Our findings suggest that the heterogeneous genetic and cellular expression patterns within glioblastoma multiforme, in part, influence anatomic and physiologic MR imaging.
Abstract
Purpose:
To determine whether magnetic resonance (MR) imaging is influenced by genetic and cellular features of glioblastoma multiforme (GBM) aggressiveness.
Materials and Methods:
In this HIPAA-compliant institutional review board–approved study, multiple enhancing and peritumoral nonenhancing stereotactic neurosurgical biopsy samples from treatment-naïve GBMs were collected prospectively, with guidance from cerebral blood volume (CBV) MR imaging measurements. By using monoclonal antibodies, tissue specimens were examined for microvascular expression, hypoxia, tumor and overall cellular density, and histopathologic features of GBM aggressiveness. Genetic expression patterns were investigated with RNA microarrays. Imaging and histopathologic variables were compared with the Welch t test and Pearson correlations. Microarray analysis was performed by using false discovery rate (FDR) statistics.
Results:
Tumor biopsy of 13 adult patients yielded 16 enhancing and 14 peritumoral nonenhancing specimens. Enhancing regions had elevated relative CBV and reduced relative apparent diffusion coefficient (ADC) measurements compared with peritumoral nonenhancing biopsy regions (P < .01). A positive correlation was found between relative CBV and all histopathologic features of aggressiveness (P < .04). An inverse correlation was found between relative ADC and all histopathologic features of aggressiveness (P < .05). RNA expression patterns between tumor regions were found to be significantly different (FDR < 0.05), with hierarchical clustering by biopsy region only.
Conclusion:
These findings suggest MR imaging is significantly influenced by GBM genetic and cellular biologic features of aggressiveness and imply physiologic MR imaging may be useful in pinpointing regions of highest malignancy within heterogeneous tissues, thus facilitating histologic grading of primary glial brain tumors.
© RSNA, 2010
Supplemental material: http://radiology.rsna.org/lookup/suppl/doi:10.1148/radiol.09090663/-/DC1
Introduction
Glioblastoma multiforme (GBM) is known to depict regional variation in magnetic resonance (MR) imaging characteristics, histopathologic features of aggressiveness, and genetic expression patterns. The heterogeneous expression of aggressive cellular features makes unguided surgical biopsy prone to sampling error, resulting in undergrading in up to 30% of cases (1–6). Assessment of GBM is primarily performed with contrast material–enhanced anatomic MR imaging; however, this imaging sequence has limited specificity in the identification of features necessary for glioma grading (6–9). The introduction of physiology based MR imaging sequences, such as diffusion-weighted (DW) imaging and T2* dynamic susceptibility weighted contrast-enhanced (DSC) perfusion-weighted (PW) imaging, hold promise in the quantitative assessment of GBM features of aggressiveness (8–20).
Apparent diffusion coefficient (ADC) is calculated to quantitatively measure relative degrees of hydrogen motion within the extracellular space (21–25). The coexistence between increased cellular nuclear-to-cytoplasmic ratio and reduced ADC has been observed in primary cerebral nervous system lymphoma; however, similar studies involving mixed-grade glioma have yielded varied results (25–29).
DSC PW MR imaging is used to quantitatively measure cerebral hemodynamic characteristics (9,30–33). Cerebral blood volume (CBV), peak height, and percentage of signal intensity recovery are three imaging measurements previously shown to be clinically useful in predicting glioma histopathologic grade and distinguishing recurrent brain tumors from radiation necrosis (6–16). Currently, DSC MR imaging measurements are not considered a valid biomarker of GBM angiogenesis because a direct comparison with tissue specimens obtained through image-guided biopsy has yet to be extensively reported (17–20).
Great strides have been made in the characterization of regional GBM genetic expression patterns. Vascular endothelial growth factor, the target of current therapeutic regimens, has been found to be up-regulated within regions of GBM angiogenesis; however, the degree to which GBM genetic expression patterns influence regional cellular features of aggressiveness and physiologic MR imaging remains poorly understood (17,18,34–38). Guided by anatomic and physiologic MR imaging characteristics, we prospectively collected stereotactic neurosurgical biopsy specimens from multiple regions of GBM to determine if the heterogeneous biology, as depicted by MR imaging, is due to underlying regional differences in intratumoral cellular and genetic expression patterns. The purpose of this preliminary study was to determine whether MR imaging is influenced by genetic and cellular features of GBM aggressiveness.
Materials and Methods
Patient Population
Adult patients referred to the University of California, San Francisco, Neurologic Surgery Service for resection of newly diagnosed, treatment-naïve, contrast material–enhancing lesions that were seen on MR images and suspected to be primary GBMs were prospectively enrolled in this Health Insurance Portability and Accountability Act–compliant, institutional review board– and committee on human research–approved study between July 2007 and May 2008. Patients younger than 18 years of age or with a medical history of a treated primary brain tumor or systemic primary cancer were not enrolled in this study. Two patients with suspected GBM were unenrolled because of tumor nonenhancement on MR images. In total, 13 adult patients (mean age, 57.5 years ± 11.0 [standard deviation]; age range, 41–74 years)—eight of whom were men (mean age, 58.6 years ± 9.0; age range, 48–74 years) and five of whom were women (mean age, 55.6 years ± 14.4; age range, 41–72 years)—were included in this study after written informed consent was obtained.
