Abstract
The Th2 cytokine gene locus has emerged as a remarkable example of coordinated gene expression, the regulation of which appears to be rooted in an extensive array of cis-regulatory regions. Using a hypothesis-generating computational approach that integrated multi-species (n=11) sequence comparisons with algorithm-based transcription factor binding site predictions, we sought to identify evolutionarily conserved non-coding regions (ECRs), and motifs shared among them, which may underlie co-regulation. Twenty-two transcription factor families were predicted to have binding sites in at least two Th2 ECRs. The ranking of these shared motifs according to their distribution and relative frequency pointed to a regulatory hierarchy among the transcription factor families. GATA sites were the most prevalent and widely distributed, consistent with the known role of GATA3 as a Th2 master switch. Unexpectedly, sites for ETS-domain proteins were also predicted within several Th2 ECRs, and the majority of these sites were found to support Ets-1 binding in vitro and in vivo. Of note, the expression of all three Th2 cytokines (Il5, Il13 and Il4) was significantly and selectively decreased in Th2 cells generated from Ets-1-deficient mice. Collectively, these data suggest that Ets-1 contributes to Th2 cytokine gene regulation by interacting with multiple cis-regulatory regions throughout the Th2 locus.
INTRODUCTION
The spatio-temporal control of eukaryotic gene expression is a complex process, orchestrated to a large extent by mechanisms which control transcription. Regulation of individual loci often relies on multiple cis-regulatory elements, located both proximal (promoters) and distal (enhancers, silencers and insulators) to the transcription start site. Clusters of functionally related genes may exhibit additional complexity and share multiple cis-regulatory elements that transduce developmental and/or environmental cues to specify sequential expression of individual genes, as in the β-globin gene cluster (1), or coordinated expression of several genes, as is the case for the T helper (Th) type-2 cytokine gene locus (2), which includes IL5, IL13 and IL4.
The Th2 cytokine genes are grouped within a 150 kb region of human chromosome 5q31 and the syntenic region of mouse chromosome 11. IL13 and IL4 are adjacent to one another, while IL5 is separated from these two by a large (85 kb) DNA repair gene, RAD50, a configuration which has been preserved for at least 300 million years (3). The regulation of this locus has been studied most extensively in the Th2 lineage, and has been shown to provide a remarkable example of coordinated gene expression (4, 5). Such coordination appears to be highly adaptive, in that the distinct components of the Th2 cytokine trio cooperate ensuring the deployment of complementary anti-parasite defense pathways (6). On the other hand, when dysregulated by genetic and/or environmental influences, coordinated Th2 cytokine expression becomes critical for the pathogenesis of allergic inflammation (7, 8).
An extensive array of highly conserved cis-regulatory elements essential for the coordinated expression of Th2 cytokine genes has been identified by a combination of DNase I hypersensitive site (HS)3 mapping and comparative sequence analysis (Figure 2A). HSS 1 and 2 (9), located between IL13 and IL4, map to conserved non-coding sequence (CNS)-1, a potent enhancer of cytokine gene expression in T cells (10–12) (Figure 2A). Two other regions, which map to the second intron of IL4 (HS II and III; IL4 intronic enhancer, IL4 IE), and downstream of IL4 (HSV/Va; CNS-2), exhibit enhancer activity in both Th2 cells and mast cells (13–15). Several HS sites (RHS 5–7) clustered within the 3' end of the DNA repair gene, RAD50 (16, 17), collectively function as a locus control region (LCR), conferring robust Th2-specific, position-independent and copy-number dependent expression to the IL4 and IL13 genes (16, 18). Finally, a single silencer located at the 3' end of IL4 (HSIV) is required to suppress IL4 expression in naïve CD4+ T and Th1 cells (14, 19). Of note, the major Th2 cis-regulatory elements typically map to extensive (300–600 bp) regions that are highly conserved between mice and humans.
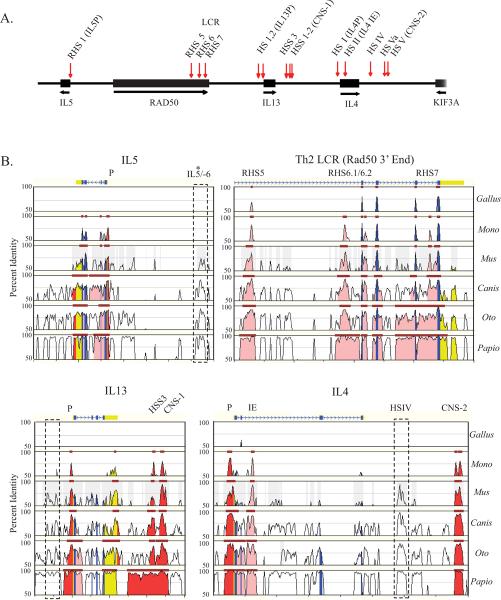
Figure 2. Th2 cis-regulatory elements pre-date the divergence of placental and non-placental mammals.
A) Map of the Th2 cytokine gene cluster with the locations of murine DNase I hypersensitive sites (vertical arrows). B) MULAN-generated graphical representation of DNA sequence alignments between the indicated species and the reference human sequence. Selected regions of the Th2 cytokine locus are shown, with gene annotations and features noted above the plots. (CNS: conserved non-coding sequence, IE: intronic enhancer, P: promoter, RHS: RAD50 hypersensitive site). Shaded regions indicate multi-species ECRs with gene features specified by color (exon: blue; UTR: yellow; intron: pink; intergenic: red). Dashed boxes denote regions that are conserved among the placental mammals but not in opossum or chicken.
Several lines of evidence point to a significant degree of interdependency among distant regulatory regions within the Th2 locus. Indeed, analysis of transgenic models has shown that no single element is sufficient to fully recapitulate Th2 cytokine gene expression at the levels driven by an intact Th2 locus (16, 18). Conversely, deletion of any enhancer region is sufficient to markedly inhibit the expression of more than one Th2 cytokine gene (11, 15, 20). Moreover, characterization of long-range intra-chromosomal interactions at the murine Th2 locus determined that in T cells, the IL5, IL13 and IL4 promoters are physically clustered with one another and with the distant regulatory elements forming a distinctive chromatin hub (21).
The advanced structural and functional characterization of the Th2 locus provides a unique opportunity to decipher molecular cues which might not only dictate the function of individual regulatory regions, but also specify coordinated gene expression. A simple model to orchestrate the co-expression of distinct genes predicts that their promoters will contain a common set of transcription factor binding sites critical for transducing relevant signals. Building on such a model, we took a comprehensive in silico approach, and complemented it with functional in vitro and in vivo studies, to decipher the regulatory logic underlying the co-expression of the Th2 cytokine gene cluster. While computational predictions of transcription factor binding sites have been hindered by considerable false-positive rates, such predictions are substantially improved by integrating motif-finding algorithms with phylogenetic comparisons (22–24), and further strengthened by functional validation. Here we show that our hypothesis-generating, multi-pronged approach succeeded in identifying Ets-1 as a novel, important regulator of coordinated Th2 cytokine gene expression.
MATERIALS AND METHODS
Multi-Species Comparative Analysis
Genomic sequences corresponding to the human Th2 cytokine gene cluster (20 kb downstream of IL5 through the 3' end of KIF3A) and the syntenic regions in Pan troglodytes (chimpanzee), Papio anubis (baboon), Callithrix jacchus (marmoset), Otolemur garnetti (bush baby), Bos taurus (cow), Canis familiaris (dog), Rattus norvegicus (rat), Mus musculus (mouse), Monodelphis domestica (opossum) and Gallus gallus (chicken) were obtained from NCBI. Some of the sequence utilized was generated by the NIH Intramural Sequencing Center (www.nisc.nih.gov).
