA-572 peptide lyse LL2-CEA targets and lysis decreases as the ratio of T cells to effector cells (LL2-CEA targets) decreases (Fig. 5e and f). These results indicate that both T-cell lines are capable of lysing CEA-expressing cells, although the T-cell line from mice vaccinated with yeast-CEA (Fig. 5f) had a higher level of activity compared to the T-cell line from mice vaccinated with rV/F-CEA/TRICOM (Fig. 5e) when LL2-CEA cell lysis was normalized to that of LL2 cells. To confirm that the cell lysis observed in Fig. 5e and f was TCR-mediated and not due to NK cell activity, CTL experiments with blocking monoclonal antibodies specific for MHC class I molecules were performed with LL2-CEA tumor targets and normalized to LL2 target cells as a control and showed that the presence of MHC class I blocking antibody abrogated cell lysis. The lack of NK cell-mediated lysis was further confirmed in a CTL using YAK1 targets which found that the various T-cell lines did not lyse YAK1 target cells. Together, these results indicate that the lytic activity of T-cell lines created from different vectors targeting the same CEA-epitope is TCR-mediated and levels of cell lysis are similar when targeting peptide-pulsed target cells, although their ability to lyse CEA-expressing tumor targets differs. Additionally, the avidity of rV/F-CEA/TRICOM-induced T-cell lines may be higher than that of T-cell lines created from yeast-CEA vaccination. These results further characterize the T-cell populations from mice vaccinated with rV-CEA/TRICOM or yeast-CEA as platform-specific.
Fig. 5.
CEA epitope-specific T-cell lines generated from mice vaccinated with rV/F-CEA/TRICOM or yeast-CEA have similar levels of lytic activity but unique avidity. T-cell lines generated from rV/F-CEA/TRICOM vaccination and specific for a CEA-526 peptide and c CEA-572 peptide were incubated with peptide-pulsed 111In-labeled EL-4 cell targets at the indicated ratios for 4 h. T-cell lines generated from yeast-CEA vaccination and specific for b CEA-526 peptide and d CEA-572 peptide were also incubated with 111In-labeled EL-4 cell targets at the indicated ratios for 4 h. 111In-labeled EL-4 cells pulsed with CEA-572 and CEA-526 peptides are represented by filled squares connected by a solid line. 111In-labeled EL-4 cells pulsed with VSVNP (negative control) are represented by open circles connected by a dotted line. Bars indicate standard error from triplicate wells. To determine T-cell avidity, (b, inset) CEA-526-specific T-cell lines from rV/F-CEA/TRICOM (filled squares) and yeast-CEA (open circles) were incubated with 111In-labeled EL-4 cells in the presence of various concentrations of CEA-526 (or HIV-gag control) peptide ranging from 1 μM to 0 μM for 4 h. T-cell lines specific for CEA-572 generated from mice vaccinated with e rV/F-CEA/TRICOM (closed bars) or f yeast-CEA (open bars) were used in cytolytic T-cell assays with 111In-labeled LL2-CEA in an overnight assay at the indicated ratios. Data were normalized to LL2 (negative control) tumor targets. Bars indicate standard error from triplicate wells