MR Imaging Protocol
All patients underwent preoperative anatomic and physiologic MR imaging with a 1.5-T Signa Horizon MR imager (GE Medical Systems, Milwaukee, Wis). All DW and DSC MR images were acquired by following the same imaging protocols. For axial DW imaging, the imaging parameters were as follows: repetition time msec/echo time msec, 10 000/99; 5-mm section thickness; no intersection gap; 24-cm field of view; and 1000 sec/mm2 b value. For T2* DSC PW gradient-echo echo-planar imaging, the parameters were as follows: 1250/54, 35° flip angle, and 5-mm section thickness. Patients underwent DSC PW MR imaging as previously described by Lupo et al (12), Cha et al (13–14), and Barajas et al (15–16). Contrast material was not administered prior to the start of the DSC PW MR imaging sequence. An echo time of 54 msec was chosen for the gradient-echo technique to maximize the effect of susceptibility changes. A 35° flip angle was selected to minimize the T1 effect during the first pass of the contrast agent.
MR Imaging Analysis and Preoperative Biopsy Site Planning
Image processing and biopsy site selection were performed in a blinded manner by one author (R.F.B., more than 3 years of experience) using commercially available software (Functool; GE Medical Systems). MR images were viewed at a standard picture archiving and communication system and scored for lesion location, presence of contrast enhancement and central necrosis, and degree of T2 edema and mass effect (39). Postcontrast three-dimensional spoiled gradient-recalled acquisition in the steady state images, T2* DSC PW images, and DW images were transferred to a commercially available image processing workstation (Advantage; GE Medical Systems). T2* DSC PW images were used in the production of CBV maps and T2* signal intensity-time curves on a voxel-by-voxel basis. Regions of signal intensity threshold values less than 100 signal intensity units or greater than 4200 signal intensity units were excluded from analysis. CBV was calculated by integrating the negative enhancement portion of the T2* signal intensity-time curve. Diffusion-weighted images were used to produce ADC maps.
Postcontrast three-dimensional spoiled gradient-recalled acquisition in the steady state images were aligned with the T2* DSC PW images, DW images, and CBV and ADC maps. Up to six enhancing and peritumoral nonenhancing regions with the highest CBV were preoperatively selected for biopsy and loaded into image-guided surgical navigation systems (Vector Vision Neuronavigation System, BrainLAB, Westchester, Ill; Stealth Station Navigation System, Medtronic, Minneapolis, Minn). We used the aligned image data set to manually define a circular 75-mm2 region of interest for each planned biopsy site, allowing for the corresponding mean, minimum, and maximum CBV and ADC values to be recorded. The size of the region of interest was selected to account for any possible inaccuracy between MR imaging measurement location, localization of biopsy sites, or intraoperative shift in brain tissue.
The T2* signal intensity-time curves generated from each region of interest were used in the quantification of peak height and percentage of signal intensity recovery imaging values (Fig 1F) (15,16). All PW and DW measurements were standardized to a 50-mm2 region of interest placed on the contralateral normal-appearing white matter to yield the following relative imaging values: rCBV, rPH, rPSR, and relative ADC (rADC). Regions of interest were approved by an attending neuroradiologist who was certified by the American Board of Radiology with a certificate-added qualification in neuroradiology and who was blinded to all patient data (S.C., more than 20 years of experience).