Generation of a multi-species alignment and identification of evolutionarily conserved regions (ECRs) was performed with the MUltiple sequence Local AligNment and visualization tool (MULAN) program (http://mulan.dcode.org/) (25). MULAN employs a local alignment strategy using the threaded blockset aligner program and utilizes the phylogenetic relationships of the sequences provided to build the multi-species alignment (25). Repeat masking was performed on all sequences with the species-appropriate filters prior to alignment. A large gap due to incomplete sequence data was detected at the 3' end of RAD50 in Callithrix jacchus. Therefore our analysis of this region (which spans RHS6.1, 6.2 and 7 within the Th2 LCR) did not include this species. Due to the very close relationship between human and marmoset, it is unlikely that exclusion of this species significantly altered either the multi-species alignment or the transcription factor binding site profiles generated for this region.
Identification of conserved transcription factor binding sites
Genomic regions (excluding exons) identified by the MULAN program as multi-species ECRs (≥ 70% identity over ≥ 100 bp, in all species examined) were examined for putative transcription factor binding sites. Ungapped sequences from each species were analyzed individually with the MatInspector program, using the matrix library version 6.0, which contains 431 position weight matrices (PWM) for vertebrate transcription factors, representing 148 different families, (Genomatix, Munich, Germany, http://www.genomatix.de/). PWM describe transcription factor binding sites in terms of the complete nucleotide distribution for each single position allowing for quantification of similarity between the weight matrix and the putative binding motif (26). Detection criteria were set to only report matches which demonstrated both a core similarity score ≥ 0.85 and a matrix similarity score that met or exceeded the optimization threshold defined for each PWM. The optimization threshold is a parameter designed to significantly reduce the rate of false-positives by limiting detection to ≤ 3 hits within 10 kb of non-regulatory sequence (26). We identified those transcription factor binding sites predicted independently in 100% of the species examined, including matches to different matrices within a family. Matrix families represent functionally related transcription factors which also have PWM that are essentially indistinguishable (26). Of these sites, those that mapped to the same location within the multi-species alignment were included in the profile. For those cases in which multiple matrix families met the conservation criteria and mapped to the same location in the alignment, all families were included in the profile.
EMSA
Single-stranded complementary oligonucleotides were annealed and PAGE purified. Annealed oligonucleotides were end-labeled with γ-32P ATP using T4 polynucleotide kinase. Binding reactions consisted of 50 ng of human recombinant Ets-1 (Alexis Biochemicals) in binding buffer (25 mM Tris-Cl, 1 mM EDTA, 60 mM KCL, 6 mM MgCl, 10 mM DTT, 1 μg Poly (dI·dC), 10 % glycerol). Competition assays were performed by pre-incubation for 30 minutes at room temperature with unlabelled oligonucleotide competitors (30 and 90-fold molar excess) prior to addition of the probe. Samples were incubated with radiolabelled probes for 30 minutes at room temperature and run on a 5% (w/v) polyacrylamide gel (20 mAmp, 4 hrs, 4°C) which was dried and subjected to autoradiography. Oligonucleotide sequences utilized as probes and/or competitors included a previously defined high affinity Ets-1 motif (SC1_ETS1 CGGCCAAGCCGGAAGTGAGTGCC; the core consensus is underlined) (27), a mutant of this motif (SC1_ETS1mut CGGCCAAGCCttAAGTGAGTGCC), and the conserved ETS sites predicted within the human Th2 cis-regulatory regions (IL5P:TGTCTTTGAGGAAATGAATAA, IL13P: GTTCGGGGAGGAAGTGGGTAG, IL4P.1:GATTTCACAGGAACATTTTAC, IL4P.2: TTTTCTCCTGGAAGAGAGGTG, IL13P:GTTCGGGGAGGAAGTGGGTAG, RHS5:GGTAACACAGGAAGTCAGCAG, IL4IE:ATGAAAACAGGAACTGAAATG, HSIV:TCTGCCACAGGATATGGGTAG, CNS2: TGGGTCACAGGAAGCCCAAGA). The intensity of the Ets-1 complexes was determined by densitometry.
Mice
Female 3–6-wk-old C57BL/6 mice were obtained from The Jackson Laboratory. Ets-1-deficient mice have been described previously (28). The animals were housed under specific pathogen-free conditions, and experiments were performed in accordance with the institutional guidelines for animal care at the University of Arizona and the Dana-Farber Cancer Institute under approved protocols.
T cell isolation and in vitro differentiation of Th2 cells
Murine Th2 cells were generated essentially as described (29). Naïve CD4+ T cells were isolated from spleens by negative selection followed by enrichment using anti-CD62L-coated magnetic beads (Miltenyi). Cells (2–5 × 106) were then cultured in the presence of anti-CD3ε antibody (145.2C11, 1 μg/ml) and anti-CD28 (37.51, 1 μg/ml) (Pharmingen) in a 25 cm2 flask coated with goat anti-hamster IgG (0.2 mg/ml), under Th2 skewing conditions (1000 U/ml IL-4, 3 μg/ml anti-IL12 and 5 μg/ml anti-IFN-γ; complete medium) for 3 days. The cells were then expanded in complete medium containing IL-2 (20 U/ml) for 7 days. Cytokine expression at the single cell level was examined by intracellular staining using the following antibodies: anti-IL4 PE (11B11), anti-IL13 Alexa (eBio13A), and anti-IFN-γ FITC (XMG1.2). For restimulation, cells were incubated with increasing concentrations (0.3–3 μg/ml) of anti-CD3 mAb, in the presence of constant amounts of anti-CD28 antibody (2 μg/ml) for 24 hrs. Cytokine secretion was quantified by ELISA (Quantikine Immunoassays, R&D Biosystems).
Chromatin Immunoprecipitation (ChIP)
Murine Th2 cells were treated with 1 % formaldehyde for 10 minutes at room temperature followed by the addition of glycine (125 mM final concentration) to halt cross-linking. Cells were harvested, washed twice with 1× PBS and resuspended in cell lysis buffer (5 mM PIPES pH 8.0, 85 mM KCl, 0.5% NP-40) supplemented with protease inhibitors (10 μg/ml Aprotinin, 1x EDTA-free complete protease inhibitor cocktail (Roche) and 1 mM PMSF). Nuclei were collected by centrifugation and lysed in nuclear lysis buffer (50 mM Tris-HCl pH 8.1, 10 mM EDTA, 1% SDS) supplemented with protease inhibitors as above. Chromatin was sheared by sonication to yield the majority of fragments in the 200 – 600 bp size range. An aliquot of chromatin (1 × 107 whole cell equivalents) was pre-cleared with a 50 % Protein A sepharose slurry (150 μl) for 20 minutes at 4°C. Pre-cleared chromatin was then diluted 10-fold in IP dilution buffer (0.01 % SDS, 1.1 % Triton X-100, 1.2 mM EDTA, 16.7 mM Tris-HCl pH 8.1, 167 mM NaCl) containing protease inhibitors. Immunoprecipitation reactions were performed with 5 × 106 whole cell equivalents overnight at 4°C with an anti-Ets-1 antibody (C-20, Santa Cruz Biotechnology) or normal rabbit IgG (10 μg). Immunocomplexes were collected with Protein A agarose beads (Millipore), washed once each with low salt wash, high salt wash and LiCl wash and washed twice with TE. Immunocomplexes were eluted with IP elution buffer (1 % SDS, 50 mM NaHCO3) and crosslinks were reversed with an overnight incubation at 65°C. DNA was purified by phenol/chloroform extraction and ethanol precipitation using glycogen as a carrier. Real time PCR was performed with the Quantitect SYBR Green PCR kit (Qiagen) on an ABI Prism 7900 Sequence Detection System. The following primer pairs were used: Il5 Promoter: 5'-ACCCTGAGTTTCAGGACTCG-3 ', 5 '-TCAGAACACTCAAGTGCAGAAG-3', Rad50HS5: 5'-GTTTGTCGTCTCAGAGT-AAAGC-3 ', 5 '-GTCACATCCTGAGCATGTTCC-3', Il13 Promoter: 5'-GTGAGAAGATGGAATCTGGC-3', 5'-CCCTACTCTTTCCTCATACTG-3', Il4 Promoter: 5'-CCTACGCTGATAAGATTAGTCTG-3', 5'-CACCAGATTGTCAGTTATTCTG-3', Il4 intronic enhancer: 5'-GATGTGACAGGCTGATAGTG-3', 5' GAGACCACTGGTAAG-TCAAGAC-3', HSIV: 5'-GATTGAAACTCATTTGCATGTC-3', 5'-GTGTTCTGACC-ATCCAAGTGTAG-3', CNS-2: 5'-GAAGCAGTGTGGGAGAGCGT-3', 5'-GGGAAA-GTGATCGTGAGGAG-3' and Cd14 Promoter: 5 '-GAATGCCCTAATGCCACTCTG-3 ' and 5 '-GCTTGTTCGACCAATTTGGC-3'. PCR reactions were performed under the following cycling conditions: 15 min at 95°C followed by 40 cycles of 15 sec at 95°C, 30 sec at 57°C and 30 sec at 72°C. Dissociation curve analysis and agarose gel electrophoresis confirmed amplification of a single product for each primer set. In addition, PCR efficiency was optimized by adjusting the primer concentration and running serial dilutions of input DNA to generate a standard curve. ChIP data are represented as the PCR signal generated for the sample immunoprecipitated with anti-Ets-1 antibody relative to normal rabbit IgG (specific enrichment).
Real-time PCR
Total RNA was extracted using Trizol and reverse-transcribed using oligo(dT) priming. Negative control reactions without reverse transcriptase were always included. Semi-quantitative RT-PCR was performed in duplicate on an ABI Prism 7900 (Applied Biosystems) using SYBR green dye detection. mRNA levels were measured using pre-designed primer sets for murine Il4, Il5, Il13, Il10, Gata3 and Gapdh (Quantitect Primer Assays, Qiagen). Products were verified by melting curve analysis. Th2 cytokine and Gata3 mRNA levels are expressed relative to Gapdh mRNA abundance within the same sample.
RESULTS
Most Th2 Cis-regulatory Elements Pre-date the Divergence Between Placental and Non-placental Mammals
To investigate the evolutionary history of the Th2 regulatory elements and establish an appropriate scope of species for subsequent binding site analyses, we first assessed to what extent the extensive array of Th2 regulatory regions characterized in humans and mice is conserved in additional placental mammals and evolutionarily more distant species. Beyond placental mammals, orthologs for IL5, IL13 and IL4 have recently been identified for a marsupial, the laboratory opossum (Monodelphis domestica) (30)and the chicken (Gallus gallus) (3). Although a draft sequence of the platypus (Ornithorhynchus anatinus) genome has recently been released (31), the genomic region expected to contain orthologs of the Th2 cytokine genes is incompletely covered, and could not be included in our analysis. Furthermore, since comparative analysis of syntenic regions and mining of expressed sequence tag libraries failed to identify these genes in earlier vertebrate species (teleosts) (32), we did not extend our analysis beyond the avian order.
Figure 1 shows the phylogenetic relationships and estimated divergence times (33–35) for all the eleven species included in our study. A multiple DNA sequence alignment for the entire Th2 cytokine gene cluster (Figure 2A) was generated with the MULAN program (25), which relies on a local alignment strategy and integrates the phylogenetic relationships among the sequences. Figure 2B shows the MULAN-generated alignment for selected regions within the Th2 locus in a subset of species representative of the clades we analyzed. The ability to detect the exons for IL5, IL13 and IL4 by multiple species alignment decreased with increasing phylogenetic distance, consistent with previous reports that the opossum (30) and chicken (3, 32) Th2 cytokines exhibit relatively low homology to the human proteins. In contrast, all exons for the DNA repair gene, RAD50, were highly conserved and readily identified in all species (Figure 2B and data not shown). As expected, all of the Th2 regulatory regions previously identified in pairwise human-mouse alignments (Figure 2A) and validated in functional studies (10, 11, 15, 17–20) were highlighted as evolutionarily conserved regions (ECRs) in the seven placental mammalian species examined (Figure 2B). In addition, this analysis identified a novel non-coding ECR, located approximately 6 kb upstream of the IL5 gene (IL5/-6). Of note, only a single non-coding element (110 bp within RHS5) was conserved in the syntenic region in chicken. In contrast, the majority of the non-coding ECR were highly conserved in the orthologous opossum locus (Figure 2B), with the exception of the functionally undefined IL5/-6, the distal region of the IL13 promoter (which encompasses a conserved GATA3 responsive element, cGRE) (36), and HSIV, which is critical for IL4 silencing in murine Th-1 cells (19).
Figure 1. Phylogenetic relationships and estimated divergence times for species used in comparative analysis.
Divergence time estimates were provided by the literature (33–35).
Collectively, these data indicate that the majority of conserved regulatory regions within the Th2 cytokine locus pre-date the divergence of marsupials and placental mammals ~ 180 million years ago (35, 37). The phylogenetic distance between chicken and mammals might preclude alignment-based detection of additional regulatory regions.
Phylogenetic Profiling of Th2 Responsive Elements
The Th2 cytokine locus ECRs identified by comparative analysis were then examined in order to search for shared transcription factor binding sites that might contribute to coordinated regulation of the Th2 locus. Since some of the Th2 ECRs were undetectable in the opossum genome, we profiled transcription factor binding sites across nine placental mammalian species, representing four major clades (primates, carnivores, artiodactyls and rodents). Multi-species comparisons of ECR increase the overall sequence divergence, which serves to refine alignments and reveal invariant positions more likely to reflect true functional constraints associated with transcription factor binding sites (22–24, 38). Indeed, the sequence diversity within the non-coding Th2 ECRs captured by the multiple species alignment was ~ 20 % greater than that obtained by pair-wise comparison between human and mouse sequences (data not shown). Th2 ECRs from placental mammals were examined for putative transcription factor binding sites using the Matinspector program (26). Searches were conducted on ungapped sequence from each species using a vertebrate position weight matrix (PWM) library and stringent match criteria. Sites predicted independently in all species and residing in the same position within the multiple species alignment were included in the profile.
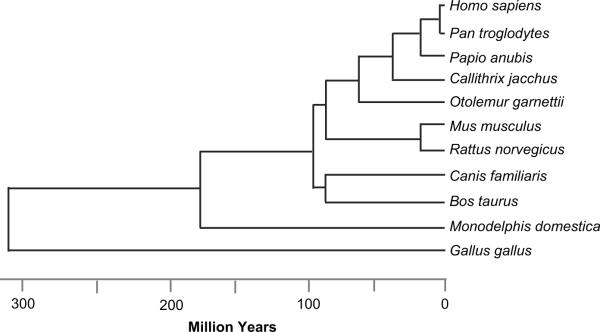
Within the Th2 ECRs, which collectively span ~ 4.8 kb, we identified 103 conserved motifs representing targets for 52 distinct TF families (data not shown). Figure 3 shows that 22 transcription factor families were predicted to have binding sites within at least two Th2 regulatory regions. When multiple sites for a particular factor were found, they typically resided in different regions, rather than clustering within one or few regulatory elements. In fact, the number of conserved sites and the number of distinct regulatory regions they are predicted to target were positively correlated (R2 = 0.81). The number of motifs common to distinct regions did not correlate with GC content nor linear proximity (data not shown). Therefore, these data are unlikely to merely reflect genomic bias. Rather, they may highlight an underlying regulatory network within the Th2 locus.
Figure 3. Common putative transcription factor binding motifs among the Th2 cis-regulatory regions.
Shown are those matrix families which exhibit conserved target sites in more than one Th2 element. Noted within each box is the number of individual conserved sites predicted for that region. Transcription factors are ranked according to the number of Th2 cis-elements that contain conserved putative binding motifs. The DNA sequence of each putative GATA and ETS motif for each species analyzed is provided as Supplemental Material.
Identification and Functional Validation of Ets-1 Binding sites in the Th2 locus
The ranking of transcription factor binding sites shared across the Th2 locus according to their distribution and relative frequency (Figure 3) pointed to a regulatory hierarchy among transcription factor families. Interestingly, the most prevalent and widely distributed motifs were those for GATA proteins, consistent with the notion that GATA3 acts as a master switch for Th2 cell differentiation and Th2 cytokine expression (36, 39–42). This finding validated our approach, and by extension raised the possibility that ETS and SORY/HMG, which like GATA proteins were predicted to target six or more Th2 cis-elements, might also play a role in the coordinated regulation of the Th2 cytokine locus.
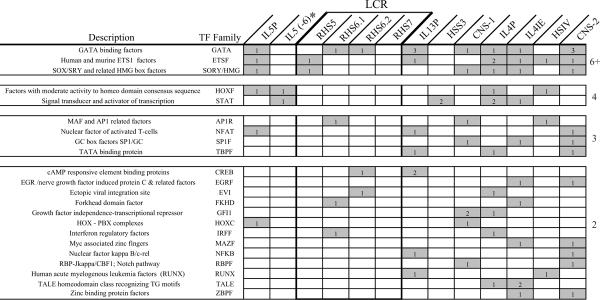
Our subsequent analysis focused on the ETS family, which had not been previously implicated in orchestrating the coordinated expression of the Th2 cytokines, and particularly on Ets-1, the family member which is predominant in T cells (43). To functionally validate the ETS motifs predicted within the Th2 ECRs, we used electrophoretic mobility shift assay (EMSA) and we compared and contrasted the ability of oligonucleotides corresponding to individual putative ETS sites to bind recombinant human Ets-1 (rhEts-1). Figure 4A shows the results of an experiment representative of this approach. Incubation of rhEts-1 with a high affinity Ets-1 consensus probe (27) led to the formation of a complex (lane 1) that was competed almost completely upon incubation with a 30-fold molar excess of unlabeled Ets-1 consensus oligonucleotide (lane 2) and was specifically supershifted by an anti-Ets-1 antibody (data not shown). Oligonucleotides spanning the putative ETS sites in HSIV (lanes 4 and 5) and the IL13 promoter (lanes 6 and 7) also competed Ets-1 binding, albeit less effectively. In contrast, an oligonucleotide encompassing one of the two putative ETS sites within the IL4 promoter (IL4P.1, lanes 12 and 13) failed to compete Ets-1 binding. Similar experiments were performed to test each predicted ETS motif, and the intensity of the relevant bands was measured by densitometry and compared to the intensity of the uncompeted Ets-1 complex. The Ets-1 consensus and an Ets-1 oligonucleotide containing two mutations in the ETS core were used as positive and negative controls throughout this study.
Figure 4. Ets-1 binds to the majority of ETS motifs predicted in the Th2 ECR.
A) Ets-1 protein/DNA interactions were characterized using a γ–32P-labeled Ets-1 consensus probe. Purified recombinant human Ets-1 (50 ng) was preincubated with an increasing molar excess of the indicated competitors before addition of a high affinity Ets-1 consensus probe. B) EMSA blots were analyzed by densitometry and results were expressed as percent inhibition of the Ets-1 band relative to the uncompeted Ets-1 complex. C) Sequences of the Ets-1 binding site core, the high affinity Ets-1 consensus probe, and the eight conserved ETS motifs predicted within the Th2 ECRs. Bold text indicates nucleotide sites within the Ets-1 core; nucleotides that differ from the defined core sequence are marked by lower case letters.
Figure 4B shows the competition curves for the eight putative ETS sites identified within the Th2 locus. The motifs within RHS5 and the IL13 promoter (IL13P) competed Ets-1 binding almost as effectively as the Ets-1 consensus oligonucleotide. The sites within HSIV, CNS2 and the IL4 intronic enhancer (IL4IE) exhibited intermediate binding ability. Competition by the IL5 promoter site (IL5P) was more modest, but still appreciable. In contrast, individual ETS motifs within the IL4 promoter (IL4P.1 and IL4P.2) failed to compete Ets-1 binding.
Figure 4C shows the core motif defined specifically for Ets-1 (27) and the sequences of the Th2 Ets-1 binding sites, ranked by relative Ets-1 binding ability. Relative to the Ets-1 core, all sequences containing no or one mismatch supported Ets-1 binding, albeit to different extents. In contrast, two mismatches appeared to compromise binding, as shown by the poor interactions detected for IL5P, IL4P.1 and IL4P.2.
Collectively, these data show that the majority of the ETS sites predicted within the Th2 ECR behave as bona fide Ets-1 binding motifs in vitro, suggesting this protein (and/or possibly other members of the ETS family) might bind at multiple locations throughout the locus, thereby participating in the concerted regulation of Th2 cytokine gene expression.
Ets-1 Binds Several Regulatory Regions in the Murine Th2 Locus in Vivo
To examine whether Ets-1 binds to the endogenous Th2 ECRs, ChIP experiments were performed on chromatin isolated from naïve CD4+ T cells cultured under Th2 skewing conditions for 7 days. Immunoprecipitation relied upon an antibody specific for Ets-1 or normal rabbit IgG as a negative control. Real-time PCR was performed using primers that target the individual Th2 ECRs previously tested for Ets-1 binding in vitro. The promoter of Cd14, a gene which is not expressed in murine Th2 cells (data not shown), was amplified for comparison.
Figure 5 shows that, relative to the Cd14 promoter, Ets-1-containing complexes were specifically enriched at all three Th2 cytokine gene promoters. The strongest occupancy was detected at the Il4 promoter, a region that contains two ETS binding motifs. Notably, the two ETS motifs in this region failed to bind Ets-1 when tested individually in vitro, suggesting the strong signal detected in vivo reflects cooperative binding. Ets-1 was also consistently found to bind distal Th2 regulatory regions, including RHS5 of the Th2 LCR, the Il4 silencer region (HSIV) and a Th2 cytokine enhancer (CNS2). An Ets-1-specific signal was detected at the Il4 intronic enhancer (IL4IE), but barely exceeded the one generated for the Cd14 promoter. Of note, our ChIP experiments detected Ets-1 and no other members of the large ETS family because specific enrichment was completely abrogated at all of the Th2 ECRs in Ets-1-deficient Th2 cells (data not shown). Overall, these data show that Ets-1 binds to multiple Th2 regulatory regions in vivo, supporting the possibility that Ets-1 participates in the coordinated regulation of the Th2 cytokine gene cluster.
Figure 5. Ets-1 binds several regulatory regions in the endogenous murine Th2 locus.
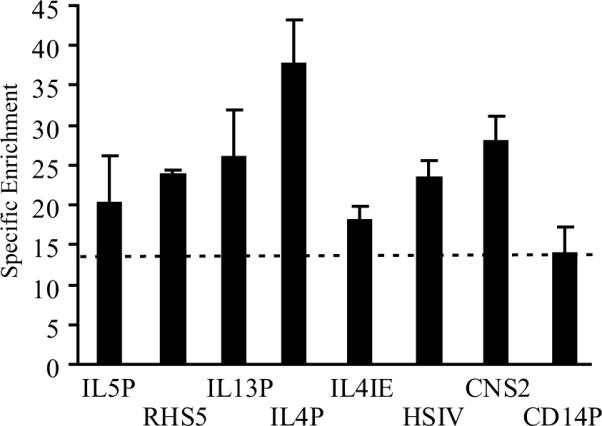
Spleens from C57BL/6 mice were harvested and naïve CD4+ T lymphocytes were isolated and cultured under Th2-skewing conditions for 7 days. Chromatin was immunoprecipitated with an anti-Ets-1 antibody or normal rabbit IgG as a negative control. The precipitated DNA was amplified by quantitative real-time PCR using primers specific for the Th2 ECRs or the Cd14 promoter as a comparison. All PCR reactions were carried out in duplicate. Results are expressed as specific enrichment for Ets-1 relative to the IgG control. Shown are mean ± SE values for two independent experiments, one of which was conducted on chromatin isolated from cells pooled from 4 mice.
Ets-1 is required for optimal expression of all three Th2 cytokine genes
In order to test the role of Ets-1 in Th2 cytokine gene expression we compared the levels of Il4, Il13 and Il5 mRNA and protein produced by Th2 cells from Ets-1+/− or Ets-1−/− mice. Naïve CD4+ T cells isolated from these mice were cultured under Th2-skewing conditions for 7 days and stimulated for 24 hr with increasing concentrations of plate-bound anti-CD3 mAb, in the presence of a constant amount of anti-CD28 antibody. mRNA levels were assessed by real-time RT-PCR. Figure 6A shows that Ets-1−/− Th2 cells exhibited a marked decrease in Il4, Il5 and Il13 mRNA relative to Ets-1+/− Th2 cells. In contrast, transcript levels for Il10, a cytokine gene expressed by Th2 cells and located on mouse chromosome 1, were comparable in Ets-1+/− and Ets-1−/− Th2 cells. Impaired Th2 cytokine transcription in cells lacking Ets-1 was unlikely to reflect a defect in Th2 cell differentiation per se because Gata3 mRNA levels in these cells were unaffected (Figure 6B).
Figure 6. Ets-1 is required for optimal expression of all three Th2 cytokine genes.
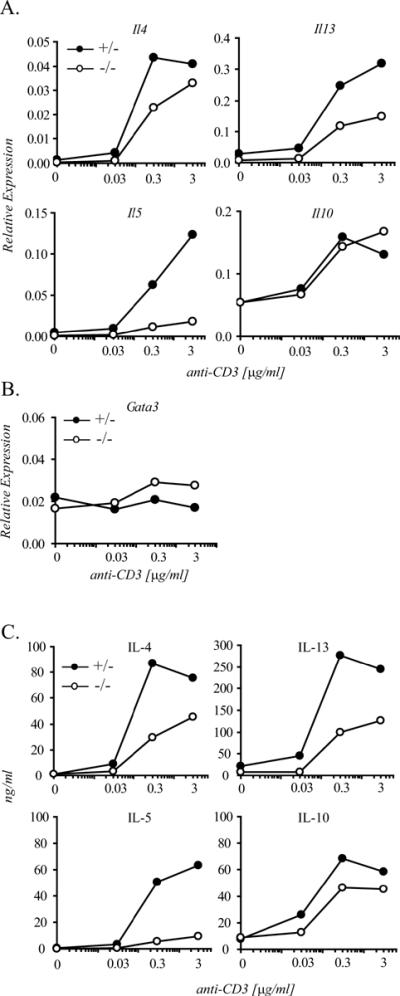
Naïve CD4+ T cells were isolated from Ets-1+/− or Ets-1−/− mice and cultured under Th2 skewing conditions in vitro. After 7 days, cells were stimulated with the indicated concentrations of plate-bound anti-CD3 mAb in the presence of a constant amount of anti-CD28 mAb (2 μg/ml) for 24 hours. A) and B) mRNA levels were assessed by quantitative real-time RT-PCR using SYBR green dye detection. All PCR reactions were carried out in duplicate. mRNA levels are expressed relative to Gapdh mRNA abundance. C) Th2 cell supernatants were collected and cytokine secretion was measured by ELISA. All samples were measured in duplicate. The figure shows one representative experiment out of three.
In parallel, we examined the impact of Ets-1 deficiency on Th2 cytokine secretion. Figure 6C shows that, consistent with the decrease observed in Th2 cytokine transcripts, Ets-1−/− Th2 cells were also impaired in their ability to release IL-4, IL-13 and IL-5, but not IL-10. Collectively, these data demonstrate that Ets-1 acts as a global positive regulator of the expression of the Th2 cytokine gene cluster, thereby providing functional validation for our phylogenetic analysis.
DISCUSSION
The Th2 cytokine gene locus typically acts as a unit, the distinct components of which (IL5, IL13 and IL4) are co-regulated and co-expressed in response to T cell activation and differentiation. This expression pattern is likely to be critical for effective immune responses, but its molecular underpinnings remain only partially understood. To investigate the molecular basis for coordinated regulation of Th2 cytokine genes, we combined comparative and functional analyses, and we focused on transcription factor binding sites that are both highly conserved across placental mammals, and common to more than one Th2 regulatory region. Our comparison of the Th2 cytokine gene locus in evolutionarily distant vertebrate species (human to chicken) revealed that while gene content, order and orientation have been preserved for over 300 million years, nearly all of the distal cis-regulatory elements known to underlie Th2 co-regulation have arisen after the split between birds and mammals. The main exception appears to be a single region conserved in chicken that corresponds to one of several murine DNase I HS sites within the Th2 LCR (RHS5). It is tempting to speculate that this region might participate in transcriptional regulation of the chicken orthologs of mammalian type-2 cytokines.
Despite remarkable conservation of the majority of Th2 regulatory regions, a few elements were not identifiable in the opossum locus. These included a GATA3-responsive element 5' of IL13 and the Th1-specific IL4 silencer, HSIV. The emergence of a Th1-selective IL4 silencer suggests a fine-tuning of the Th1/Th2 dichotomy may have occurred during mammalian evolution. Resistance to certain mammalian pathogens (e.g., Leishmania) does require extremely polarized Th cell responses (44, 45). Thus, regulatory mechanisms that place increasingly tighter restrictions on alternative cytokine responses might be adaptive to the host.
Perhaps most importantly, our identification of highly conserved Ets-1 binding sites in multiple cis-regulatory regions in the Th2 locus, and the finding that expression of Il5, Il13 and Il4 transcripts and protein were substantially and selectively impaired in Ets-1-deficient murine Th2 cells, for the first time define Ets-1 as a global regulator of coordinated Th2 cytokine gene expression. Previously, GATA3, the Th2 master regulator, was the only transcription factor known to have a broad influence on Th2 locus regulation, directly controlling both chromatin structure and transcriptional activity (46). The phenotype of Ets-1 deficiency (global reduction of Th2 cytokine expression) is less severe than the phenotype of GATA-3 deficiency (complete inhibition of Th2 cell development) (47), but is similar in scope. Of note, GATA-3 and Ets-1 were shown to regulate the Il5 promoter cooperatively by binding to adjacent sites (48–50). This finding raises the possibility that the Th2 cytokine defect detected in Ets-1 knockout mice results from an impairment of the functional synergism between GATA-3 and Ets-1. However, whilst GATA and ETS sites coexist in all but two of the Ets-1 binding ECRs, these sites are in close proximity only in the Il5 promoter and in CNS2, the enhancer located 3' of Il4. This arrangement suggests that altered cooperativity between Ets-1 and GATA-3 may contribute to, but not be the only mechanism for, the locus-wide defect in Th2 cytokine expression observed in Ets-1−/− mice.
The consequences of Ets-1 deficiency are not limited to Th2 cells. Ets-1−/− Th1 cells exhibit a significant defect in the establishment of polarized cytokine expression programs, with defective IFN-γ production and inappropriate expression of Il4 (51). The latter mirrors the phenotype of CD4+ T cells lacking the Il4 silencer, HSIV (19, 51). Interestingly, our bioinformatic analysis identified a highly conserved ETS binding site within HSIV that did support Ets-1 binding in vitro and in vivo. Loss of Ets-1 binding at HSIV may play a role in the abnormal regulation of Il4 expression detected in the absence of Ets-1. Overall our study highlighted specific functional elements that Ets proteins may target to orchestrate Th2 cytokine expression. Interestingly, our analysis also revealed a remarkable heterogeneity in the sequence of Ets-1 binding motifs, particularly their flanking regions. This heterogeneity might result in a functional hierarchy among Ets-1 binding sites, particularly under conditions of limited Ets availability, as well as in differential interaction between Ets-1 and its protein partners.
Synergism between homotypic factors that occupy distant DNA elements is a regulatory arrangement that has been appreciated since early studies conducted with phage λ (52). The highly conserved, common motifs we have identified across the Th2 locus may provide a molecular basis for functional cooperation between distant Th2 regulatory regions. Indeed, data describing the three-dimensional architecture of the murine Th2 locus highlight the potential for direct communication among distant Th2 cis-elements. In naïve CD4+ T cells, the Il5, Il13 and Il4 promoters are physically clustered with one another and with distant Th2 cis-regulatory regions (21). Analogous to a single regulatory element containing tandem arrays of identical binding sites, three-dimensional clustering of homotypic sites may serve as a strategy for the Th2 cytokine genes to effectively vie for limited amounts of trans-acting factors and in turn, foster expression of the locus as a whole.
Very little is known about how long-range chromatin contacts are initiated and/or maintained, yet the earliest evidence supporting a DNA looping model pointed toward the homo-oligomerization of transcriptional activators and/or repressors bound to distant target sites (52–54). These early studies were conducted on artificial DNA templates, but more recent experiments in a native chromosomal context also link sequence-specific DNA binding proteins to chromosomal looping and implicate distant, matching binding sites in this process. For example, the erythroid-specific Krüppel-like factor (EKLF) directly contributes to the formation of an active chromatin hub in the β-globin locus by interacting with multiple cis-regulatory elements distributed across the locus (55). Similarly, androgen receptor-mediated regulation of the prostate specific antigen gene proceeds via independent recruitment of androgen receptors to both a distant upstream enhancer and the proximal promoter, which subsequently form a stable chromosomal loop (56). The distribution of the highly conserved GATA motifs across the Th2 locus seems to reflect the role that GATA3 plays in higher-order chromatin architecture, since ectopic expression of GATA3 in fibroblasts induces chromatin contacts between the Th2 LCR and the Il4 and Il13 promoters (21). On the other hand, these associations were relatively weak, suggesting that additional T cell lineage-restricted factors may be required to reinforce and/or maintain these long-range interactions (21). Our finding that functional binding sites for Ets-1 are distributed across the Th2 locus raises the intriguing possibility that the contribution of Ets-1 to the coordinated regulation of Th2 cytokine genes may extend to the higher-order chromatin architecture of the locus.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant 5RO1HL66391 (to D. V.) and a National Science Foundation IGERT Fellowship in Genomics (to J. M. S.).
Footnotes
Abbreviations used in this paper: ChIP, chromatin immunoprecipitation; CNS, conserved non-coding region; ECR, evolutionarily conserved region; HS, hypersensitive site; LCR, locus control region; MULAN, multiple sequence local alignment and visualization tool; PWM, position weight matrix.
REFERENCES
- 1.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee GR, Kim ST, Spilianakis CG, Fields PE, Flavell RA. T helper cell differentiation: regulation by cis elements and epigenetics. Immunity. 2006;24:369–379. doi: 10.1016/j.immuni.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 3.Avery S, Rothwell L, Degen WD, Schijns VE, Young J, Kaufman J, Kaiser P. Characterization of the first nonmammalian T2 cytokine gene cluster: the cluster contains functional single-copy genes for IL-3, IL-4, IL-13, and GM-CSF, a gene for IL-5 that appears to be a pseudogene, and a gene encoding another cytokinelike transcript, KK34. J. Interferon Cytokine Res. 2004;24:600–610. doi: 10.1089/jir.2004.24.600. [DOI] [PubMed] [Google Scholar]
- 4.Kelly BL, Locksley RM. Coordinate regulation of the IL-4, IL-13, and IL-5 cytokine cluster in Th2 clones revealed by allelic expression patterns. J. Immunol. 2000;165:2982–2986. doi: 10.4049/jimmunol.165.6.2982. [DOI] [PubMed] [Google Scholar]
- 5.Cousins DJ, Lee TH, Staynov DZ. Cytokine coexpression during human Th1/Th2 cell differentiation: direct evidence for coordinated expression of Th2 cytokines. J Immunol. 2002;169:2498–2506. doi: 10.4049/jimmunol.169.5.2498. [DOI] [PubMed] [Google Scholar]
- 6.Anthony RM, Rutitzky LI, Urban JF, Jr., Stadecker MJ, Gause WC. Protective immune mechanisms in helminth infection. Nat. Rev. Immunol. 2007;7:975–987. doi: 10.1038/nri2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wills-Karp M, Luyimbazi J, Xu X, Schofield B, Neben TY, Karp CL, Donaldson DD. Interleukin-13: Central mediator of allergic asthma. Science (New York, N.Y. 1998;282:2258–2261. doi: 10.1126/science.282.5397.2258. [DOI] [PubMed] [Google Scholar]
- 8.Shi H, Qin S, Huang G, Chen Y, Xiao C, Xu H, Liang G, Xie Z, Qin X, Wu J, Li G, Zhang C. Infiltration of eosinophils into the asthmatic airways caused by interleukin 5. Am. J. Respir. Cell Mol. Biol. 1997;16:220–224. doi: 10.1165/ajrcmb.16.3.9070605. [DOI] [PubMed] [Google Scholar]
- 9.Takemoto N, Koyano-Nakagawa N, Yokota T, Arai N, Miyatake S, Arai K-I. Th2-specific DNase I-hypersensitive sites in the murine IL-13 and IL-4 intergenic region. Int. Immunol. 1998;10:1981–1985. doi: 10.1093/intimm/10.12.1981. [DOI] [PubMed] [Google Scholar]
- 10.Loots GG, Locksley RM, Blankenspoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparison. Science (New York, N.Y. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 11.Mohrs M, Blankespoor CM, Wang ZE, Loots GG, Afzal V, Hadeiba H, Shinkai K, Rubin EM, Locksley RM. Deletion of a coordinate regulator of type 2 cytokine expression in mice. Nat. Immunol. 2001;2:842–847. doi: 10.1038/ni0901-842. [DOI] [PubMed] [Google Scholar]
- 12.Lee GR, Fields PE, Flavell RA. Regulation of IL-4 gene expression by distal regulatory elements and GATA-3 at the chromatin level. Immunity. 2001;14:447–459. doi: 10.1016/s1074-7613(01)00125-x. [DOI] [PubMed] [Google Scholar]
- 13.Henkel G, Weiss DL, McCoy R, Deloughery T, Tara D, Brown MA. A DNase I-hypersensitive site in the second intron of the murine IL-4 gene defines a mast cell-specific enhancer. J. Immunol. 1992;149:3229–3246. [PubMed] [Google Scholar]
- 14.Agarwal S, Rao A. Modulation of chromatin structure regulates cytokine gene expression during T cell differentiation. Immunity. 1998;9:765–775. doi: 10.1016/s1074-7613(00)80642-1. [DOI] [PubMed] [Google Scholar]
- 15.Solymar DC, Agarwal S, Bassing CH, Alt FW, Rao A. A 3' enhancer in the IL-4 gene regulates cytokine production by TH2 cells and mast cells. Immunity. 2002;17:41–50. doi: 10.1016/s1074-7613(02)00334-5. [DOI] [PubMed] [Google Scholar]
- 16.Fields PE, Lee GR, Kim ST, Bartsevich V, Flavell RA. Th2-specific chromatin remodeling and enhancer activity in the Th2 cytokine locus control region. Immunity. 2004;21:865–876. doi: 10.1016/j.immuni.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 17.Lee DU, Rao A. Molecular analysis of a locus control region in the T helper 2 cytokine gene cluster: A target for STAT6 but not GATA3. Proc. Natl. Acad. Sci. USA. 2004;101:16010–16015. doi: 10.1073/pnas.0407031101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee G, Fields P, Griffin T, Flavell R. Regulation of the Th2 cytokine locus by a locus control region. Immunity. 2003;19:145–153. doi: 10.1016/s1074-7613(03)00179-1. [DOI] [PubMed] [Google Scholar]
- 19.Ansel KM, Greenwald RJ, Agarwal S, Bassing CH, Monticelli S, Interlandi J, Djuretic IM, Lee DU, Sharpe AH, Alt FW, Rao A. Deletion of a conserved Il4 silencer impairs T helper type 1-mediated immunity. Nat. Immunol. 2004;5:1251–1259. doi: 10.1038/ni1135. [DOI] [PubMed] [Google Scholar]
- 20.Lee GR, Spilianakis CG, Flavell RA. Hypersensitive site 7 of the TH2 locus control region is essential for expressing TH2 cytokine genes and for long-range intrachromosomal interactions. Nat. Immunol. 2005;6:42–48. doi: 10.1038/ni1148. [DOI] [PubMed] [Google Scholar]
- 21.Spilianakis CG, Flavell RA. Long-range intrachromosomal interactions in the T helper type 2 cytokine locus. Nat. Immunol. 2004;5:1017–1027. doi: 10.1038/ni1115. [DOI] [PubMed] [Google Scholar]
- 22.Loots GG, Ovcharenko I. rVISTA 2.0: evolutionary analysis of transcription factor binding sites. Nucl. Acids Res. 2004;32:W217–221. doi: 10.1093/nar/gkh383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lenhard B, Sandelin A, Mendoza L, Engstrom P, Jareborg N, Wasserman W. Identification of conserved regulatory elements by comparative genome analysis. J. Biology. 2003;2:13. doi: 10.1186/1475-4924-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sumiyama K, Kim CB, Ruddle FH. An efficient cis-element discovery method using multiple sequence comparisons based on evolutionary relationships. Genomics. 2001;71:260–262. doi: 10.1006/geno.2000.6422. [DOI] [PubMed] [Google Scholar]
- 25.Ovcharenko I, Loots GG, Giardine BM, Hou M, Ma J, Hardison RC, Stubbs L, Miller W. Mulan: multiple-sequence local alignment and visualization for studying function and evolution. Genome Res. 2005;15:184–194. doi: 10.1101/gr.3007205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, Frisch M, Bayerlein M, Werner T. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–2942. doi: 10.1093/bioinformatics/bti473. [DOI] [PubMed] [Google Scholar]
- 27.Nye JA, Petersen JM, Gunther CV, Jonsen MD, Graves BJ. Interaction of murine ets-1 with GGA-binding sites establishes the ETS domain as a new DNA-binding motif. Genes Dev. 1992;6:975–990. doi: 10.1101/gad.6.6.975. [DOI] [PubMed] [Google Scholar]
- 28.Barton K, Muthusamy N, Fischer C, Ting CN, Walunas TL, Lanier LL, Leiden JM. The Ets-1 transcription factor is required for the development of natural killer cells in mice. Immunity. 1998;9:555–563. doi: 10.1016/s1074-7613(00)80638-x. [DOI] [PubMed] [Google Scholar]
- 29.Avni O, Lee D, Macian F, Szabo SJ, Glimcher LH, Rao A. T(H) cell differentiation is accompanied by dynamic changes in histone acetylation of cytokine genes. Nat. Immunol. 2002;3:643–651. doi: 10.1038/ni808. [DOI] [PubMed] [Google Scholar]
- 30.Wong ES, Young LJ, Papenfuss AT, Belov K. In silico identification of opossum cytokine genes suggests the complexity of the marsupial immune system rivals that of eutherian mammals. Immunome Res. 2006;2:4. doi: 10.1186/1745-7580-2-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Warren WC, Hillier LW, Marshall Graves JA, Birney E, Ponting CP, Grutzner F, Belov K, Miller W, Clarke L, Chinwalla AT, Yang SP, Heger A, Locke DP, Miethke P, Waters PD, Veyrunes F, Fulton L, Fulton B, Graves T, Wallis J, Puente XS, Lopez-Otin C, Ordonez GR, Eichler EE, Chen L, Cheng Z, Deakin JE, Alsop A, Thompson K, Kirby P, Papenfuss AT, Wakefield MJ, Olender T, Lancet D, Huttley GA, Smit AF, Pask A, Temple-Smith P, Batzer MA, Walker JA, Konkel MK, Harris RS, Whittington CM, Wong ES, Gemmell NJ, Buschiazzo E, Vargas Jentzsch IM, Merkel A, Schmitz J, Zemann A, Churakov G, Kriegs JO, Brosius J, Murchison EP, Sachidanandam R, Smith C, Hannon GJ, Tsend-Ayush E, McMillan D, Attenborough R, Rens W, Ferguson-Smith M, Lefevre CM, Sharp JA, Nicholas KR, Ray DA, Kube M, Reinhardt R, Pringle TH, Taylor J, Jones RC, Nixon B, Dacheux JL, Niwa H, Sekita Y, Huang X, Stark A, Kheradpour P, Kellis M, Flicek P, Chen Y, Webber C, Hardison R, Nelson J, Hallsworth-Pepin K, Delehaunty K, Markovic C, Minx P, Feng Y, Kremitzki C, Mitreva M, Glasscock J, Wylie T, Wohldmann P, Thiru P, Nhan MN, Pohl CS, Smith SM, Hou S, Nefedov M, de Jong PJ, Renfree MB, Mardis ER, Wilson RK. Genome analysis of the platypus reveals unique signatures of evolution. Nature. 2008;453:175–183. doi: 10.1038/nature06936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huising MO, Kruiswijk CP, Flik G. Phylogeny and evolution of class-I helical cytokines. J. Endocrinol. 2006;189:1–25. doi: 10.1677/joe.1.06591. [DOI] [PubMed] [Google Scholar]
- 33.Goodman M. Molecular Evolution '99 - The genomic record of humankind's evolutionary roots. Am. J. Hum. genet. 1999;64:31–39. doi: 10.1086/302218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy WJ, Eizirik E, Johnson WE, Zhang YP, Ryder OA, O'Brien SJ. Molecular phylogenetics and the origins of placental mammals. Nature. 2001;409:614–618. doi: 10.1038/35054550. [DOI] [PubMed] [Google Scholar]
- 35.Hedges SB, Kumar S. Genomic clocks and evolutionary timescales. Trends Genet. 2003;19:200–206. doi: 10.1016/S0168-9525(03)00053-2. [DOI] [PubMed] [Google Scholar]
- 36.Yamashita M, Ukai-Tadenuma M, Kimura M, Omori M, Inami M, Taniguchi M, Nakayama T. Identification of a conserved GATA3 response element upstream proximal of the IL-13 gene locus. J. Biol. Chem. 2002;277:42399–42408. doi: 10.1074/jbc.M205876200. [DOI] [PubMed] [Google Scholar]
- 37.Mikkelsen TS, Wakefield MJ, Aken B, et al. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature. 2007;447:167–177. doi: 10.1038/nature05805. [DOI] [PubMed] [Google Scholar]
- 38.Cooper GM, Brudno M, Green ED, Batzoglou S, Sidow A. Quantitative estimates of sequence divergence for comparative analyses of mammalian genomes. Genome Res. 2003;13:813–820. doi: 10.1101/gr.1064503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zheng W-P, Flavell RA. The transcription factor GATA-3 is necessary and sufficient for Th2 cytokine gene expression in CD4 T cells. Cell. 1997;89:587–596. doi: 10.1016/s0092-8674(00)80240-8. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D-H, Cohn L, Ray P, Bottomly K, Ray A. Transcription factor GATA-3 is differentially expressed in murine Th1 and Th2 cells and controls Th2-specific expression of the Interleukin-5 gene. J. Biol. Chem. 1997;272:21597–21603. doi: 10.1074/jbc.272.34.21597. [DOI] [PubMed] [Google Scholar]
- 41.Kishikawa H, Sun J, Choi A, Miaw SC, Ho IC. The cell type-specific expression of the murine IL-13 gene is regulated by GATA-3. J. Immunol. 2001;167:4414–4420. doi: 10.4049/jimmunol.167.8.4414. [DOI] [PubMed] [Google Scholar]
- 42.Lavenu-Bombled C, Trainor CD, Makeh I, Romeo P-H, Max-Audit I. IL-13 gene expression is regulated by GATA-3 in T cells: Role of a critical association of a GATA and two GATG motifs. J. Biol. Chem. 2002;277:18313–18321. doi: 10.1074/jbc.M110013200. [DOI] [PubMed] [Google Scholar]
- 43.Hollenhorst PC, Jones DA, Graves BJ. Expression profiles frame the promoter specificity dilemma of the ETS family of transcription factors. Nucl. Acids Res. 2004;32:5693–5702. doi: 10.1093/nar/gkh906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Heinzel FP, Sadick MD, Holaday BJ, Coffman RL, Locksley RM. Reciprocal expression of interferon gamma or interleukin 4 during the resolution or progression of murine leishmaniasis. Evidence for expansion of distinct helper T cell subsets. J Exp Med. 1989;169:59–72. doi: 10.1084/jem.169.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Noben-Trauth N, Kropf P, Müller I. Susceptibility to leishmania major infection in interleukin-4-deficient mice. Science (New York, N.Y. 1996;271:987–990. doi: 10.1126/science.271.5251.987. [DOI] [PubMed] [Google Scholar]
- 46.Wilson CB, Rowell E, Sekimata M. Epigenetic control of T-helper-cell differentiation. 2009;9:91–105. doi: 10.1038/nri2487. [DOI] [PubMed] [Google Scholar]
- 47.Pai SY, Truitt ML, Ho IC. GATA-3 deficiency abrogates the development and maintenance of T helper type 2 cells. Proc Natl Acad Sci U S A. 2004;101:1993–1998. doi: 10.1073/pnas.0308697100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blumenthal SG, Aichele G, Wirth T, Czernilofsky AP, Nordheim A, Dittmer J. Regulation of the human Interleukin-5 promoter by Ets transcription factors. Ets1 and Ets2, but not ELF-1, cooperate with GATA3 and HTLV-1 Tax1. J. Biol. Chem. 1999;274:12910–12916. doi: 10.1074/jbc.274.18.12910. [DOI] [PubMed] [Google Scholar]
- 49.Wang J, Shannon MF, Young IG. A role for Ets1, synergizing with AP-1 and GATA-3 in the regulation of IL-5 transcription in mouse Th2 lymphocytes. Int. Immunol. 2006;18:313–323. doi: 10.1093/intimm/dxh370. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Young IG. Eosinophilic inflammation: mechanisms regulating IL-5 transcription in human T lymphocytes. Allergy. 2007;62:1131–1138. doi: 10.1111/j.1398-9995.2007.01510.x. [DOI] [PubMed] [Google Scholar]
- 51.Grenningloh R, Kang BY, Ho I-C. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005;201:615–626. doi: 10.1084/jem.20041330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ptashne M. A Genetic Switch. Blackwell Scientific Publications and Cell Press; Cambridge, Massachusetts: 1992. [Google Scholar]
- 53.Su W, Jackson S, Tjian R, Echols H. DNA looping between sites for transcriptional activation: self- association of DNA-bound Sp1. Genes Dev. 1991;5:820–826. doi: 10.1101/gad.5.5.820. [DOI] [PubMed] [Google Scholar]
- 54.Knight JD, Li R, Botchan M. The activation domain of the bovine papillomavirus E2 protein mediates association of DNA-bound dimers to form DNA loops. Proc. Natl. Acad. Sci. U S A. 1991;88:3204–3208. doi: 10.1073/pnas.88.8.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drissen R, Palstra RJ, Gillemans N, Splinter E, Grosveld F, Philipsen S, de Laat W. The active spatial organization of the beta-globin locus requires the transcription factor EKLF. Genes Dev. 2004;18:2485–2490. doi: 10.1101/gad.317004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.