Combining rV/F-CEA/TRICOM and yeast-CEA is an efficacious antitumor therapy in a murine orthotopic pulmonary metastasis model
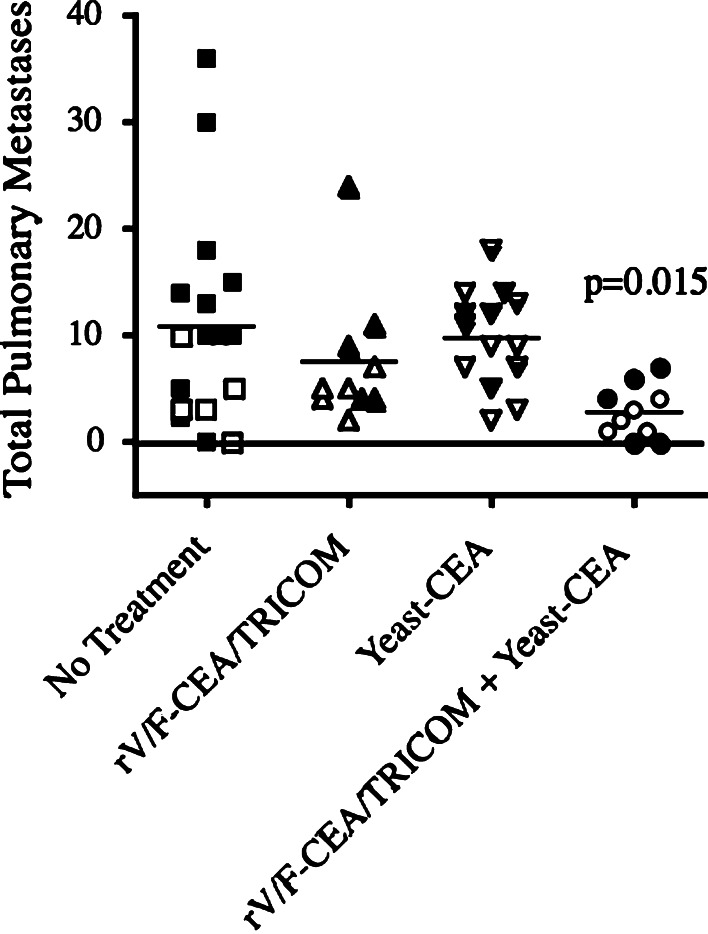
We next conducted studies to determine if concurrent administration of the two vaccines would generate antitumor activity superior to vaccination with either vaccine platform alone. CEA-Tg mice were injected i.v. with murine lung carcinoma cells expressing CEA (LL2-CEA cells). On day 4, mice were primed with rV-CEA/TRICOM, or yeast-CEA, or a combination of both vaccines. A group of mice was left untreated as a control. Mice were boosted with rF-CEA/TRICOM, or yeast-CEA, or the combination, respectively, 7 days later and every week until the duration of the experiment. On day 45, mice were killed, lungs were inflated and fixed as described in “Materials and methods”, and the number of metastases per animal was calculated (Fig. 6). Untreated mice had an average of 10.84 tumors per mouse (SE ± 2.41). Mice vaccinated with rV/F-CEA/TRICOM had an average of 7.50 metastases per mouse (±2.02), and mice vaccinated with yeast-CEA had an average of 9.71 metastases per mouse (±1.22). However, mice vaccinated with the combination of rV/F-CEA/TRICOM and yeast-CEA had 2.80 metastases per mouse (±0.77); this combination group was the only group with a significantly lower number of metastases compared to the untreated control (p = 0.015). Also, the maximum number of metastases per mouse for the untreated, rV/F-CEA/TRICOM, and yeast-CEA groups was 36, 24, and 18, respectively, while the maximum number of metastases in the combination group was 7. Moreover, the log-rank test (mice bearing >9 pulmonary tumor nodules on day 45; assumed to have ≤1 week to live) showed statistical significance in tumor number between untreated mice and the mice that received the combination of rV/F-CEA/TRICOM and yeast-CEA (p = 0.0027). Also, mice treated with rV/F-CEA/TRICOM alone versus concurrent vaccination with rV/F-CEA/TRICOM and yeast-CEA showed statistical significance in tumor number (p = 0.0293). In addition, there was statistical significance between mice treated with yeast-CEA alone versus concurrent vaccination with rV/F-CEA/TRICOM and yeast-CEA (p = 0.0017). These results, taken together, indicate that concurrent administration of rV/F-CEA/TRICOM and yeast-CEA vaccines can increase antitumor efficacy.
Fig. 6.
Combining rV/F-CEA/TRICOM and yeast-CEA vaccines in an orthotopic pulmonary metastasis model increases antitumor efficacy. CEA-Tg mice were injected i.v. with LL2-CEA tumor cells. On day 4, mice were primed with rV-CEA/TRICOM (n = 10), yeast-CEA (n = 14), or rV-CEA/TRICOM and yeast-CEA (n = 10); a control group (n = 17) received no treatment. Mice were boosted every 7 days for the duration of the experiment. The rV/F-CEA/TRICOM group was boosted with rF-CEA/TRICOM. The yeast-CEA group was boosted with yeast-CEA only. The combination group was boosted with rF-CEA/TRICOM and yeast-CEA. For these studies, rV/F-CEA/TRICOM was injected s.c. on the dorsal right flank while 1 YU yeast-CEA was delivered s.c. to each inner leg and shoulder blade to target multiple draining lymph nodes. On day 45, mice were killed and lungs were harvested, stained, and fixed. Data represent the number of lung metastases per mouse from two separate experiments (indicated by open vs. closed symbols). The bar indicates the average number of metastases per mouse. p = 0.015 comparing untreated mice with the rV/F-CEA/TRICOM and yeast-CEA combination group
Discussion
We have previously reported that both the rV/F-CEA/TRICOM and yeast-CEA vaccines induce robust immune responses against CEA in self-antigen CEA-Tg mice [10, 11, 15, 16]. This study was undertaken to focus on the distinct T-cell populations induced by each platform and their combined use. Millar et al. [5] previously showed that the functionality of T-cell populations induced by two different platforms (rV and recombinant adenovirus) targeting the same antigen (OVA) did not differ. Our study provides evidence that both the vaccine platform and the antigen can affect the functionality of the T-cell population induced by rV/F-CEA/TRICOM and yeast-CEA vaccines. To investigate the platform- and antigen-specific induction of T-cell functionality and phenotype, we compared the T-cell populations induced by both vaccines in terms of cytokine production, gene expression, and TCR profiling. Our studies found that rV-CEA/TRICOM induces a Th1 response and CD8+ T cells with a Tc1 phenotype, while yeast-CEA induces a mixed Th1/Th2 response and CD8+ T cells with a mixed Tc1/Tc2 phenotype (Figs. 1 and 3). These differences in cytokine response could also be due to differences in affinity for peptide (Fig. 5b, inset), magnitude of the response (Fig. 5e and f), or timing of activation, which may differ due to the differences in antigen processing and presentation after administration of either vaccine. We observed up-regulation of genes involved in immune cell migration and TCR signaling and T-cell proliferation by both vaccines (Table 1). Although these vaccines each elicit distinct gene expression profiles for genes involved in cellular pathways important in T-cell activation and chemotaxis of immune cells, both elicit antigen-specific immune responses and antitumor efficacy, presumably through either Th1 or a mixed Th1/Th2 response. We also observed that the T-cell populations induced by either vaccine have both shared and unique Vα and Vβ TCR gene usage (Fig. 2) and that T-cell lines created from vaccinated CEA-Tg mice, specific for one of two CEA epitopes, demonstrate differential avidity and antigen-specific cytolytic activity (Fig. 5). Taken together, these studies demonstrate that the two vaccines induce distinct T-cell populations and the differences in the phenotype and function of these T-cell populations may be attributed to both the platform and the antigen. The mode of antigen delivery by either vaccine platform may influence the generation of these distinct responses. The mechanism by which yeast-CEA predominantly activates the immune response is through the uptake of the CEA-expressing yeast and subsequent processing and presentation of the CEA antigen by DCs [15], while the rV/F-CEA/TRICOM vectors infect cells, inducing intracellular expression that allows the CEA antigen to be processed and presented [10]. Another difference observed is that rV/F-CEA/TRICOM vaccination induces the production of CEA-specific antibody while yeast-CEA does not (data not shown). Such mechanistic differences in the activation of the cellular and humoral arms of the immune system may also influence the platform- and antigen-specific induction of distinct T-cell populations.
Our studies led us to hypothesize that employing the two vaccines concurrently could lead to a more polyclonal T-cell population that would be advantageous in an anticancer setting while several studies have documented that the induction of a more diverse T-cell population is advantageous in mounting an immune response in various models of disease, including cancer [26–33]. While there are no reports of concurrent use of vaccines that target the same antigen, we have previously documented the advantages of a diversified prime–boost vaccination strategy with recombinant vaccinia and fowlpox vectors targeting CEA [34, 35]. This strategy was employed because immune response to the first vaccine has been shown to reduce the effects of subsequent vaccinations with the same vector [3, 12, 34, 36]. Therefore, diversified prime–boost strategies avoid any immune response against the first vaccine platform while expanding the antigen-specific T-cell population with each boost with the second vaccine platform. Similar results demonstrating the clear advantages of a diversified prime–boost strategy have been described in a variety of cancer and other disease models, including HIV and malaria [36–41]. The enhanced responses observed in these studies have been attributed to amplification of the population of antigen-specific T cells. However, as our study found that rV/F-CEA/TRICOM and yeast-CEA induced distinct T-cell populations, we hypothesized that concurrent administration of the vaccines may induce a more diverse T-cell population consisting of T cells generated from both vaccines, making a diversified prime–boost schedule unnecessary. Our study differs from diversified prime–boost studies because we are trying to maximize the immune response beginning with the initial vaccination by inducing a more diverse T-cell population that is then boosted and expanded in magnitude with each subsequent vaccination. Such a strategy would be efficacious in cancer patients because a more diverse T-cell population would be induced early in their treatment. We therefore investigated the antitumor effects of combining rV/F-CEA/TRICOM and yeast-CEA delivered concurrently. We found that combining two vaccines targeting the same antigen induces distinct T-cell populations and results in significantly higher antitumor immunity in a murine orthotopic pulmonary metastasis model (Fig. 6). Given the various shared phenotypic and functional characteristics of the T-cell populations induced by either vaccine alone, a diversified prime–boost strategy may still be useful when combining rF/V-CEA/TRICOM and yeast-CEA. Further studies examining the efficacy of diversified prime and boost as well as concurrent vaccination should be performed in this and other tumor models.
This study showed for the first time that two vaccine platforms targeting the same antigen could be concurrently administrated due to their induction of distinct T-cell populations. Furthermore, our data show that concurrent administration of the two vaccines results in significantly increased antitumor effects, due to the induction of a more diverse T-cell population targeting the same antigen. These findings provide a rationale for future studies concurrently combining vaccines to increase antigen-specific immunity.
Acknowledgments
The authors acknowledge the excellent technical assistance of Marion Taylor and Anais Kasten-Sportes, and the editorial assistance of Bonnie L. Casey in the preparation of this manuscript.
References
- 1.Weide B, Garbe C, Rammensee HG, Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol Lett. 2008;115(1):33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 2.Riezebos-Brilman A, Walczak M, Regts J, Rots MG, Kamps G, Dontje B, et al. A comparative study on the immunotherapeutic efficacy of recombinant Semliki Forest virus and adenovirus vector systems in a murine model for cervical cancer. Gene Ther. 2007;14(24):1695–1704. doi: 10.1038/sj.gt.3303036. [DOI] [PubMed] [Google Scholar]
- 3.Naslund TI, Uyttenhove C, Nordstrom EK, Colau D, Warnier G, Jondal M, et al. Comparative prime-boost vaccinations using Semliki Forest virus, adenovirus, and ALVAC vectors demonstrate differences in the generation of a protective central memory CTL response against the P815 tumor. J Immunol. 2007;178(11):6761–6769. doi: 10.4049/jimmunol.178.11.6761. [DOI] [PubMed] [Google Scholar]
- 4.Mylin LM, Schell TD, Roberts D, Epler M, Boesteanu A, Collins EJ, et al. Quantitation of CD8(+) T-lymphocyte responses to multiple epitopes from simian virus 40 (SV40) large T antigen in C57BL/6 mice immunized with SV40, SV40 T-antigen-transformed cells, or vaccinia virus recombinants expressing full-length T antigen or epitope minigenes. J Virol. 2000;74(15):6922–6934. doi: 10.1128/JVI.74.15.6922-6934.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millar J, Dissanayake D, Yang TC, Grinshtein N, Evelegh C, Wan Y, et al. The magnitude of the CD8+ T cell response produced by recombinant virus vectors is a function of both the antigen and the vector. Cell Immunol. 2007;250(1–2):55–67. doi: 10.1016/j.cellimm.2008.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Hodge JW, Poole DJ, Aarts WM, Gomez Yafal A, Gritz L, Schlom J. Modified vaccinia virus Ankara recombinants are as potent as vaccinia recombinants in diversified prime and boost vaccine regimens to elicit therapeutic antitumor responses. Cancer Res. 2003;63(22):7942–7949. [PubMed] [Google Scholar]
- 7.Chan T, Sami A, El-Gayed A, Guo X, Xiang J. HER-2/neu-gene engineered dendritic cell vaccine stimulates stronger HER-2/neu-specific immune responses compared to DNA vaccination. Gene Ther. 2006;13(19):1391–1402. doi: 10.1038/sj.gt.3302797. [DOI] [PubMed] [Google Scholar]
- 8.Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, et al. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77(11):6305–6313. doi: 10.1128/JVI.77.11.6305-6313.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bos R, van Duikeren S, van Hall T, Lauwen MM, Parrington M, Berinstein NL, et al. Characterization of antigen-specific immune responses induced by canarypox virus vaccines. J Immunol. 2007;179(9):6115–6122. doi: 10.4049/jimmunol.179.9.6115. [DOI] [PubMed] [Google Scholar]
- 10.Hodge JW, Sabzevari H, Yafal AG, Gritz L, Lorenz MG, Schlom J. A triad of costimulatory molecules synergize to amplify T-cell activation. Cancer Res. 1999;59(22):5800–5807. [PubMed] [Google Scholar]
- 11.Hodge JW, Grosenbach DW, Aarts WM, Poole DJ, Schlom J. Vaccine therapy of established tumors in the absence of autoimmunity. Clin Cancer Res. 2003;9(5):1837–1849. [PubMed] [Google Scholar]
- 12.Grosenbach DW, Barrientos JC, Schlom J, Hodge JW. Synergy of vaccine strategies to amplify antigen-specific immune responses and antitumor effects. Cancer Res. 2001;61(11):4497–4505. [PubMed] [Google Scholar]
- 13.Greiner JW, Zeytin H, Anver MR, Schlom J. Vaccine-based therapy directed against carcinoembryonic antigen demonstrates antitumor activity on spontaneous intestinal tumors in the absence of autoimmunity. Cancer Res. 2002;62(23):6944–6951. [PubMed] [Google Scholar]
- 14.Arlen PM, Gulley JL, Madan RA, Hodge JW, Schlom J. Preclinical and clinical studies of recombinant poxvirus vaccines for carcinoma therapy. Crit Rev Immunol. 2007;27(5):451–462. doi: 10.1615/critrevimmunol.v27.i5.40. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein MB, Chakraborty M, Wansley EK, Guo Z, Franzusoff A, Mostbock S, et al. Recombinant Saccharomyces cerevisiae (yeast-CEA) as a potent activator of murine dendritic cells. Vaccine. 2008;26(4):509–521. doi: 10.1016/j.vaccine.2007.11.033. [DOI] [PubMed] [Google Scholar]
- 16.Wansley EK, Chakraborty M, Hance KW, Bernstein MB, Boehm AL, Guo Z, et al. Vaccination with a recombinant Saccharomyces cerevisiae expressing a tumor antigen breaks immune tolerance and elicits therapeutic antitumor responses. Clin Cancer Res. 2008;14(13):4316–4325. doi: 10.1158/1078-0432.CCR-08-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robbins PF, Kantor JA, Salgaller M, Hand PH, Fernsten PD, Schlom J. Transduction and expression of the human carcinoembryonic antigen gene in a murine colon carcinoma cell line. Cancer Res. 1991;51(14):3657–3662. [PubMed] [Google Scholar]
- 18.Kass E, Panicali DL, Mazzara G, Schlom J, Greiner JW. Granulocyte/macrophage-colony stimulating factor produced by recombinant avian poxviruses enriches the regional lymph nodes with antigen-presenting cells and acts as an immunoadjuvant. Cancer Res. 2001;61(1):206–214. [PubMed] [Google Scholar]
- 19.Haller AA, Lauer GM, King TH, Kemmler C, Fiolkoski V, Lu Y, et al. Whole recombinant yeast-based immunotherapy induces potent T cell responses targeting HCV NS3 and core proteins. Vaccine. 2007;25(8):1452–1463. doi: 10.1016/j.vaccine.2006.10.035. [DOI] [PubMed] [Google Scholar]
- 20.Pannetier C, Cochet M, Darche S, Casrouge A, Zoller M, Kourilsky P. The sizes of the CDR3 hypervariable regions of the murine T-cell receptor beta chains vary as a function of the recombined germ-line segments. Proc Natl Acad Sci USA. 1993;90(9):4319–4323. doi: 10.1073/pnas.90.9.4319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arden B, Klotz JL, Siu G, Hood LE. Diversity and structure of genes of the alpha family of mouse T-cell antigen receptor. Nature. 1985;316(6031):783–787. doi: 10.1038/316783a0. [DOI] [PubMed] [Google Scholar]
- 22.Hodge JW, Chakraborty M, Kudo-Saito C, Garnett CT, Schlom J. Multiple costimulatory modalities enhance CTL avidity. J Immunol. 2005;174(10):5994–6004. doi: 10.4049/jimmunol.174.10.5994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Derby M, Alexander-Miller M, Tse R, Berzofsky J. High-avidity CTL exploit two complementary mechanisms to provide better protection against viral infection than low-avidity CTL. J Immunol. 2001;166(3):1690–1697. doi: 10.4049/jimmunol.166.3.1690. [DOI] [PubMed] [Google Scholar]
- 24.Wunderlich J, Shearer G. Induction and measurement of cytotoxic T lymphocyte activity. In: Coligan J, Kruisbeek A, Margulies D, Shevach E, Strober W, editors. Current protocols in immunology. Hoboken, NJ: Wiley; 1994. [Google Scholar]
- 25.Wexler H. Accurate identification of experimental pulmonary metastases. J Natl Cancer Inst. 1966;36(4):641–645. doi: 10.1093/jnci/36.4.641. [DOI] [PubMed] [Google Scholar]
- 26.Dudley ME, Ngo LT, Westwood J, Wunderlich JR, Rosenberg SA. T-cell clones from melanoma patients immunized against an anchor-modified gp100 peptide display discordant effector phenotypes. Cancer J. 2000;6(2):69–77. [PubMed] [Google Scholar]
- 27.Dutoit V, Rubio-Godoy V, Dietrich PY, Quiqueres AL, Schnuriger V, Rimoldi D, et al. Heterogeneous T-cell response to MAGE-A10(254–262): high avidity-specific cytolytic T lymphocytes show superior antitumor activity. Cancer Res. 2001;61(15):5850–5856. [PubMed] [Google Scholar]
- 28.Echchakir H, Vergnon I, Dorothee G, Grunenwald D, Chouaib S, Mami-Chouaib F. Evidence for in situ expansion of diverse antitumor-specific cytotoxic T lymphocyte clones in a human large cell carcinoma of the lung. Int Immunol. 2000;12(4):537–546. doi: 10.1093/intimm/12.4.537. [DOI] [PubMed] [Google Scholar]
- 29.Ferradini L, Roman-Roman S, Azocar J, Avril MF, Viel S, Triebel F, et al. Analysis of T-cell receptor alpha/beta variability in lymphocytes infiltrating a melanoma metastasis. Cancer Res. 1992;52(17):4649–4654. [PubMed] [Google Scholar]
- 30.Messaoudi I, Guevara Patino JA, Dyall R, LeMaoult J, Nikolich-Zugich J. Direct link between MHC polymorphism, T cell avidity, and diversity in immune defense. Science. 2002;298(5599):1797–1800. doi: 10.1126/science.1076064. [DOI] [PubMed] [Google Scholar]
- 31.Nikolich-Zugich J, Slifka MK, Messaoudi I. The many important facets of T-cell repertoire diversity. Nat Rev Immunol. 2004;4(2):123–132. doi: 10.1038/nri1292. [DOI] [PubMed] [Google Scholar]
- 32.Sportes C, Hakim FT, Memon SA, Zhang H, Chua KS, Brown MR, et al. Administration of rhIL-7 in humans increases in vivo TCR repertoire diversity by preferential expansion of naive T cell subsets. J Exp Med. 2008;205(7):1701–1714. doi: 10.1084/jem.20071681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou X, Jun DY, Thomas AM, Huang X, Huang LQ, Mautner J, et al. Diverse CD8+ T-cell responses to renal cell carcinoma antigens in patients treated with an autologous granulocyte-macrophage colony-stimulating factor gene-transduced renal tumor cell vaccine. Cancer Res. 2005;65(3):1079–1088. doi: 10.1158/0008-5472.CAN-04-3509. [DOI] [PubMed] [Google Scholar]
- 34.Hodge JW, McLaughlin JP, Kantor JA, Schlom J. Diversified prime and boost protocols using recombinant vaccinia virus and recombinant non-replicating avian pox virus to enhance T-cell immunity and antitumor responses. Vaccine. 1997;15(6–7):759–768. doi: 10.1016/S0264-410X(96)00238-1. [DOI] [PubMed] [Google Scholar]
- 35.Marshall JL, Hoyer RJ, Toomey MA, Faraguna K, Chang P, Richmond E, et al. Phase I study in advanced cancer patients of a diversified prime-and-boost vaccination protocol using recombinant vaccinia virus and recombinant nonreplicating avipox virus to elicit anti-carcinoembryonic antigen immune responses. J Clin Oncol. 2000;18(23):3964–3973. doi: 10.1200/JCO.2000.18.23.3964. [DOI] [PubMed] [Google Scholar]
- 36.Wu L, Kong WP, Nabel GJ. Enhanced breadth of CD4 T-cell immunity by DNA prime and adenovirus boost immunization to human immunodeficiency virus Env and Gag immunogens. J Virol. 2005;79(13):8024–8031. doi: 10.1128/JVI.79.13.8024-8031.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pancholi P, Liu Q, Tricoche N, Zhang P, Perkus ME, Prince AM. DNA prime-canarypox boost with polycistronic hepatitis C virus (HCV) genes generates potent immune responses to HCV structural and nonstructural proteins. J Infect Dis. 2000;182(1):18–27. doi: 10.1086/315646. [DOI] [PubMed] [Google Scholar]
- 38.Barnett SW, Klinger JM, Doe B, Walker CM, Hansen L, Duliege AM, et al. Prime–boost immunization strategies against HIV. AIDS Res Hum Retrovir. 1998;14(Suppl 3):S299–S309. [PubMed] [Google Scholar]
- 39.Dunachie SJ, Hill AV. Prime-boost strategies for malaria vaccine development. J Exp Biol. 2003;206(Pt 21):3771–3779. doi: 10.1242/jeb.00642. [DOI] [PubMed] [Google Scholar]
- 40.McMichael AJ. HIV vaccines. Annu Rev Immunol. 2006;24:227–255. doi: 10.1146/annurev.immunol.24.021605.090605. [DOI] [PubMed] [Google Scholar]
- 41.Moore AC, Hill AV. Progress in DNA-based heterologous prime-boost immunization strategies for malaria. Immunol Rev. 2004;199:126–143. doi: 10.1111/j.0105-2896.2004.00138.x. [DOI] [PubMed] [Google Scholar]