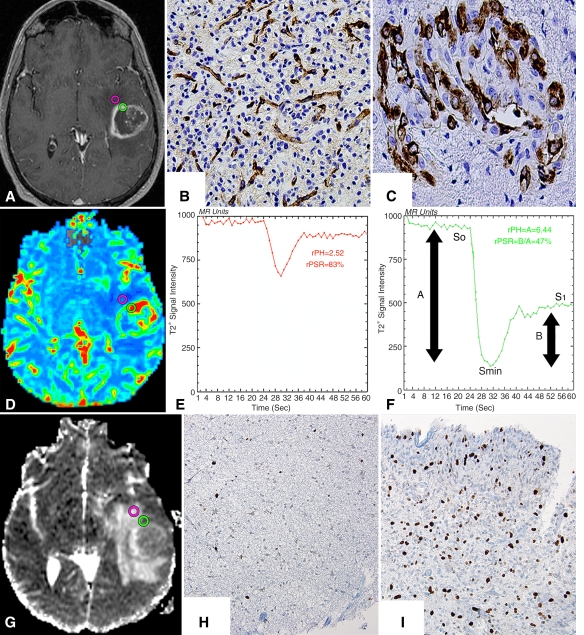
Figure 1:
Histopathologic correlation of MR imaging with stereotactic biopsy specimens in patient 10, a 51-year-old man with a left temporal GBM. A, Axial three-dimensional spoiled gradient-recalled acquisition in the steady state T1-weighted contrast-enhanced image with, D, an aligned CBV map, and, G, an ADC map show preoperatively selected nonenhancing (pink circle) and enhancing (green circle) biopsy regions. B, C, Tissue specimens (corresponding to A) were stained with factor VIII monoclonal antibody and show normal delicate microvasculature (brown-stained cells) and complex hyperplastic microvasculature within, B, nonenhancing and, C, enhancing biopsy samples. (Factor VIII monoclonal antibody stain; original magnification ×20.) E, F, T2* signal intensity time curves (corresponding to D) from, E, nonenhancing and, F, enhancing regions show increased relative CBV (rCBV) and relative peak height (rPH) with reduced relative percentage of signal intensity recovery (rPSR) within the enhancing region. Peak height (PH) was calculated with the following equation: PH = S0−Smin. Percentage of signal intensity recovery (PSR) was calculated with the following equation: PSR = (S1−Smin)/(So−Smin), where S0 is the baseline precontrast signal intensity, Smin is the lowest signal intensity during the first passage of the intravascular contrast bolus, and S1 is the signal baseline during the recirculation steady-state phase after administration of the intravascular contrast material bolus. H, I, Histopathologic slides (corresponding to G) of, H, nonenhancing and, I, enhancing tissue samples stained for cellular mitosis show both biopsy samples infiltrated with malignant cells associated with increased mitotic activity (brown-stained cells) and overall cellularity within, I, enhancing regions correlating to areas of reduced ADC. (Mindbomb homolog 1 monoclonal antibody stain; original magnification ×20.)
Image-guided Biopsy Specimen Processing
All biopsy specimens were collected with image-guided stereotactic biopsy techniques by one of three authors (A.T.P, M.W.M, M.S.B; each with more than 20 years of experience). Screen shots and MR imaging coordinates of biopsy locations were recorded to confirm the accuracy of biopsy sampling from preplanned locations. One half of the biopsy specimen was used for RNA microarray analysis. The second half underwent histopathologic analysis.
Cellular density was quantified on mindbomb homolog 1 monoclonal antibody–stained tissue slices with a standardized proliferation index (40–45). Overall tumor cellular density was manually quantified as the average of the total number of cells within three high-power fields at a magnification of ×20 (1.0 mm2). Microvascular expression and hypoxia were measured qualitatively with factor VIII monoclonal antibody– and carbonic anhydrase IX monoclonal antibody–stained sections and use of a four-tier ordinal scale (0, no immunoreactivity; 3, intense immunoreactivity) within three high-power fields at a magnification of ×20. Microvascular morphology was graded as delicate (resembling normal), simple hyperplasia (hyperplastic with definitive lumen), or glomeruloid (complex hyperplasia) type. An attending neuropathologist (S.R.V, more than 20 years of experience) who was blinded to the results of MR imaging performed all quantitative and qualitative assessments.
Microarray Analysis
Sample preparation, labeling, and array hybridizations were performed in accordance with standard protocols (46,47). Total RNA quality was assessed with a Pico Chip on an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, Calif). Cyanine cytidine 5-triphosphate–labeled RNA was assessed with Nandrop ND-100 (Nanodrop Technologies, Wilmington, Del). Equal amounts of Cy3-labeled target were hybridized to Agilent whole mouse genome 4 × 44 K ink jet arrays (Agilent Technologies). Arrays were scanned with the microarray scanner (Agilent Technologies), and raw signal intensities were extracted with Feature Extraction software (version 9.1; Agilent Technologies). The microarray data set was normalized with the quantile normalization method (48) by using the Linear Models for Microarray Data (LIMMA) R package (Bioconductor, Cambridge, England) (49). Additionally, genetic expression patterns from six samples of nonneoplastic human brain gliosis, previously obtained in a separate cohort of six adult patients, were examined to help differentiate gene expression due to astrocyte repair or scarring.
Statistical Analysis
Separate comparison of MR imaging and histopathologic variables was investigated with a linear fit model, treating all ordinal variables as continuous and taking into account subject effects. Pearson correlation was used to analyze the relationships between imaging and histopathologic variables.
To compare gene expression between contrast-enhancing and nonenhancing biopsy sites, a linear model was fit to the comparison to enable us to estimate the mean fold change (M value) for each gene and calculate the moderated t statistic, B statistic (log10 posterior odds ratios), false discovery rate (FDR), and adjusted P value for each gene comparison of interest (50–52). All gene expression analysis was performed by using the aforementioned LIMMA software (49,53). Functional annotation of microarray expression data was performed with the Database for Annotation, Visualization, and Integrated Discovery (54,55).
Results
Patient Population
Table 1 summarizes the patient population included in this study. Thirty biopsy specimens were obtained; 16 were from contrast-enhancing regions, and 14 were from peritumoral nonenhancing regions (Table E1 [online]). Malignant tumor cells infiltrated all tissue specimens. In 85% of patients, all lesions had contrast enhancement on MR images that was associated with the presence of central necrosis. In 77% of patients, a moderate to severe degree of T2 prolongation was observed in the setting of mild to absent mass effect.
Table 1.
Patient Population Data and Clinical Outcome
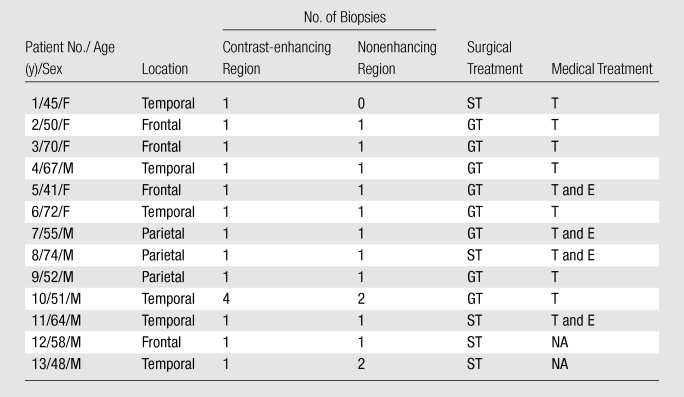
Note.—GBM was independently confirmed in all patients with clinical examination of separate biopsy specimens. E= enzastaurin, GT= gross total surgical resection, NA= patient declined treatment or medical therapy not clinically indicated, ST= subtotal surgical resection, T= temozolamide.
Comparison of Regional PW and DW Imaging Characteristics
Table 2 summarizes regional PW and DW imaging measurements. Contrast-enhancing regions had rCBV and rPH values that were significantly elevated (P < .01) compared with those in peritumoral nonenhancing regions selected for biopsy (Table 2). The rPSR and rADC values within contrast-enhancing regions were found to be significantly lower (P < .01) than those in peritumoral nonenhancing regions (Table 2). Correlation analysis revealed a strong correlation between all physiologic MR imaging measurements (P < .02), with the exception of the rPH and rADC comparison (P = .14, Fig E1 [online]).
Table 2.
Regional PW and DW MR Imaging Findings
Note.—Data are means ± standard deviations. Unless otherwise indicated, all P values were less than .01.
P = .02.
Comparison of Regional Cellular Features of GBM Aggressiveness
Table 3 summarizes the regional histopathologic findings of this study. Contrast-enhancing biopsy sites had significantly elevated factor VIII expression (P < .01), with increased expression of simple and complex hyperplastic microvasculature (P < .01 and P = .03, respectively; Table 3). Enhancing biopsy sites were also found to have a significantly higher expression of cellular mitosis (mindbomb homolog 1 monoclonal antibody), overall cellularity, and hypoxia (carbonic anhydrase IX monoclonal antibody) compared with peritumoral nonenhancing biopsy regions (P < .01, Table 3).
Table 3.
Regional Immunohistochemical Pathologic Findings

Note.—Data are means ± standard deviations. Unless otherwise indicated, all P values were less than .01.
Microvascular expression (factor VII) graded on a scale from 0 to 9.
Vascular hyperplasia (factor VIII) graded on a scale from 0 to 3.
P = .03.
Hypoxia (carbonic anhydrase IX monoclonal antibody) graded on a scale from 0 to 3.
Cellular density is equal to a percentage of mindbomb homolog 1 expression.
Tumor density is equal to the number of cells within four ×20 magnification fields.
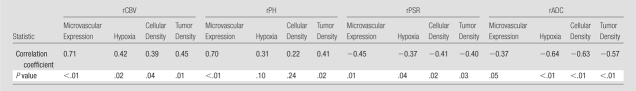
Correlation of Physiologic MR Imaging and Cellular Features of GBM Aggressiveness
Table 4 lists the results of correlation analysis between DSC PW and DW imaging values and the cellular histopathologic features of GBM aggressiveness for all biopsy sites. A positive correlation between rCBV and microvascular expression, hypoxia, cellular mitosis, and overall cellularity was observed (P ≤ .04, Table 4, Fig E2 [online]). In addition, rADC and rPSR showed an inverse correlation with microvascular expression, hypoxia, cellular mitosis, and overall cellularity (P ≤ .05, Table 4, Fig E3 [online]).
Table 4.
Correlation Analysis of Mean DSC PW and DW Imaging Values and Histopathologic Characteristics
Note.—Correlation values were calculated by using measurements from all 30 biopsy locations.
MR Imaging Predictive of GBM Genetic Expression Patterns
RNA from a patient in whom one contrast-enhancing sample and one peritumoral nonenhancing sample were obtained was of insufficient quality to be hybridized to the microarray platform. As a result, RNA extracted from 12 of 13 patients (28 of 30 biopsies) was of sufficient quality to be included in the microarray analysis of this study. RNA expression patterns from six gliosis (previously obtained from a separate cohort of adult patients), 15 enhancing, and 13 nonenhancing biopsy sites were found to be significantly different (FDR < 0.05, Table 5).
Table 5.
Genes Differentially Expressed between Biopsy Locations

Note.—Data are numbers of genes. FDR is less stringent than adjusted P value in controlling the false-positive rates. B statistics are more powerful in the detection of weak gene differential gene expression but are not comparable with the other two methods in terms of the false-positive rate.
An adjusted P value of .05 indicates that the declared differential gene expression set has a 5% chance to have at least one false-positive finding for each gene comparison of interest.
FDR is the percentage of falsely declared differential gene expression among the set of declared differentially expressed genes. An FDR cutoff of 0.05 indicates that 5% of the declared differentially expressed genes are expected to be false-positive findings.
B statistic (log10 posterior odds ratios) greater than 0 indicates equal probability or better that a gene is differentially expressed as opposed to nondifferentially expressed between sample groups.
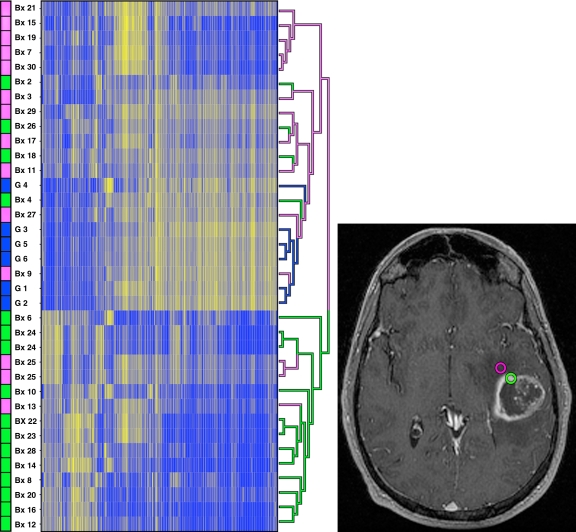
The largest degree of differential gene expression occurred between contrast-enhancing samples and gliosis samples (11389 genes, FDR < 0.05). Hierarchical clustering of the 500 most variant genes across arrays revealed clustering only by biopsy sample (Fig 2). Clustering by array set, hybridization date, and patients was not observed, suggesting that the arrays were successful in depicting a large degree of regional biologic variability.
Figure 2:
Presence of contrast enhancement is predictive of GBM genetic expression pattern. Right: Axial three-dimensional spoiled gradient-recalled acquisition in the steady state T1-weighted contrast-enhanced MR image obtained in the same patient as in Figure 1. Purple and green circles indicate nonenhancing and enhancing biopsy regions, respectively. Left: Genetic expression map of all biopsy samples with hierarchical clustering of the 500 most variant genes shows clustering only by biopsy sample. Purpleindicates nonenhancing samples; blue, gliosis samples; and green, enhancing samples. In the left column, biopsy location and sample number are provided for cross reference purposes with Table E1 [online]. Gliosis samples 1 through 6 were included as control samples to enable us to differentiate between gene expression due to astrocyte repair and scarring. These are not included in Table E1 [online]. RNA extracted from biopsy samples 1 and 5 was of insufficient quality to be hybridized to the microarray platform; therefore, these samples were not included in this analysis. Separate distinct sample locations from biopsies 24 and 25 were used twice in this genetic analysis. Bx = biopsy sample, G = gliosis sample.
To further explore differential gene expression between contrast-enhancing and nonenhancing biopsy locations, we compared the average expression of 36866 probe sets (genes) between enhancing and nonenhancing samples. Overall, 6653 genes were differentially expressed between contrast-enhancing and nonenhancing biopsy sites (FDR < 0.05). A total of 359 genes (302 unique UniGene identification numbers) were significantly overexpressed at least twofold on average in contrast-enhancing samples relative to peritumoral nonenhancing samples (M > 1, B > 0, FDR < 0.05; Table E2 [online]). Conversely, 684 probe sets (544 unique UniGene identification numbers) were significantly overexpressed at least twofold on average in peritumoral nonenhancing samples relative to enhancing samples (M > 1, B > 0, FDR < 0.05; Table E3 [online]).
To determine the biologic processes associated with the observed genetic expression patterns, we assessed their gene ontology classifications with the Database for Annotation, Visualization, and Integrated Discovery (54,55). Consistent with the significant increase in mindbomb homolog 1 monoclonal antibody staining in enhancing regions, gene ontology classification revealed highly significant enrichment (P < .01, FDR < 0.01) for 30 genes associated with mitosis within enhancing regions (Tables E4, E5 [online]). A total of 19 genes associated with angiogenesis (P < .01, FDR = 3.58) and apoptosis (P = .03, FDR = 46) also tended to be highly expressed within enhancing regions; however, this tendency was not significant.
As a cohort, vascular endothelial growth factor (UniGene identification number, 73793) expression level was found to be significantly elevated within enhancing samples (average log2 intensity, 11.35 ± 0.92) compared with that within nonenhancing samples (average log2 intensity, 8.86 ± 0.77; FDR < 0.03). Eight patients had more than a threefold increase (range, 3–34) in vascular endothelial growth factor expression levels within enhancing regions relative to that within nonenhancing regions.
To further characterize the mitosis gene signature associated with contrast-enhancing tumor regions, we assessed expression of 41 mitosis-related genes overexpressed in GBM specimens previously propagated as subcutaneous xenografts. This revealed that 33 of the 41 mitosis-related genes were significantly overexpressed within contrast-enhancing samples (FDR < 0.05, Table 6). All 33 genes were among the most highly expressed genes (98th percentile) in contrast-enhancing samples after rank ordering all 36866 microarray probes on the basis of differential gene expression between the two sample groups.
Table 6.
Mitotic Genetic Signatures Prospectively Identified as Significantly Overexpressed within Contrast-enhancing Regions
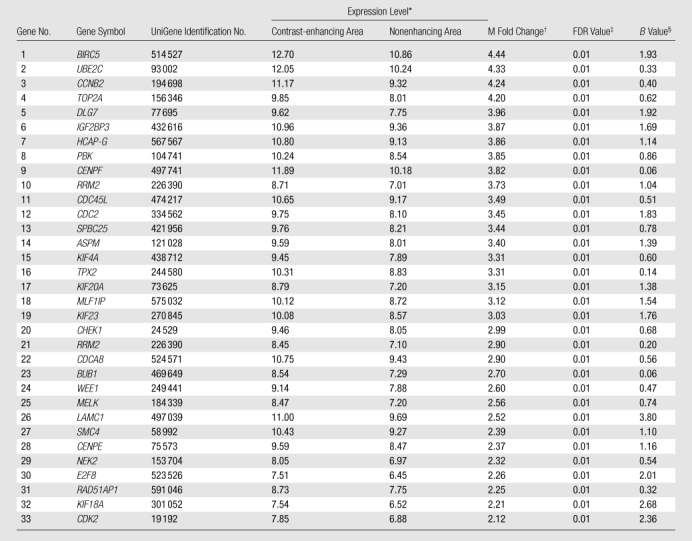
Expression level is the average log2-based intensity of the same probe across all arrays and serves as a proxy for gene expression level.
M fold change is the log2-based fold change of contrast-enhancing lesion over non-contrast-enhancing lesion. If M is equal to 1, it means there is a twofold increase in contrast-enhancing lesion compared with non-contrast-enhancing lesion. If M is equal to 2, it means there is a fourfold up-regulation.
FDR is the percentage of falsely declared genes among the set of declared genes. An FDR cutoff of 0.05 indicates that 5% of the declared genes are expected to be false-positive findings.
B value (log10 posterior odds ratios) is the ratio between the probability that a given gene is differentially expressed over the probability that a given gene is not differentially expressed.
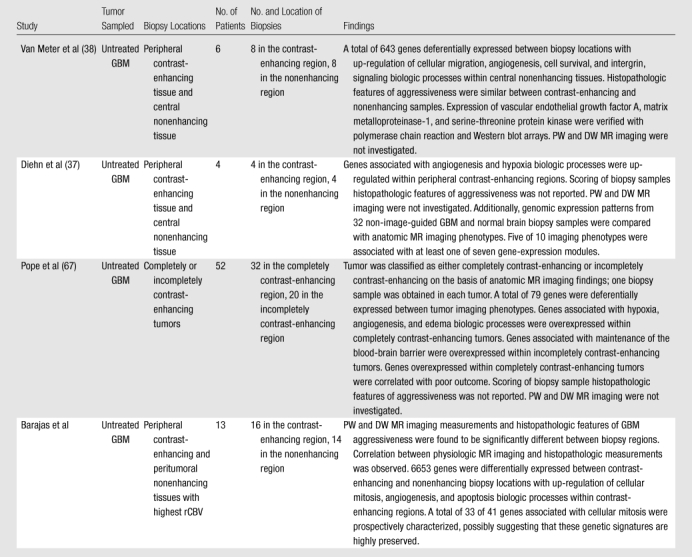
compare and contrast the findings of our study with findings of previous MR-guided investigations of GBM intra- and intertumoral genetic expression patterns.
Table 7.
Comparison of MR-guided Studies Performed to Investigate Intra- and Intertumoral Genetic Expression Patterns
Discussion
In this study, we collected tissue samples from prospectively defined intraoperative biopsy sites within multiple regions of treatment-naïve GBM and used MR-guided stereotactic neurosurgical techniques to investigate how inter- and intratumoral regional differences in genetic and cellular expression patterns influence anatomic and physiologic MR imaging. We found that DSC PW and DW imaging measurements were significantly different between biopsy regions. Additionally, DSC PW and DW imaging measurements were found to correlate with GBM histopathologic features of aggressiveness. Finally, a significant difference in genetic expression patterns was observed between biopsy regions. Up-regulated genes were associated with similar cellular malignant biologic processes observed to correlate with anatomic- and physiologic-based MR imaging measurements.
In our study, we observed an inverse correlation between rADC measurements and the histopathologic features of GBM aggressiveness. Previous studies have yielded mixed findings, likely due to the complex interactions of water diffusivity within the heterogeneous inter- and intracellular environment of gliomas (17,26–29). Many factors—including nuclear-to-cytoplasmic ratio, intra- and extracellular edema, and cellular density—have been proposed to influence the degree to which water molecular motion is reduced (26–29). The results of our study lend further evidence to the theory that regions of low rADC within GBM represent sites of neoplastic angiogenesis and elevated cellular density.
Our results show that there is a correlation between rCBV, rPH, and the histopathologic features of glioma aggressiveness. The correlation between rCBV, rPH, and neoplastic angiogenesis is likely due to regional variation in microvascular morphologic expression within GBM (17,56–60). The regions of highest aggressiveness contain complex and simple microvascular hyperplasia (58–60). In our study, complex and simple microvascular hyperplasia were significantly overexpressed within contrast-enhancing regions and corresponded to a significant increase in rCBV and rPH measurements. Vascular proliferation has long been recognized as a biomarker of glioma aggressiveness (1–3,59–62). Broadly elevated microvascular density has been shown to correlate with elevated tumor cellularity, malignant progression, and extensive tumor invasiveness, resulting in poor prognostic outcomes (59–63). Discrepancies between the presence of contrast enhancement and regions of GBM aggressiveness have been previously documented; however, the sampling of contrast-enhancing tissues for diagnostic specimens remains the current standard of care (62–66). While contrast-enhancing tissues on anatomic MR images may appear homogeneous, they are often found to be histopathologically heterogeneous. The unguided sampling of contrast-enhancing tissues can result in undergrading. The results of this and other studies indicate that physiologic MR imaging measurements may prove useful in supplementing anatomic MR imaging when intraoperatively selecting diagnostic biopsy regions. Specifically, the sampling of contrast-enhancing regions with the highest rCBV may prove to reduce sampling error if only enhancing regions are to be sampled (59–66).
In light of the finding that histologic features of GBM aggressiveness were overexpressed within biopsy specimens obtained in the enhancing region, we hypothesized that the genetic expression patterns between enhancing and nonenhancing regions would be significantly different. This hypothesis was confirmed when we found that 359 genes significantly overexpressed within contrast-enhancing samples tended to cluster into biologic processes associated with regulation of mitosis, angiogenesis, and apoptosis. Furthermore, the ability to prospectively characterize mitotic gene signatures previously propagated from GBM subcutaneous xenografts may suggest that these mitotic genes are highly preserved. The findings of regional variation in vascular endothelial growth factor expression, correlation of cellular and genetic expression patterns with physiologic MR imaging measures, and ability to prospectively characterize mitotic gene signatures possibly highlights therapeutic opportunities that could be used to test targeted treatment strategies—the effects of which, in light of our findings—possibly could be monitored with MR imaging.
The findings of our study are consistent with the findings of previous MR-guided investigations of GBM intra- and intertumoral genetic expression patterns (37,38,67). Similar to previous researchers, we found that contrast-enhancing tissues have significant up-regulation of genes associated with cellular proliferative or infiltrative processes, hypoxia, and angiogenesis biologic processes. Validated up-regulation of these biologic processes may further influence future diagnostic and therapeutic strategies.
Our study differed from previous investigations by Van Meter et al, Diehn et al, and Pope et al (37,38,67) in several aspects. First, recent investigations have addressed correlations between anatomic MR imaging and tumor genetic expression patterns without exploring associations with histopathology, which is the reference standard for glioma diagnosis and staging. In our study, we directly correlated MR imaging findings with clinically important histopathologic and molecular genetic markers of cellular mitosis and angiogenesis, possibly providing biologic and molecular correlates for much of the structural, physiologic, and functional information encoded within the MR images of GBM. Second, previous researchers have reported genetic differential expression between regions of central nonenhancing and adjacent enhancing tissues (37,38). While knowledge of genetic expression patterns of GBM central nonenhancement is undoubtably important, we believe investigating peritumoral nonenhancing tumor regions with deranged DSC PW and DW MR imaging is clinically pragmatic, as these are the areas most at risk for future tumor progression. Third, we report the prospective validation of 33 mitotic gene signatures within enhancing tumors that may emphasize potential therapeutic opportunities which, in light of our findings, may be spatially resolved with MR imaging.
Our findings suggest that the heterogeneous genetic and cellular expression patterns within GBM, in part, influence anatomic and physiologic MR imaging. These findings have several clinical implications, if confirmed through further prospective study. First, physiologic MR imaging may provide useful information that complements conventional contrast-enhanced MR imaging, making it possible to noninvasively identify regions of glioma aggressiveness and allowing for the intraoperative collection of tumor specimens of highest aggressive cellular features. Second, the synthesis of MR imaging, histopathologic, and functional genetic analysis furthers the validation of imaging biomarkers that in the future maybe used to noninvasively identify, prognosticate, and monitor clinically important features of GBM aggressiveness (37,38,67).
Our report was the result of a preliminary exploratory analysis; therefore, it had several limitations. First, given the limited sample size, we advocate further prospective investigation to confirm the efficacy of the techniques described in this study. Second, the potential for a mismatch in registration between the biopsy site and MR images on the neurosurgical navigation system because of intraoperative shifts in brain tissues theoretically exists. We sought to minimize substantial amounts of brain shift by (a) performing accurate intraoperative neuronavigational system registration to the patient’s facial anatomy, (b) avoiding substantial loss of cerebrospinal fluid, (c) watching for intraoperative brain swelling, (d) testing registration accuracy against visible cortical landmarks immediately prior to biopsy sampling, (e) obtaining biopsy specimens from MR homogeneous tissues (entirely enhancing or nonenhancing tissue) located 1 cm apart prior to resection, and (f) using standardized regions of interest of sufficient size to compensate for any minimal shift in brain location. We believe that any minimal amount of brain shift that may have occurred prior to biopsy sampling did not result in significant sampling error or adversely affect the results of this study; however, in further prospective studies, researchers may consider the use of closed brain biopsies, when clinically prudent, to negate this potential limitation.
In conclusion, the results of our preliminary study, albeit one with a small sample size and in need of prospective independent validation with a larger cohort of patients, suggest MR imaging is significantly influenced by GBM genetic and cellular biologic features of aggressiveness and imply physiologic-based MR imaging may be useful to clinicians by pinpointing regions of highest malignancy within a heterogeneous tumor facilitating histologic grading of primary glial brain tumors.
Advances in Knowledge.
In this preliminary study, regional physiologic MR imaging measurements, genetic expression patterns, and cellular features of glioma aggressiveness were found to differ significantly within glioblastoma multiforme (GBM) on the basis of the presence or absence of contrast enhancement.
Up-regulated cellular and genetic expression of angiogenic and mitotic signatures within GBM contrast-enhancing regions correlate with physiologic MR imaging measurements.
Anatomic and physiologic MR imaging is significantly influenced by GBM genetic and cellular biologic features of aggressiveness.
Implications for Patient Care.
Physiologic MR imaging provides useful information that complements contrast-enhanced MR imaging in the noninvasive identification of regions of glioma aggressiveness prior to surgical intervention.
Physiologic MR imaging may prove successful in providing important information about the stratification of grades of malignancy within gliomas during intraoperative biopsy collection, allowing for selection of tumor specimens of the highest aggressive cellular features, thereby reducing common sampling errors.
Supplementary Material
Acknowledgments
The first author thanks Alvin Au, King Chiu, Cynthia Cowdrey, Celeste Thomas, and Bethany Barajas for their tissue processing assistance and helpful comments regarding this manuscript. In addition, the first author acknowledges the Sandler Asthma Basic Research Center Functional Genomics Core Facility and staff (Rebecca Barbeau, Chris Eisley, Andrea Barczak, and David Erle) for their gene analysis assistance and expertise.
Received April 16, 2009; revision requested June 2; revision received July 22; accepted August 12; final version accepted August 27.
Authors stated no financial relationship to disclose.
Funding:This research was supported by the National Institutes of Health(grants NS045013, TL1 RR024129-01, and UL1RR024131).
Abbreviations:
- ADC
- apparent diffusion coefficient
- CBV
- cerebral blood volume
- DSC
- dynamic susceptibility weighted contrast enhanced
- DW
- diffusion weighted
- FDR
- false discovery rate
- GBM
- glioblastoma multiforme
- PW
- perfusion weighted
- rADC
- relative ADC
- rCBV
- relative CBV
- rPH
- relative peak height
- rPSR
- relative percentage of signal intensity recovery
References
- 1.Fulling KH, Garcia DM. Anaplastic astrocytoma of the adult cerebrum: prognostic value of histologic features. Cancer 1985;55:928–931 [DOI] [PubMed] [Google Scholar]
- 2.Kleihues P, Soylemezoglu F, Schäuble B, Scheithauer BW, Burger PC. Histopathology, classification, and grading of gliomas. Glia 1995;15:211–221 [DOI] [PubMed] [Google Scholar]
- 3.Gilles FH, Brown WD, