Abstract
The Rheb1 and Rheb2 small GTPases and their effector mTOR are aberrantly activated in human cancer and are attractive targets for anti-cancer drug discovery. Rheb is targeted to endomembranes via its C-terminal CAAX (C = cysteine, A = aliphatic, X = terminal amino acid) motif, a substrate for posttranslational modification by a farnesyl isoprenoid. Following farnesylation, Rheb undergoes two additional CAAX-signaled processing steps, Rce1-catalyzed cleavage of the AAX residues and Icmt-mediated carboxylmethylation of the farnesylated cysteine. However, whether these post-prenylation processing steps are required for Rheb signaling through mTOR is not known. We found that Rheb1 and Rheb2 localize primarily to the endoplasmic reticulum and Golgi apparatus. We determined that Icmt and Rce1 processing is required for Rheb localization, but is dispensable for Rheb-induced activation of the mTOR substrate p70 S6 kinase (S6K). Finally, we evaluated whether farnesylthiosalicylic acid (FTS) blocks Rheb localization and function. Surprisingly, FTS prevented S6K activation induced by a constitutively active mTOR mutant, indicating that FTS inhibits mTOR at a level downstream of Rheb. We conclude that inhibitors of Icmt and Rce1 will not block Rheb function, but FTS could be a promising treatment for Rheb- and mTOR-dependent cancers.
Keywords: Rheb; mTOR; Ras converting enzyme 1 (Rce1); Isoprenylcysteine carboxyl methyltransferase (Icmt); S-trans, trans-farnesylthiosalicylic acid (FTS)
Introduction
Rheb is a member of the Ras branch of the Ras superfamily of small GTPases and is evolutionarily conserved from yeast to humans (Aspuria and Tamanoi, 2004). The two mammalian Rheb isoforms, Rheb1 (Rheb) and Rheb2 (RhebL1; 51% identity), function as GTP/GDP regulated binary switches. Although both are expressed in many tissues and tumor types, current knowledge of Rheb comes largely from the study of Rheb1. Like Ras, Rheb proteins cycle between activated, GTP-bound and inactive GDP-bound states. Rheb is negatively regulated by the tumor suppressor TSC1 (hamartin)-TSC2 (tuberin) complex which functions as a GTPase activating protein (GAP) towards Rheb (Huang and Manning, 2008).
Rheb is a critical component of the phosphatidylinositol 3-kinase (PI3K)-Akt-TSC-mTOR pathway which is frequently hyperactivated in cancer. PI3K activates Akt, which phosphorylates and inactivates TSC2, leading to an increase in GTP-bound Rheb (Huang and Manning, 2008). Rheb-GTP is an activator of the rapamycin-sensitive mTOR complex 1 (mTORC1) (Long et al., 2005), which regulates the initiation of protein translation, nutrient sensing, and cell growth. mTOR controls protein synthesis by phosphorylating proteins involved in translation initiation, such as the p70 ribosomal S6 kinase (S6K).
Loss-of-function germline mutations in TSC1 or TSC2 cause Rheb hyperactivation and tuberous sclerosis complex disease which is characterized by the formation of hamartomas in a variety of organs (Astrinidis and Henske, 2005; Inoki et al., 2005). Rheb and mTOR are also hyperactivated in many sporadic cancers due to mutations in PI3K and PTEN. Rheb1 and Rheb2 are overexpressed in a variety of tumors, including glioma, oral squamous cell carcinoma, and breast, prostate, lung, colon, and ovarian cancer (Basso et al., 2005; Chakraborty et al., 2008; Gromov et al., 1995; Jiang and Vogt, 2008; Nardella et al., 2008). In addition, a Rheb mutation has been found in colon cancer (Wood et al., 2007), and the RHEB gene is amplified in some prostate cancers (Nardella et al., 2008). Therefore, Rheb inhibition may be therapeutically beneficial for a variety of cancers.
Like Ras, Rheb terminates in a C-terminal CAAX motif (C = cysteine, A = aliphatic, X = terminal amino acid), a substrate for farnesyltransferase (FTase)-catalyzed posttranslational modification by a C15 farnesyl isoprenoid lipid (Aspuria and Tamanoi, 2004). Two additional CAAX-signaled posttranslational processing steps, proteolytic cleavage of the AAX residues [catalyzed by Ras converting enzyme (Rce1)] and carboxylmethylation [(catalyzed by isoprenylcysteine carboxyl methyltransferase (Icmt)], are required for Rheb localization (Takahashi et al., 2005). However, whether these processing steps are also required for Rheb signaling through mTOR is not yet known.
The CAAX-signaled modifications are necessary but not sufficient for the proper membrane association and subcellular localization of a majority of Ras and Rho family small GTPases (Cox and Der, 2002; Sebti and Der, 2003). In addition, a second membrane-targeting signal positioned in sequences immediately upstream of the CAAX motif is required. For example, in H-Ras, the second signal is comprised of two palmitoylated cysteines upstream of the CAAX motif, whereas in K-Ras4B and Rac1, it is comprised of polybasic-rich sequences. Rheb1 and Rheb2 lack either of these, which may account for the absence of any plasma membrane-associated Rheb. However, other unidentified sequence elements may provide a second signal (Chenette et al., 2006), so it remains possible that Rheb subcellular localization is not dictated solely by CAAX-signaled modifications.
FTase inhibitors (FTIs) are a class of anti-cancer agents that were originally developed to inhibit Ras farnesylation and membrane association. While FTIs have shown anti-tumor activity, this activity is not due to inhibition of Ras, but rather to inhibition of other FTase substrates (Cox and Der, 2002; Sebti and Der, 2003), possibly including Rheb (Basso et al., 2005; Castro et al., 2003; Mavrakis et al., 2008). Rce1 and Icmt inhibitors, such as cysmethynil, are being developed as potential inhibitors of Ras membrane association and function (Konstantinopoulos et al., 2007; Winter-Vann and Casey, 2005), but whether they block Rheb signaling and function is not known. Additionally, farnesyl-containing small molecules, such as S-trans, trans-farnesylthiosalicylic acid (FTS; salirasib), have been developed as potential inhibitors of Ras membrane association and are currently undergoing clinical evaluation (Blum et al., 2008). Intriguingly, FTS has also exhibited properties of an mTOR inhibitor (McMahon et al., 2005; Yue et al., 2007; Yue et al., 2005), but whether this is due to inhibition of farnesylated Rheb is not known.
In this study, we determined whether inhibition of CAAX-signaled modifications could be used to block aberrant Rheb signaling in cancer. We found that Rheb1 and Rheb2 both localized to the endoplasmic reticulum (ER) and Golgi apparatus and that the CAAX motif of Rheb1, but not of Rheb2, was sufficient for proper localization. While Rce1- and Icmt-catalyzed modifications were required for proper Rheb localization, surprisingly, they were dispensable for Rheb-mediated activation of mTOR. Finally, we found that FTS directly blocked mTOR-induced activation of S6K independently of inhibition of Rheb or other prenylated proteins, suggesting that FTS is a novel type of mTOR inhibitor.
Results
Rheb1 and Rheb2 localize to the ER and Golgi
Previous reports indicated that ectopically expressed Rheb1 localizes to the Golgi apparatus (Buerger et al., 2006), endoplasmic reticulum (ER) (Jiang and Vogt, 2008), and vesicular structures (Buerger et al., 2006; Saito et al., 2005; Sancak et al., 2008), and that endogenous Rheb localizes to mitochondria (Ma et al., 2008). The precise subcellular localization of Rheb2 has not been described. Therefore, we first wanted to confirm and extend the previous disparate observations on Rheb1 and additionally to evaluation Rheb2 localization.
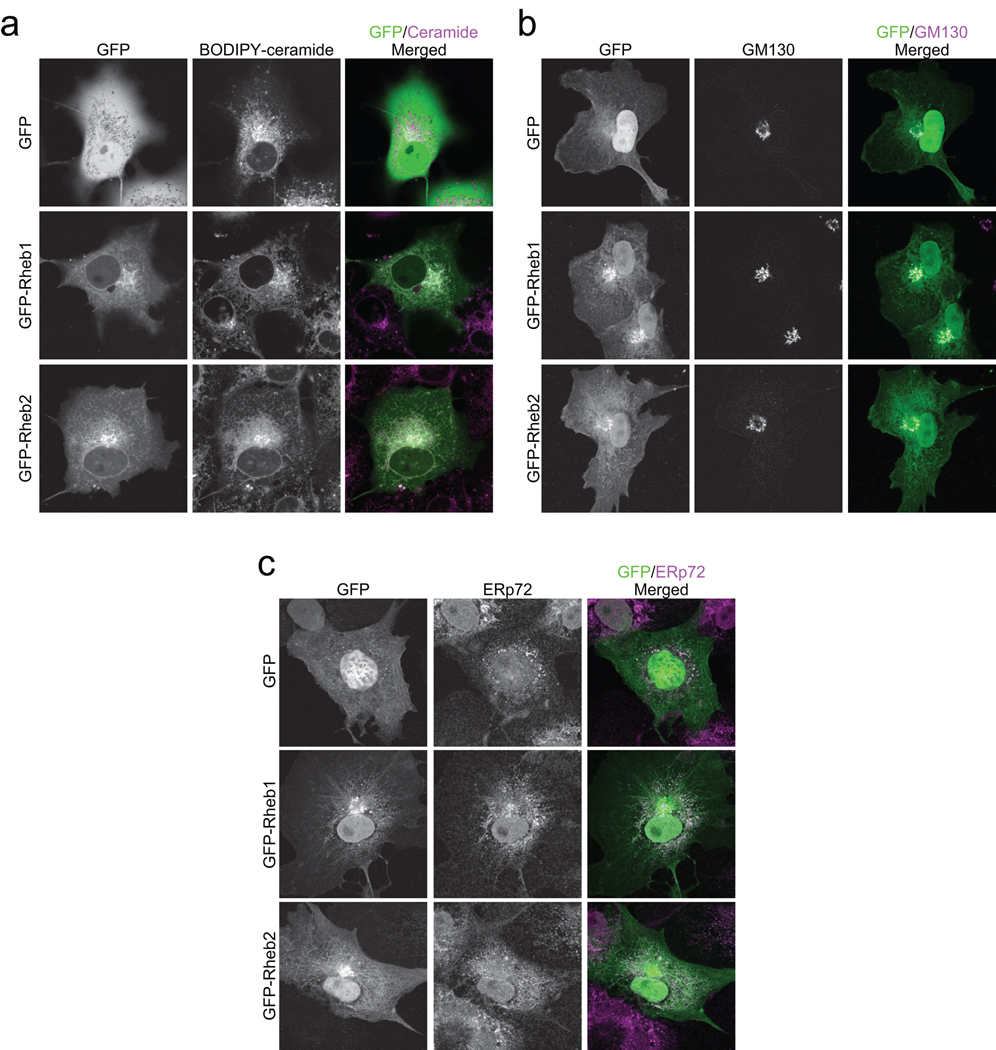
Because no Rheb2 antibody was available for evaluation of endogenous protein, we utilized green fluorescent protein (GFP)-tagged Rheb proteins for our analyses. Previous studies with similar GFP-tagged Ras family small GTPases have validated this approach for accurate determination of endogenous protein subcellular distribution (Choy et al., 1999; Heo and Meyer, 2003; Michaelson et al., 2001). We transiently transfected COS-7 epithelial cells with either GFP alone or with GFP-tagged Rheb1 and Rheb2 and treated live cells with BODIPY TR C5 ceramide, which detects cis- and trans-Golgi and associated vesicles, as well as with MitoTracker, a mitochondrial marker, and LysoTracker, a lysosome marker. In addition, we fixed cells and stained with markers of the following intracellular compartments: ERp72 (ER), GM130 (cis-Golgi), EEA1 (early endosomes), and LAMP2 (late endosomes). GFP-Rheb1 and GFP-Rheb2 both exhibited strong colocalization with BODIPY TR C5-ceramide (live cells) and GM130 (fixed cells), confirming that they localize to the Golgi apparatus (Figures 1a and 1b). GFP-Rheb1 and GFP-Rheb2 also colocalized with BODIPY-C5 ceramide in web-like structures in the cytoplasm in live cells (Figure 1a), indicative of ER localization (Choy et al., 1999). We also observed colocalization with ERp72 in fixed cells, confirming ER localization (Figure 1c). We did not observe colocalization of either Rheb1 or Rheb2 with EEA1, LAMP2, LysoTracker, or MitoTracker (Supplementary Figure 1), suggesting that Rheb1 and Rheb2 do not localize to early endosomes, late endosomes, lysosomes, or mitochondria. We confirmed these results in NIH 3T3 mouse fibroblasts (data not shown) and we observed similar localization of GFP-Rheb in fixed and live cells, although the web-like ER structure was disrupted in fixed cells, as observed previously (Choy et al., 1999).
Figure 1. Rheb1 and Rheb2 are localized primarily to the ER and Golgi. COS-7 cells were transiently transfected with pEGFP vector or with pEGFP encoding GFP-tagged Rheb1 or Rheb2.
(a) Rheb localization in cis- and trans-Golgi and associated endomembranes. Live cells were imaged after staining with BODIPY TR C5 ceramide. (b) Rheb localization in cis-Golgi. Cells were fixed and immunostained with mouse anti-GM130 antibody followed by Alexa Fluor 594-conjugated anti-mouse secondary antibody. (c) Rheb localization in the ER. Cells were fixed and immunostained with rabbit anti-ERp72 antibody followed by Alexa Fluor 594-conjugated anti-rabbit secondary antibody. All images were acquired on a Zeiss confocal microscope using LSM software. Data shown are representative of more than 10 images from at least two independent experiments.
The CAAX motif of Rheb1, but not Rheb2, is sufficient for proper localization
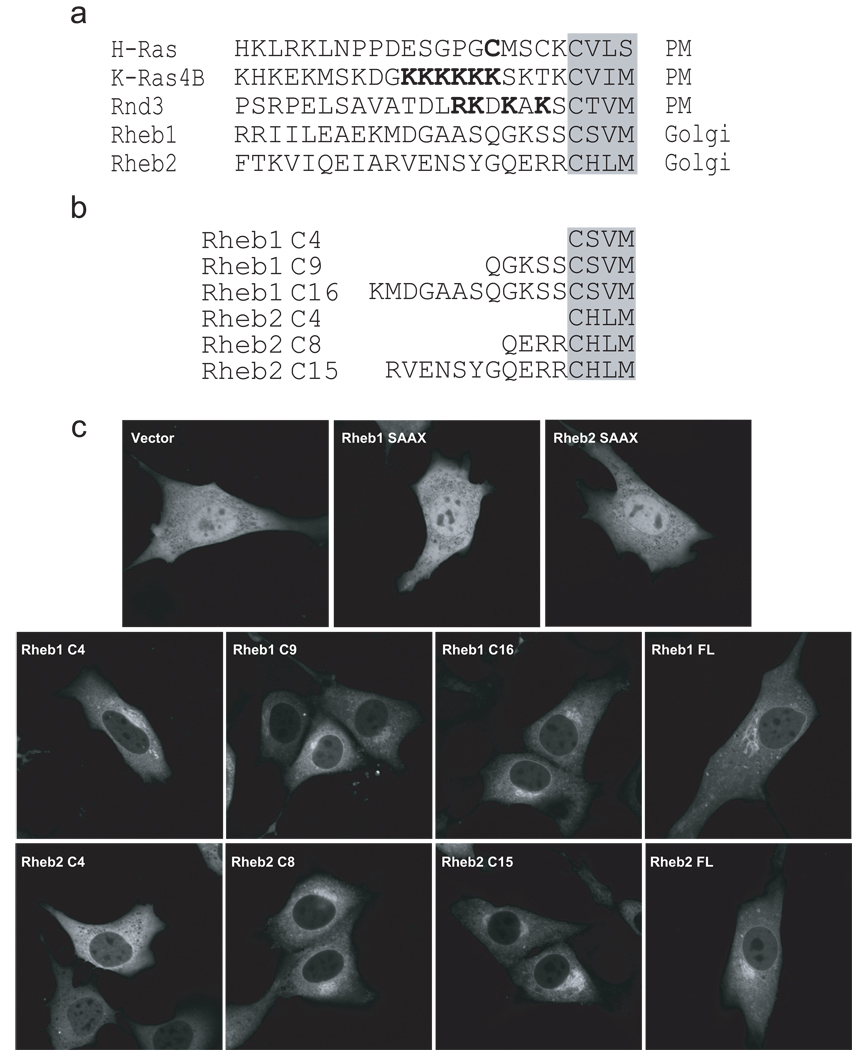
Next, we determined the minimal C-terminal amino acid sequences sufficient for proper Rheb localization. Since Rheb proteins lack either a cysteine or a polybasic amino acid-rich stretch upstream of their CAAX motifs (Figure 2a), we hypothesized that they lack a “second signal” for localization and that the CAAX motif alone would dictate proper subcellular localization. To address this possibility, we engineered expression constructs encoding the last 4 amino acids of either Rheb1 (CSVM; Rheb1 C4) or Rheb2 (CHLM; Rheb2 C4) fused to GFP (Figure 2b). We also created similar constructs with progressively longer C-terminal Rheb sequences to determine the minimum membrane targeting sequence of Rheb and transiently transfected them into NIH 3T3 cells. As controls, we used GFP-tagged full-length (FL) Rheb1 and Rheb2, as well as GFP-tagged Rheb1 C181S and Rheb2 C180S (“SAAX” mutants), which cannot be farnesylated, and hence display a nuclear and cytoplasmic localization identical to GFP alone (Figure 2c). Rheb1 C4, C9, and C16 all exhibited a subcellular localization indistinguishable from that of full length Rheb1, suggesting that the CAAX motif-signaled modifications alone are sufficient for proper Rheb1 localization (Figure 2c). Surprisingly, Rheb2 C4 did not localize to the distinct perinuclear region as seen with full length Rheb2, but instead exhibited a more diffuse cytoplasmic localization, and indicating that it was partially mislocalized, but still distinct from the localization of the unfarnesylated SAAX mutant. In contrast, Rheb2 C8 displayed a localization identical to the full length protein, suggesting that amino acid residues immediately upstream of the CAAX motif are required for proper Rheb2 localization to the Golgi.
Figure 2. CAAX-signaled modifications alone are sufficient for Rheb1 but not Rheb2 subcellular localization to the ER and Golgi.
(a) Comparison of C-terminal membrane targeting sequence elements of H-Ras, K-Ras4B, Rnd3, Rheb1, and Rheb2. Palmitoylated cysteines (in H-Ras) and polybasic residues (in K-Ras 4B and Rnd3) are underlined. The C-terminal CAAX motifs are shaded. (b) GFP fusion proteins terminating in the Rheb1 or Rheb2 C-terminal sequences shown were encoded by cDNA sequences subcloned into the pEGFP vector. (c) NIH 3T3 cells were transiently transfected with pEGFP vector, or with pEGFP encoding the following proteins: GFP-tagged full-length Rheb1 C181S (Rheb1 SAAX), full-length Rheb2 C180S (Rheb2 SAAX), the indicated C-terminal Rheb1 or Rheb2 sequences, or full-length (FL) Rheb1 or Rheb2. Live cells were imaged by confocal microscopy. Data shown are representative of two independent experiments.
Localization of farnesylated Rheb is more dependent on Rce1- and Icmt-catalyzed modifications than geranylgeranylated Rheb
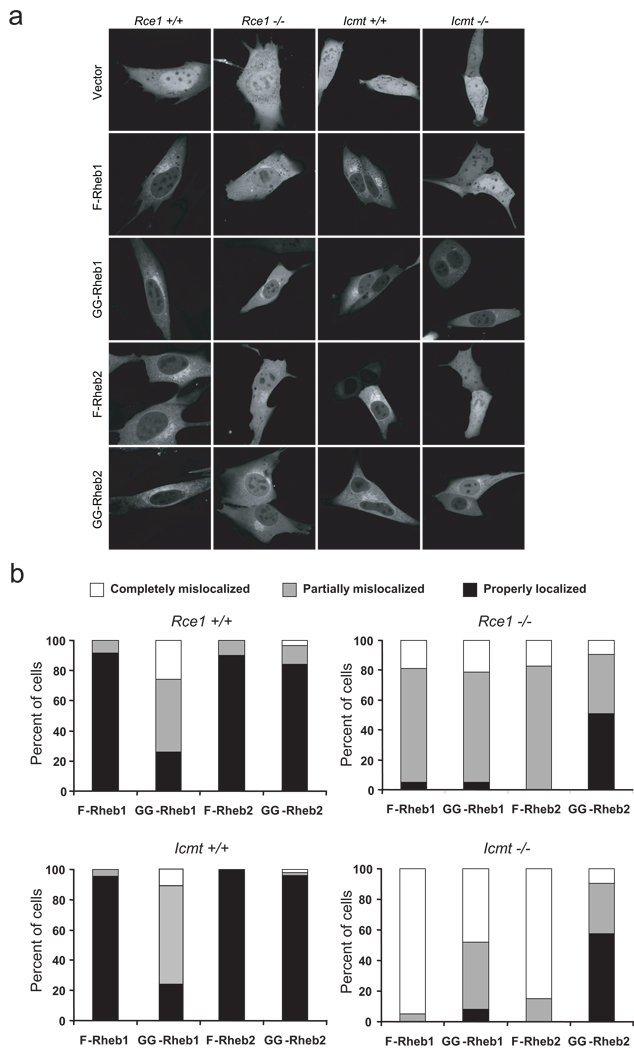
A previous study showed that Rheb1 localization is impaired in MEFs lacking Rce1 or Icmt, but whether Rheb1 function was impaired in these MEFs was not addressed (Takahashi et al., 2005). To determine if Rheb1-mediated activation of mTORC1 was dependent on these modifications, we first sought to confirm these localization findings, and to extend them to Rheb2. We transiently transfected wild-type (Rce1 +/+ and Icmt +/+), Rce1-deficient (Rce1 −/−), and Icmt-deficient (Icmt −/−) MEFs with expression vectors encoding GFP-Rheb1 and GFP-Rheb2. We recently utilized this set of MEFs and verified that the subcellular localication of GFP-tagged versions of Ras and Rho proteins accurately corresponded to the localization of their endogenously-expressed counterparts (Roberts et al., 2008). The localization of GFP-Rheb1 and GFP-Rheb2 was identical in two independent isolates of wild-type MEFs (Figure 3a). Consistent with previous results for Rheb1 (Takahashi et al., 2005), we found that the normal localization of both Rheb1 and Rheb2 was completely impaired in Icmt −/− MEFs. Both Rheb1 and Rheb2 showed significant nuclear accumulation and were indistinguishable from the subcellular distribution of GFP alone (Figures 3a and 3b). Rce1 deficiency is expected to prevent the subsequent Icmt modification. However, Rheb1 and Rheb2 were only partially mislocalized in Rce1 −/− MEFs: Rheb1 and Rheb2 accumulated in the nucleus and cytosol, but Golgi localization was still visible (Figures 3a and 3b). Thus, Rheb1 and Rheb2 are more dependent on Icmt-mediated methylation than on Rce1-mediated AAX cleavage, a pattern that we have observed with other farnesylated small GTPases (Roberts et al., 2008). We also observed an increase in the cytosolic fraction of endogenous Rheb1 in Rce1 −/− and Icmt −/− MEFs by subcellular fractionation (data not shown).
Figure 3. Rheb subcellular localization is dependent on Rce1- and Icmt-catalyzed modifications.
(a) Rce1 +/+, Rce1 −/−, Icmt +/+, and Icmt −/− MEFs were transiently transfected with pEGFP vector or with pEGFP encoding GFP-tagged Rheb1 (F-Rheb1), Rheb1 M184L (GG-Rheb1), Rheb2 (F-Rheb2), or Rheb2 M183L (GG-Rheb2). Live cells were imaged by confocal microscopy. (b) Quantification of the data shown in (a). At least 30 cells in each condition were scored.
The CAAX motifs of some GTPases, particularly Rho GTPases, are substrates for geranylgeranyltransferase-I (GGTase-I)-catalyzed addition of a longer C20 geranylgeranyl isoprenoid lipid. A previous study found that the localization of geranylgeranylated proteins is less dependent on Rce1- and Icmt-catalyzed modifications than farnesylated protein localization (Michaelson et al., 2005). Whether modification by the more hydrophobic geranylgeranyl group also reduces Rheb sensitivity to Icmt and Rce1 loss was not known. To obtain geranylgeranylated Rheb (GG-Rheb) mutants, we mutated the C-terminal amino acid of Rheb1 and Rheb2 from methionine to leucine (Rheb1 M184L and Rheb2 M183L). Similar CAAX mutants of S. pombe and human Rheb showed FTase-independent function (Basso et al., 2005; Gau et al., 2005; Mavrakis et al., 2008; Nakase et al., 2006). We utilized pharmacologic inhibitors of FTase and GGTase-I (GGTI) and confirmed that GG-Rheb was less sensitive to inhibition by FTI than wild-type farnesylated Rheb (F-Rheb; data not shown).
To determine whether localization of GG-Rheb also depends on Rce1- and Icmt-catalyzed modifications, we transiently transfected wild-type, Rce1 −/−, and Icmt −/− MEFs with GFP-tagged GG-Rheb1 and GG-Rheb2. As expected, in wild-type MEFs, GG-Rheb2 displayed identical subcellular localization to F-Rheb. However, we were surprised to find that GG-Rheb1 localization was similar to that seen with wild type F-Rheb1 in only a subset of cells. Instead, a majority (74–77%) of GG-Rheb1 expressing cells showed partially (48–66%) or completely (11–26%) mislocalized distributions (Figures 3a and 3b). Thus, unlike most Ras and Rho small GTPases, where the type of isoprenoid modification did not alter protein localization, geranylgeranyl modification did not substitute for farnesylation to fully support Rheb1 subcellular localization.
In Rce1 −/− MEFs, F-Rheb1 and GG-Rheb1 were partially mislocalized (74–76%) (Figure 3b). However, when expressed in Icmt −/− MEFs, a much lower percentage (48%) of GG-Rheb1 was completely mislocalized when compared to F-Rheb1 (95%). Similarly, a majority of GG-Rheb2 (>50%) retained proper localization in both Rce1 −/− and Icmt −/− cells, although all of the F-Rheb2 was partially or completely mislocalized in these cells. Thus, modification by a geranylgeranyl group reduces the requirement of both Rheb1 and Rheb2 for the subsequent CAAX modifications, particularly by Icmt.
Rheb activation of mTOR is more dependent on farnesylation than on postprenylation CAAX processing
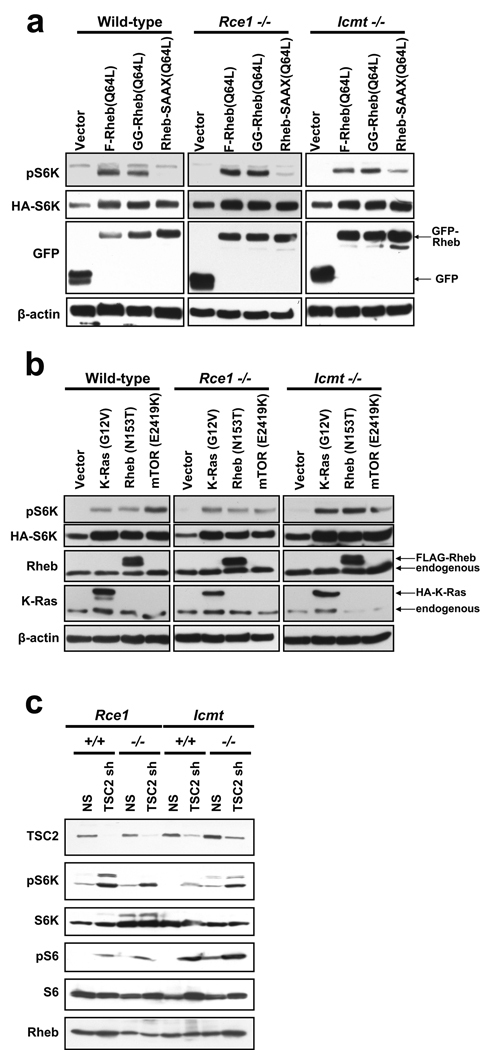
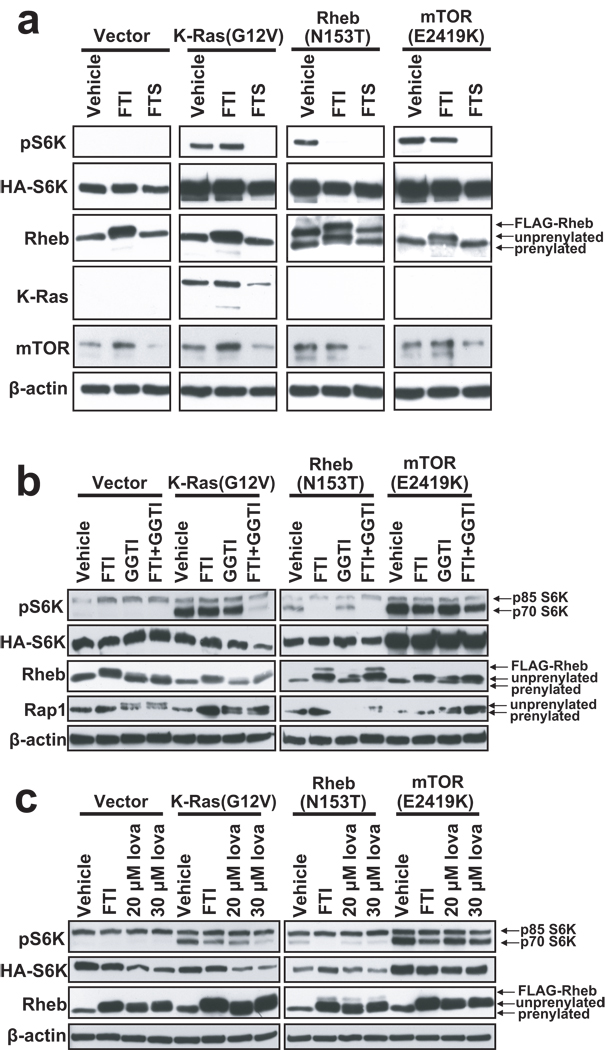
Since Rheb1 and Rheb2 are mislocalized in the absence of Rce1 and Icmt, we asked if Rheb signaling was impaired in Rce1 −/− and Icmt −/− MEFs. First, we wanted to confirm in our assays that Rheb activation of mTOR is dependent on farnesylation (Buerger et al., 2006). We transfected wild-type or knockout MEFs with constructs encoding GFP-tagged fusion proteins of an activated mutant of Rheb1 (Q64L) with an intact CAAX sequence or with two CAAX missense mutants: a geranylgeranylated variant (X=L; M184L), designated GG-Rheb1, and a nonfarnesylated mutant (C=S; C181S), designated Rheb1-SAAX. As expected, both F-Rheb and GG-Rheb efficiently induced S6K phosphorylation in wild-type Icmt +/+ MEFs (Figure 4a). We obtained similar results in Rce1 +/+ MEFs (data not shown). In agreement with previous observations (Basso et al., 2005; Buerger et al., 2006; Clark et al., 1997), we found that the nonfarnesylated Rheb1-SAAX mutant was greatly impaired in stimulating S6K phosphorylation.
Figure 4. Rce1- and Icmt-catalyzed modifications are not required for Rheb1 activation of S6K.
(a) Wild-type (Icmt +/+) Rce1 −/−, and Icmt −/− mouse embryo fibroblasts were transiently transfected with pEGFP vector or with pEGFP encoding Rheb1 Q64L (F-Rheb1 64L), Rheb1 M184L/Q64L (GG-Rheb1 64L), or Rheb1 C181S/Q64L (Rheb1-SAAX 64L), along with pRK7 HA-S6K1, and serum- and amino-acid starved as described in Materials & Methods. Cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies to phospho-S6K, HA, GFP, or β-actin (loading control). (b) Wild-type (Icmt +/+), Rce1 −/−, and Icmt −/− MEFs were transiently transfected with pcDNA3 vector, or with pcDNA3 encoding HA-tagged K-Ras G12V, FLAG-tagged Rheb1 N153T, or AU1-tagged mTOR E2419K, along with pRK7 HA-tagged S6K1, and serum- and amino acid-starved as in (a). Cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies to phospho-S6K, HA, Rheb1, K-Ras, or β-actin (loading control). (c) Rce1 +/+, Rce1 −/−, Icmt +/+, and Icmt −/− MEFs were stably infected with either non-specific shRNA (NS) or shRNA targeting mouse TSC2 (TSC2 sh). Cells were lysed 5 d post-infection and lysates were immunoblotted with antibodies to TSC2, phospho-S6K, total S6K, phospho-S6, total S6, or Rheb1. Data shown are representative of at least two independent experiments.
Since Rheb1-SAAX fails to undergo all three CAAX-signaled modifications, we next determined a role for the Rce1- and Icmt-catalyzed modifications in Rheb1 function. Surprisingly, both F-Rheb and GG-Rheb retained the ability to stimulate S6K phosphorylation when expressed in either Rce1 −/− or Icmt −/− MEFs (Figure 4a).
Since these observations were unexpected, we also compared the activity of Rheb1 with a different N-terminal tag (FLAG) and activating mutation (N153T) (Urano et al., 2005) with that of constitutively activated K-Ras4B(G12V), which is expected to require these modifications, and with that of constitutively activated mTOR (E2419K; a kinase-domain mutation) (Urano et al., 2007), which is not a substrate for either CAAX modifying enzyme. We transiently transfected wild-type, Rce1 −/−, and Icmt −/− MEFs with either pcDNA3 empty vector, or with pcDNA3 encoding constitutively activate mutants of K-Ras (HA-K-Ras G12V), Rheb1 (FLAG-Rheb N153T), or mTOR (AU1-mTOR E2419K), along with pRK7 HA-S6K1, and used western blot analysis to determine S6K phosphorylation. Consistent with previous studies (Sato et al., 2008; Urano et al., 2005; Urano et al., 2007), we observed that activated K-Ras4B, Rheb1, and mTOR all efficiently induced phosphorylation of S6K in wild type MEFs. As expected, since mTOR is not a substrate for Rce1 and Icmt, constitutively active mTOR also induced S6K phosphorylation in Rce1 −/− and Icmt −/− cells (Figure 4b). Unexpectedly, K-Ras4B also did not show impaired S6K phosphorylation when expressed in Rce1 −/− or Icmt −/− MEFs. However, this result is consistent with our recent observation that K-Ras4B subcellular localization and plasma membrane association were not impaired to any significant degree in Rce1- or Icmt-null cells (Roberts et al., 2008). Consistent with GFP-tagged Rheb1, FLAG-tagged Rheb-induced S6K phosphorylation was also not impaired in Rce1 −/− or Icmt −/− MEFs. Thus, while these CAAX modifications are necessary for proper subcellular localization, Rheb activation of mTOR is not impaired in the absence of Rce1 or Icmt function. Taken together, these data suggest that Rheb activation of mTOR is more dependent on farnesylation than on Rce1- and Icmt-catalyzed modifications.
Postprenylation CAAX processing is not required for activation of mTOR signaling by endogenous Rheb
The above results suggested that Rce1- and Icmt-catalyzed modifications are required for Rheb localization, but not for Rheb activation of mTOR. However, the above experiments were performed using ectopically expressed Rheb protein, which may not accurately reflect endogenous Rheb activity. Therefore, to determine whether endogenous Rheb activation is impaired in the absence of Rce1 and Icmt, we stably infected wild-type, Rce1 −/−, and Icmt −/− MEFs with a previously described short hairpin RNA (shRNA) targeting mouse TSC2 (Peterson et al., 2009), encoding the Rheb GAP catalytic subunit. The TSC2 shRNA strongly reduced endogenous TSC2 expression in all MEFs (Figure 4c). As expected, in both isolates of wild-type MEFs, knocking down TSC2 increased phosphorylation of S6K and its substrate, the ribosomal subunit S6 (Figure 4c). Similarly, phosphorylation of S6K and S6 was increased in Rce1 −/− and Icmt −/− MEFs expressing TSC2 shRNA. These results are consistent with our experiments performed using ectopically expressed Rheb and suggest that Rce1- and Icmt-catalyzed modifications are not required for activation of endogenous Rheb-mTOR signaling.
FTS does not disrupt the subcellular localization of Rheb
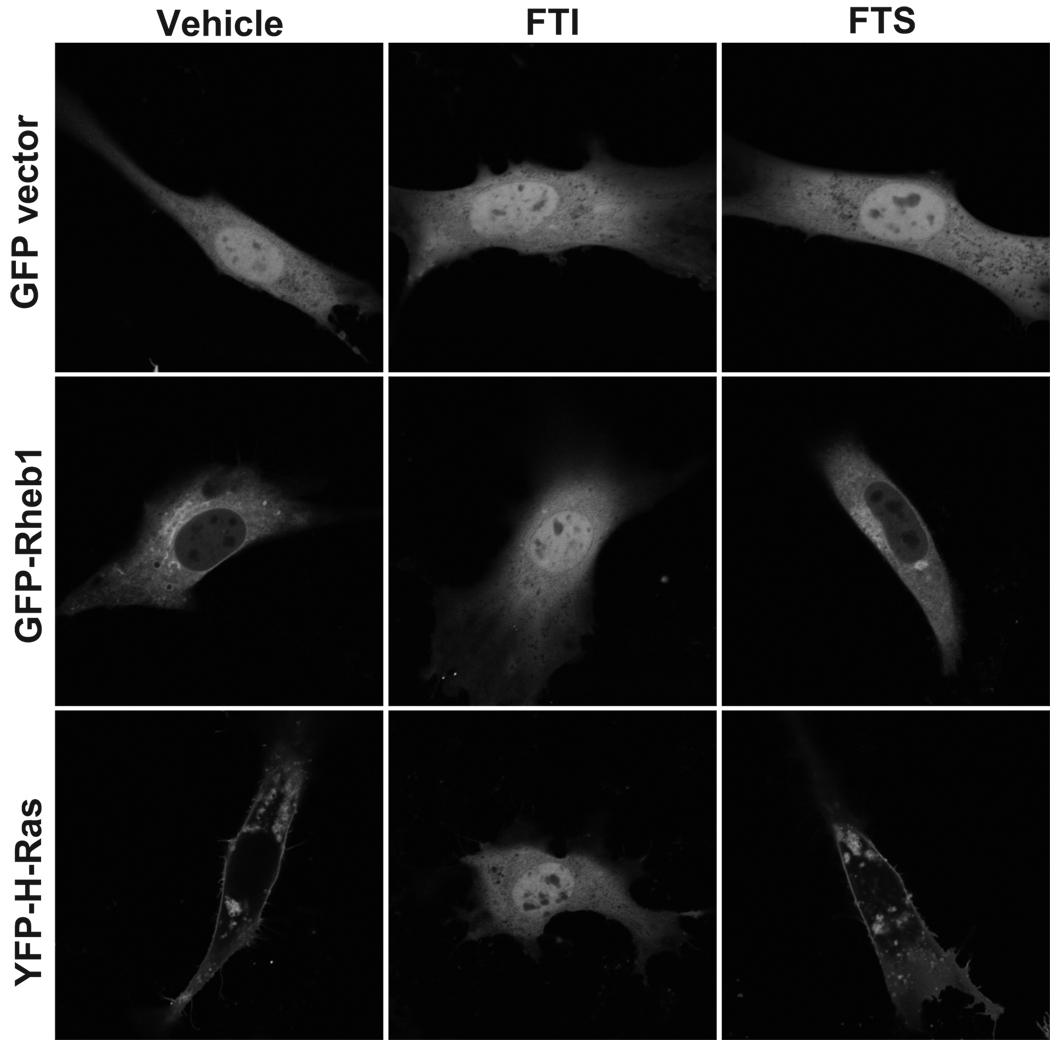
FTS (also called salirasib) is a farnesylcysteine mimetic that is proposed to dislodge Ras proteins from the plasma membrane and thus inhibit Ras activity (Blum et al., 2008; Kloog and Cox, 2004). However, whether FTS also displaces and inhibits the function of other farnesylated proteins, such as Rheb, is not known. FTS has also been shown to reduce phosphorylation of the mTOR substrates S6K and 4E-BP1 (Yue et al., 2007; Yue et al., 2005), but whether this is due to Rheb inhibition was not investigated. To determine if FTS inhibits Rheb localization, we transiently expressed GFP-tagged Rheb1 or YFP-tagged H-Ras in NIH 3T3 cells and treated them with FTS. As a control, we treated cells with FTI-2153, since FTIs are known to impair Rheb and H-Ras localization (Buerger et al., 2006; Roberts et al., 2008; Takahashi et al., 2005). Indeed, GFP-Rheb and YFP-H-Ras were completely mislocalized in FTI-treated cells (Figure 5). Surprisingly, FTS did not cause mislocalization of either H-Ras or Rheb. We obtained similar results with endogenous Rheb and Ras subcellular localization when evaluated by subcellular fractionation (data not shown). These results suggest that FTS does not detectably disrupt the membrane association and subcellular localization of Rheb or Ras proteins under these conditions.
Figure 5. FTS does not disrupt Rheb or H-Ras subcellular localization.
(a) NIH 3T3 cells were transiently transfected with pEGFP vector, pEGFP encoding GFP-tagged Rheb1, or pEYFP encoding YFP-tagged H-Ras and treated with DMSO (Vehicle), 5 µM FTI-2153, or 75 µM FTS overnight. Live cells were imaged by confocal microscopy.
FTS inhibits mTOR-induced S6K activation and reduces mTOR protein levels
Since it remained possible that FTS may disrupt Rheb interaction with mTOR, we next asked if FTS inhibits Rheb-induced activation of mTOR. Activated K-Ras4B, Rheb1, and mTOR all efficiently induced S6K phosphorylation in vehicle (DMSO)-treated cells (Figure 6a). As expected, FTI treatment strongly inhibited Rheb1-induced S6K phosphorylation and resulted in a shift in Rheb mobility as seen by SDS-PAGE, indicating the accumulation of unprenylated Rheb1. We also observed an increase in Rheb protein levels in FTI-treated cells, in agreement with previous studies (Buerger et al., 2006). Importantly, FTI treatment did not block mTOR-induced S6K phosphorylation, since mTOR E2419K is constitutively active and is not dependent on Rheb. In contrast, FTS strongly inhibited K-Ras, Rheb, and mTOR-induced S6K phosphorylation, suggesting that FTS inhibits S6K activation at the level of mTOR, and not by inhibiting upstream activators.
Figure 6. FTS inhibits mTOR-induced S6K activation by a mechanism independent of prenylated proteins.
(a) NIH 3T3 cells were transiently transfected with pcDNA3 vector, or with pcDNA3 encoding HA-tagged K-Ras G12V, FLAG-tagged Rheb1 N153T, or AU1-tagged mTOR E2419K, along with pRK7 HA-tagged S6K1. Three h following transfection, medium was replaced with complete medium containing either DMSO (Vehicle) or 5 µM FTI-2153. Twenty-four h following transfection, cells were serum-starved in DMEM supplemented with 0.1% BSA containing DMSO, 5 µM FTI-2153, or 75 µM FTS overnight, and then amino-acid starved in PBS containing the indicated compound for 1 h. Cell lysates were resolved by SDS-PAGE and immunoblotting was performed with antibodies to phospho-S6K, HA, Rheb1, K-Ras, or β-actin (loading control). Data are representative of three independent experiments. (b) NIH 3T3 cells were transiently transfected with the indicated plasmids, as described above. Three h following transfection, medium was replaced with complete medium supplemented with DMSO, 10 µM FTI-2153, 20 µM GGTI-2417, or 10 µM FTI + 20 µM GGTI. Cells were serum- and amino-acid starved as in (a). Cell lysates were resolved by SDS-PAGE and immunoblotting was performed with antibodies to phospho-S6K, HA, Rheb1, K-Ras, Rap1, or β-actin (loading control). (c) NIH 3T3 cells were transiently transfected with the indicated plasmids. Three h following transfection, medium was replaced with complete medium supplemented with DMSO (Vehicle), 10 µM FTI-2153, 20 µM lovastatin, or 30 µM lovastatin. Serum- and amino acid-starved cell lysates were resolved by SDS-PAGE and immunoblotting was performed with antibodies to phospho-S6K, HA, Rheb1, K-Ras, H-Ras, or β-actin (loading control). Data shown are representative of at least two independent experiments.
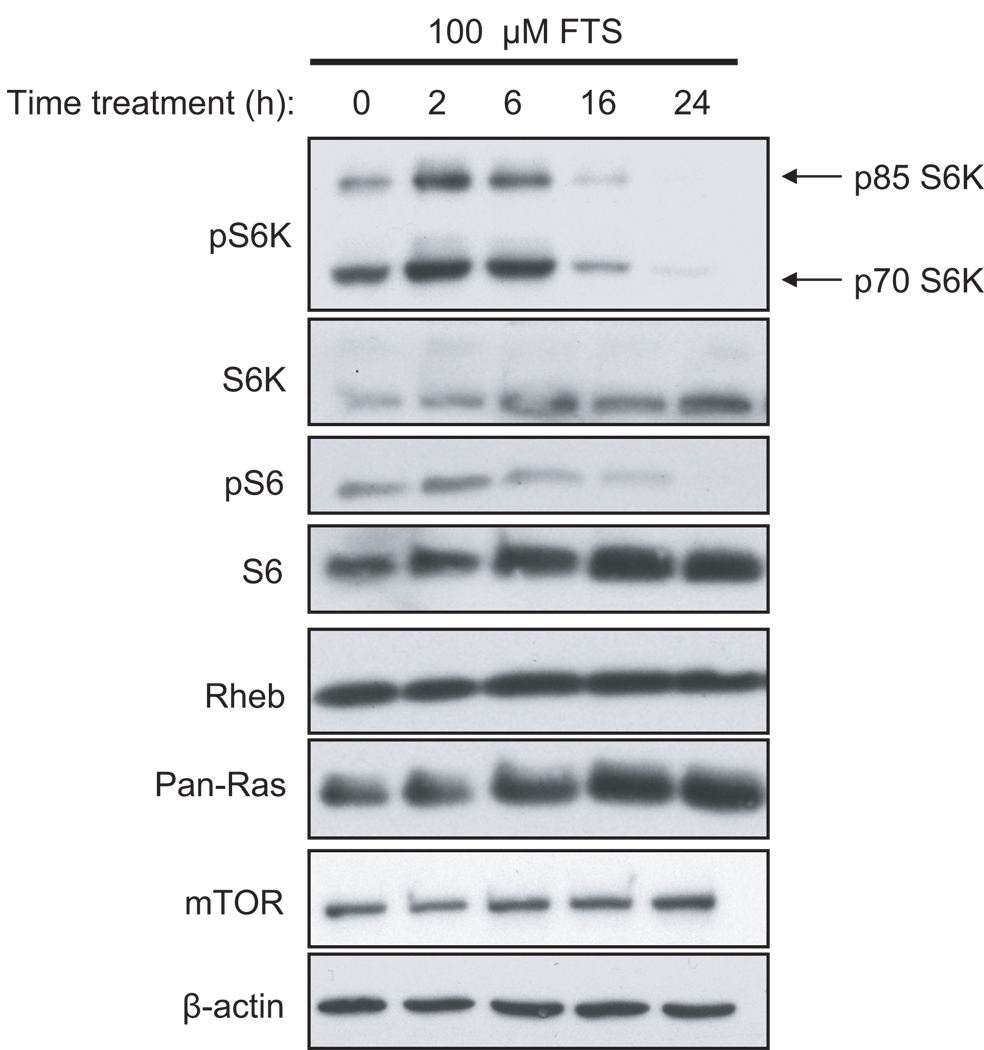
Surprisingly, we found that FTS treatment of NIH 3T3 cells decreased levels of both endogenous and ectopically expressed mTOR protein (Figure 6a), but did not decrease levels of K-Ras, Rheb, or β-actin. Thus, FTS inhibition of mTOR may be due, in part, to promoting the loss of mTOR protein. However, FTS treatment did not decrease endogenous mTOR in MCF-7 human breast carcinoma cells, whereas it did decrease phospho-S6K in these cells (Figure 7, discussed below), suggesting that the FTS-induced reduction in mTOR levels is not the primary cause of the reduction in S6K phosphorylation.
Figure 7. FTS blocks endogenous S6K activation in MCF-7 cells.
MCF-7 cells growing in complete medium were treated with 100 µM FTS in complete medium for 0, 2, 6, 16, or 24 h. Cell lysates were resolved by SDS-PAGE and immunoblotted with antibodies to phospho-S6K, total S6K, phospho-S6, total S6, mTOR, and β-actin (loading control). Data shown are representative of two independent experiments.
FTS inhibition of mTOR does not require protein prenylation
One explanation for the above results could be that FTS is blocking the function of an unidentified prenylated protein required for mTOR-induced S6K phosphorylation. For example, Cdc42 and Rac1, which are geranylgeranylated, have previously been implicated in S6K phosphorylation (Chou and Blenis, 1996). In addition, RalA, another geranylgeranylated protein, was recently shown to be required for Rheb-induced S6K phosphorylation (Maehama et al., 2008). To determine whether prenylated proteins are required for mTOR-induced S6K phosphorylation, we transiently transfected NIH 3T3 cells as described above, and treated cells with FTI, a GGTase-I inhibitor (GGTI), or both. We verified that the GGTI was functional and inhibited modification of Rap1A (Figure 6b). As expected, FTI inhibited Rheb1-induced, but not mTOR-induced, S6K phosphorylation (Figure 6b). Combined treatment with FTI and GGTI efficiently inhibited K-Ras4B-induced S6K phosphorylation, presumably as a consequence of blocking alternative prenylation of K-Ras4B (Rowell et al., 1997; Whyte et al., 1997). In contrast, FTI and GGTI in combination failed to prevent mTOR-induced S6K phosphorylation, suggesting that prenylated proteins modified by either farnesyltransferase or GGTase-I are not required for mTOR activation of S6K.
To confirm these results and to exclude the possibility that prenylated GGTase-II substrates (such as Rab GTPases) are required for mTOR activation of S6K, we treated cells with lovastatin, which blocks all protein prenylation. As expected, lovastatin treatment caused a mobility shift in Rheb, and reduced both Rheb- and K-Ras-induced S6K phosphorylation (Figure 6c). However, lovastatin failed to inhibit mTOR-induced S6K phosphorylation. Thus, FTS inhibition of mTOR does not involve inhibition of a prenylated protein.
FTS inhibits endogenous S6K activity in breast cancer cells
As described previously (Yue et al., 2007), we confirmed that FTS inhibited endogenous S6K activity in MCF-7 cells, but treatment did not affect mTOR or Rheb1 protein levels (Figure 7). These data are consistent with the hypothesis that FTS is an mTOR inhibitor.
Discussion
Rheb functions as a key intermediate in the PI3K-AKT-TSC-Rheb-mTOR network that is aberrantly activated in a majority of human cancers (Cully et al., 2006). While pharmacologic inhibitors of PI3K, AKT and mTOR are currently in Phase I/II clinical trials, Rheb may also be a useful target. A key goal of our study was to determine if specific approaches can block Rheb function by disrupting the membrane association and subcellular distribution of Rheb essential for its biological activity. We assessed two different strategies for blocking Rheb function. The first strategy involves preventing the CAAX-mediated posttranslational modifications that promote Rheb membrane association. We found that both Rheb1 and Rheb2 localization, but not mTOR activation, was disrupted in the absence of the post-prenylation processing enzymes Rce1 or Icmt, suggesting that inhibitors of these enzymes will not block Rheb signaling and oncogenic activity. The second strategy involves inhibition of Rheb endomembrane association by FTS. Unexpectedly, we found that FTS did not effectively impair Rheb or Ras protein membrane association. Instead, we found that FTS blocked Rheb activation of mTOR, but this effect was independent of inhibition of Rheb or of any other prenylated protein. Rather, we suggest that a potent mechanism for FTS-mediated growth inhibition may involve direct inhibition of mTOR.
Although the subcellular localization of Rheb proteins has been addressed in multiple previous studies, conflicting observations were made (Buerger et al., 2006; Jiang and Vogt, 2008; Ma et al., 2008; Sancak et al., 2008; Takahashi et al., 2005). Furthermore, the precise subcellular localization of Rheb2 had not been determined. We confirmed that Rheb1 is primarily localized to the Golgi and ER endomembranes, in agreement with others (Buerger et al., 2006; Jiang and Vogt, 2008). We also found that the localization of Rheb2 is indistinguishable from that of Rheb1, and that Rheb localization overlaps with the observed localization of Rheb binding partners (Drenan et al., 2004; Du et al., 2003; Wienecke et al., 1996). The essentially identical subcellular localization of Rheb1 and Rheb2 contrasts with the situation seen with closely related isoforms of other Ras family small GTPases. For example, although RalA and RalB share 82% overall sequence identity, their distinct subcellular distributions result in strikingly distinct biological roles (Bodemann and White, 2008).
We found that, unlike most Ras and Rho small GTPases, the CAAX motif alone of Rheb1 was sufficient for proper localization. These data are consistent with Choy et al., who showed that the CAAX motif of K-Ras and N-Ras alone promoted localization to the ER and Golgi (Choy et al., 1999). In contrast, the CAAX motif alone was not sufficient for proper localization of Rheb2, which required amino acids immediately upstream of the CAAX motif, suggesting that the targeting motifs of Rheb1 and Rheb2 are not functionally identical.
We also found that geranylgeranylated mutants of Rheb1 and Rheb2 (Rheb1 M184L and Rheb2 M183L), like their farnesylated wild-type counterparts, localized to the ER and Golgi. However, the localization of GG-Rheb was less dependent on Icmt- and Rce-1 catalyzed modifications, in agreement with previous studies showing that the localization of geranygeranylated Ras mutants was less dependent on these modifications than farnesylated Ras (Michaelson et al., 2005).
Farnesylation of Rheb was required for its ability to activate mTOR, as shown by the inability of the nonfarnesylated SAAX mutant of Rheb1 to induce S6K phosphorylation. These observations are consistent with several studies showing that FTI impairs S6K or S6 phosphorylation (Basso et al., 2005; Gau et al., 2005; Law et al., 2000; Mavrakis et al., 2008). In contrast, although Rce and Icmt were required for proper Rheb localization, neither of the post-prenyl processing steps was required for Rheb-induced mTOR activation. These results are surprising, since they suggest that Rheb does not need to be localized properly in order to activate mTOR. Future experiments should determine whether co-immunoprecipitation of endogenous Rheb and mTOR is disrupted in the absence of Rce1 and Icmt. Although signaling pathway activation can vary substantially between independently immortalized MEF cell lines, we observed identical results in two different wild-type MEF isolates. Furthermore, we found that the Icmt inhibitor cysmethynil did not block Rheb activation of mTOR (data not shown). These data are consistent with those of Buerger et al., who showed that the methyltransferase inhibitor AFC does not block Rheb-induced S6K activation (Buerger et al., 2006). Therefore, we predict that inhibitors of Rce1 and Icmt will not inhibit Rheb activity.
We found that FTS strongly inhibited both Rheb- and mTOR-induced S6K activation. FTS is proposed to compete with farnesylated Ras for membrane binding sites and dislodge Ras from the membrane, thereby disrupting its function (Haklai et al., 1998; Marom et al., 1995; Rotblat et al., 2008; Weisz et al., 1999). However, we failed to observe detectable disruption of H-Ras or Rheb membrane association. Instead, our data strongly suggest that the FTS-mediated inhibition of mTOR-induced S6K activation is not due to inhibition of a prenylated protein. Rather, our data are consistent with the observation that FTS disrupts the association of mTOR with raptor (McMahon et al., 2005). We failed to observe a concomitant decrease in Akt phosphorylation on S473 in FTS-treated cells (data not shown), suggesting that FTS inhibits mTORC1 but not mTOR complex 2 (mTORC2) activity.
In summary, we have found that inhibition of Rce1- and Icmt-catalyzed modifications does not prevent Rheb activation of mTOR, and that FTS inhibits mTOR through a mechanism that does not involve Rheb inhibition. Although FTS is not a direct Rheb inhibitor, mTOR is essential for Rheb oncogenic activity (Jiang and Vogt, 2008; Mavrakis et al., 2008); hence, FTS would be expected to disrupt both Rheb- and mTOR-induced transformation. Since FTS is currently in clinical trials for cancer treatment, our results have significant implications for the selection of patients who may benefit from FTS therapy and suggest that S6K phosphorylation may be a useful biomarker to monitor FTS activity.
Materials and methods
See supplementary information for complete methods.
Supplementary Material
GFP-Rheb1 and GFP-Rheb2 do not localize to early endosomes, late endosomes, lysosomes, or mitochondria. COS-7 cells were transiently transfected with pEGFP vector or with pEGFP encoding GFP-tagged Rheb1 or Rheb2. (a) Live cells were imaged after staining with 500 nM MitoTracker dye (500 nM). (b) Prior to fixation, cells were stained with 75 nM LysoTracker dye for 2 h. (c–f) Cells were fixed and immunostained with (c) mouse anti-EEA1 antibody followed by Alexa Fluor 594-conjugated anti-mouse secondary antibody, (d) mouse anti-LAMP2 antibody followed by Alexa Fluor 594-conjugated anti-mouse secondary antibody, (e) Alexa Fluor 594-conjugated anti-mouse secondary antibody alone, (f) Alexa Fluor 594-conjugated anti-rabbit secondary antibody alone. All images were acquired on a Zeiss confocal microscope using LSM software.
Acknowledgements
We thank Yoel Kloog, Saïd Sebti, and Andrew Hamilton for inhibitors, Lawrence Quilliam and Mark Philips for providing plasmid constructs, Stephen Young for MEF cell lines deficient in Rce1 and Icmt, and the UNC Michael Hooker Microscopy Facility for imaging assistance.
Funding: Our research was supported by grants from the National Institutes of Health to CJD (CA042978), ADC (CA109550), CJD and ADC (CA67771), and FT (CA41996). ABH was supported by a Department of Defense Breast Cancer Research Program predoctoral fellowship (BC061107).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Aspuria PJ, Tamanoi F. The Rheb family of GTP-binding proteins. Cell Signal. 2004;16:1105–1112. doi: 10.1016/j.cellsig.2004.03.019. [DOI] [PubMed] [Google Scholar]
- Astrinidis A, Henske EP. Tuberous sclerosis complex: linking growth and energy signaling pathways with human disease. Oncogene. 2005;24:7475–7481. doi: 10.1038/sj.onc.1209090. [DOI] [PubMed] [Google Scholar]
- Basso AD, Mirza A, Liu G, Long BJ, Bishop WR, Kirschmeier P. The farnesyl transferase inhibitor (FTI) SCH66336 (lonafarnib) inhibits Rheb farnesylation and mTOR signaling. Role in FTI enhancement of taxane and tamoxifen anti-tumor activity. J Biol Chem. 2005;280:31101–31108. doi: 10.1074/jbc.M503763200. [DOI] [PubMed] [Google Scholar]
- Blum R, Cox AD, Kloog Y. Inhibitors of chronically active ras: potential for treatment of human malignancies. Recent Patents Anticancer Drug Discov. 2008;3:31–37. doi: 10.2174/157489208783478702. [DOI] [PubMed] [Google Scholar]
- Bodemann BO, White MA. Ral GTPases and cancer: linchpin support of the tumorigenic platform. Nat Rev Cancer. 2008;8:133–140. doi: 10.1038/nrc2296. [DOI] [PubMed] [Google Scholar]
- Buerger C, DeVries B, Stambolic V. Localization of Rheb to the endomembrane is critical for its signaling function. Biochem Biophys Res Commun. 2006;344:869–880. doi: 10.1016/j.bbrc.2006.03.220. [DOI] [PubMed] [Google Scholar]
- Castro AF, Rebhun JF, Clark GJ, Quilliam LA. Rheb binds tuberous sclerosis complex 2 (TSC2) and promotes S6 kinase activation in a rapamycin- and farnesylation-dependent manner. J Biol Chem. 2003;278:32493–32496. doi: 10.1074/jbc.C300226200. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Mohiyuddin SA, Gopinath KS, Kumar A. Involvement of TSC genes and differential expression of other members of the mTOR signaling pathway in oral squamous cell carcinoma. BMC Cancer. 2008;8:163. doi: 10.1186/1471-2407-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chenette EJ, Mitin NY, Der CJ. Multiple sequence elements facilitate Chp Rho GTPase subcellular location, membrane association, and transforming activity. Mol Biol Cell. 2006;17:3108–3121. doi: 10.1091/mbc.E05-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MM, Blenis J. The 70 kDa S6 kinase complexes with and is activated by the Rho family G proteins Cdc42 and Rac1. Cell. 1996;85:573–583. doi: 10.1016/s0092-8674(00)81257-x. [DOI] [PubMed] [Google Scholar]
- Choy E, Chiu VK, Silletti J, Feoktistov M, Morimoto T, Michaelson D, et al. Endomembrane trafficking of ras: the CAAX motif targets proteins to the ER and Golgi. Cell. 1999;98:69–80. doi: 10.1016/S0092-8674(00)80607-8. [DOI] [PubMed] [Google Scholar]
- Clark GJ, Kinch MS, Rogers-Graham K, Sebti SM, Hamilton AD, Der CJ. The Ras-related protein Rheb is farnesylated and antagonizes Ras signaling and transformation. J Biol Chem. 1997;272:10608–10615. doi: 10.1074/jbc.272.16.10608. [DOI] [PubMed] [Google Scholar]
- Cox AD, Der CJ. Farnesyltransferase inhibitors: promises and realities. Curr Opin Pharmacol. 2002;2:388–393. doi: 10.1016/s1471-4892(02)00181-9. [DOI] [PubMed] [Google Scholar]
- Cully M, You H, Levine AJ, Mak TW. Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat Rev Cancer. 2006;6:184–192. doi: 10.1038/nrc1819. [DOI] [PubMed] [Google Scholar]
- Drenan RM, Liu X, Bertram PG, Zheng XF. FKBP12-rapamycin-associated protein or mammalian target of rapamycin (FRAP/mTOR) localization in the endoplasmic reticulum and the Golgi apparatus. J Biol Chem. 2004;279:772–778. doi: 10.1074/jbc.M305912200. [DOI] [PubMed] [Google Scholar]
- Du G, Altshuller YM, Vitale N, Huang P, Chasserot-Golaz S, Morris AJ, et al. Regulation of phospholipase D1 subcellular cycling through coordination of multiple membrane association motifs. J Cell Biol. 2003;162:305–315. doi: 10.1083/jcb.200302033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gau CL, Kato-Stankiewicz J, Jiang C, Miyamoto S, Guo L, Tamanoi F. Farnesyltransferase inhibitors reverse altered growth and distribution of actin filaments in Tsc-deficient cells via inhibition of both rapamycin-sensitive and -insensitive pathways. Mol Cancer Ther. 2005;4:918–926. doi: 10.1158/1535-7163.MCT-04-0347. [DOI] [PubMed] [Google Scholar]
- Gromov PS, Madsen P, Tomerup N, Celis JE. A novel approach for expression cloning of small GTPases: identification, tissue distribution and chromosome mapping of the human homolog of rheb. FEBS Lett. 1995;377:221–226. doi: 10.1016/0014-5793(95)01349-0. [DOI] [PubMed] [Google Scholar]
- Haklai R, Weisz MG, Elad G, Paz A, Marciano D, Egozi Y, et al. Dislodgment and accelerated degradation of Ras. Biochemistry. 1998;37:1306–1314. doi: 10.1021/bi972032d. [DOI] [PubMed] [Google Scholar]
- Heo WD, Meyer T. Switch-of-function mutants based on morphology classification of Ras superfamily small GTPases. Cell. 2003;113:315–328. doi: 10.1016/s0092-8674(03)00315-5. [DOI] [PubMed] [Google Scholar]
- Huang J, Manning BD. The TSC1-TSC2 complex: a molecular switchboard controlling cell growth. Biochem J. 2008;412:179–190. doi: 10.1042/BJ20080281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoki K, Corradetti MN, Guan KL. Dysregulation of the TSC-mTOR pathway in human disease. Nat Genet. 2005;37:19–24. doi: 10.1038/ng1494. [DOI] [PubMed] [Google Scholar]
- Jiang H, Vogt PK. Constitutively active Rheb induces oncogenic transformation. Oncogene. 2008;27:5729–5740. doi: 10.1038/onc.2008.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloog Y, Cox AD. Prenyl-binding domains: potential targets for Ras inhibitors and anti-cancer drugs. Semin Cancer Biol. 2004;14:253–261. doi: 10.1016/j.semcancer.2004.04.004. [DOI] [PubMed] [Google Scholar]
- Konstantinopoulos PA, Karamouzis MV, Papavassiliou AG. Post-translational modifications and regulation of the RAS superfamily of GTPases as anticancer targets. Nat Rev Drug Discov. 2007;6:541–555. doi: 10.1038/nrd2221. [DOI] [PubMed] [Google Scholar]
- Law BK, Norgaard P, Moses HL. Farnesyltransferase inhibitor induces rapid growth arrest and blocks p70s6k activation by multiple stimuli. J Biol Chem. 2000;275:10796–10801. doi: 10.1074/jbc.275.15.10796. [DOI] [PubMed] [Google Scholar]
- Long X, Lin Y, Ortiz-Vega S, Yonezawa K, Avruch J. Rheb binds and regulates the mTOR kinase. Curr Biol. 2005;15:702–713. doi: 10.1016/j.cub.2005.02.053. [DOI] [PubMed] [Google Scholar]
- Ma D, Bai X, Guo S, Jiang Y. The switch I region of Rheb is critical for its interaction with FKBP38. J Biol Chem. 2008;283:25963–25970. doi: 10.1074/jbc.M802356200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maehama T, Tanaka M, Nishina H, Murakami M, Kanaho Y, Hanada K. RalA functions as an indispensable signal mediator for the nutrient-sensing system. J Biol Chem. 2008;283:35053–35059. doi: 10.1074/jbc.M805822200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marom M, Haklai R, Ben-Baruch G, Marciano D, Egozi Y, Kloog Y. Selective inhibition of Ras-dependent cell growth by farnesylthiosalisylic acid. J Biol Chem. 1995;270:22263–22270. doi: 10.1074/jbc.270.38.22263. [DOI] [PubMed] [Google Scholar]
- Mavrakis KJ, Zhu H, Silva RL, Mills JR, Teruya-Feldstein J, Lowe SW, et al. Tumorigenic activity and therapeutic inhibition of Rheb GTPase. Genes Dev. 2008;22:2178–2188. doi: 10.1101/gad.1690808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon LP, Yue W, Santen RJ, Lawrence JC., Jr Farnesylthiosalicylic acid inhibits mammalian target of rapamycin (mTOR) activity both in cells and in vitro by promoting dissociation of the mTOR-raptor complex. Mol Endocrinol. 2005;19:175–183. doi: 10.1210/me.2004-0305. [DOI] [PubMed] [Google Scholar]
- Michaelson D, Ali W, Chiu VK, Bergo M, Silletti J, Wright L, et al. Postprenylation CAAX processing is required for proper localization of Ras but not Rho GTPases. Mol Biol Cell. 2005;16:1606–1616. doi: 10.1091/mbc.E04-11-0960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaelson D, Silletti J, Murphy G, D'Eustachio P, Rush M, Philips MR. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakase Y, Fukuda K, Chikashige Y, Tsutsumi C, Morita D, Kawamoto S, et al. A defect in protein farnesylation suppresses a loss of Schizosaccharomyces pombe tsc2+, a homolog of the human gene predisposing to tuberous sclerosis complex. Genetics. 2006;173:569–578. doi: 10.1534/genetics.106.056895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nardella C, Chen Z, Salmena L, Carracedo A, Alimonti A, Egia A, et al. Aberrant Rheb-mediated mTORC1 activation and Pten haploinsufficiency are cooperative oncogenic events. Genes Dev. 2008;22:2172–2177. doi: 10.1101/gad.1699608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. DEPTOR is an mTOR inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. 2009;137:873–886. doi: 10.1016/j.cell.2009.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts PJ, Mitin N, Keller PJ, Chenette EJ, Madigan JP, Currin RO, et al. Rho Family GTPase modification and dependence on CAAX motif-signaled posttranslational modification. J Biol Chem. 2008;283:25150–25163. doi: 10.1074/jbc.M800882200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotblat B, Ehrlich M, Haklai R, Kloog Y. The ras inhibitor farnesylthiosalicylic Acid (salirasib) disrupts the spatiotemporal localization of active ras: a potential treatment for cancer. Methods Enzymol. 2008;439:467–489. doi: 10.1016/S0076-6879(07)00432-6. [DOI] [PubMed] [Google Scholar]
- Rowell CA, Kowalczyk JJ, Lewis MD, Garcia AM. Direct demonstration of geranylgeranylation and farnesylation of Ki-Ras in vivo. J Biol Chem. 1997;272:14093–14097. doi: 10.1074/jbc.272.22.14093. [DOI] [PubMed] [Google Scholar]
- Saito K, Araki Y, Kontani K, Nishina H, Katada T. Novel role of the small GTPase Rheb: its implication in endocytic pathway independent of the activation of mammalian target of rapamycin. J Biochem (Tokyo) 2005;137:423–430. doi: 10.1093/jb/mvi046. [DOI] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, et al. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Umetsu A, Tamanoi F. Characterization of the Rheb-mTOR Signaling Pathway in Mammalian Cells: Constitutive Active Mutants of Rheb and mTOR. Methods Enzymol. 2008;438:307–320. doi: 10.1016/S0076-6879(07)38021-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebti SM, Der CJ. Opinion: Searching for the elusive targets of farnesyltransferase inhibitors. Nat Rev Cancer. 2003;3:945–951. doi: 10.1038/nrc1234. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Nakagawa M, Young SG, Yamanaka S. Differential membrane localization of ERas and Rheb, two Ras-related proteins involved in the phosphatidylinositol 3-kinase/mTOR pathway. J Biol Chem. 2005;280:32768–32774. doi: 10.1074/jbc.M506280200. [DOI] [PubMed] [Google Scholar]
- Urano J, Comiso MJ, Guo L, Aspuria PJ, Deniskin R, Tabancay AP, Jr, et al. Identification of novel single amino acid changes that result in hyperactivation of the unique GTPase, Rheb, in fission yeast. Mol Microbiol. 2005;58:1074–1086. doi: 10.1111/j.1365-2958.2005.04877.x. [DOI] [PubMed] [Google Scholar]
- Urano J, Sato T, Matsuo T, Otsubo Y, Yamamoto M, Tamanoi F. Point mutations in TOR confer Rheb-independent growth in fission yeast and nutrient-independent mammalian TOR signaling in mammalian cells. Proc Natl Acad Sci U S A. 2007;104:3514–3519. doi: 10.1073/pnas.0608510104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz B, Giehl K, Gana-Weisz M, Egozi Y, Ben-Baruch G, Marciano D, et al. A new functional Ras antagonist inhibits human pancreatic tumor growth in nude mice. Oncogene. 1999;18:2579–2588. doi: 10.1038/sj.onc.1202602. [DOI] [PubMed] [Google Scholar]
- Whyte DB, Kirschmeier P, Hockenberry TN, Nunez-Oliva I, James L, Catino JJ, et al. K- and N-Ras are geranylgeranylated in cells treated with farnesyl protein transferase inhibitors. J Biol Chem. 1997;272:14459–14464. doi: 10.1074/jbc.272.22.14459. [DOI] [PubMed] [Google Scholar]
- Wienecke R, Maize JC, Jr, Shoarinejad F, Vass WC, Reed J, Bonifacino JS, et al. Co-localization of the TSC2 product tuberin with its target Rap1 in the Golgi apparatus. Oncogene. 1996;13:913–923. [PubMed] [Google Scholar]
- Winter-Vann AM, Casey PJ. Post-prenylation-processing enzymes as new targets in oncogenesis. Nat Rev Cancer. 2005;5:405–412. doi: 10.1038/nrc1612. [DOI] [PubMed] [Google Scholar]
- Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- Yue W, Fan P, Wang J, Li Y, Santen RJ. Mechanisms of acquired resistance to endocrine therapy in hormone-dependent breast cancer cells. J Steroid Biochem Mol Biol. 2007;106:102–110. doi: 10.1016/j.jsbmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Wang J, Li Y, Fan P, Santen RJ. Farnesylthiosalicylic acid blocks mammalian target of rapamycin signaling in breast cancer cells. Int J Cancer. 2005;117:746–754. doi: 10.1002/ijc.21222. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
GFP-Rheb1 and GFP-Rheb2 do not localize to early endosomes, late endosomes, lysosomes, or mitochondria. COS-7 cells were transiently transfected with pEGFP vector or with pEGFP encoding GFP-tagged Rheb1 or Rheb2. (a) Live cells were imaged after staining with 500 nM MitoTracker dye (500 nM). (b) Prior to fixation, cells were stained with 75 nM LysoTracker dye for 2 h. (c–f) Cells were fixed and immunostained with (c) mouse anti-EEA1 antibody followed by Alexa Fluor 594-conjugated anti-mouse secondary antibody, (d) mouse anti-LAMP2 antibody followed by Alexa Fluor 594-conjugated anti-mouse secondary antibody, (e) Alexa Fluor 594-conjugated anti-mouse secondary antibody alone, (f) Alexa Fluor 594-conjugated anti-rabbit secondary antibody alone. All images were acquired on a Zeiss confocal microscope using LSM software.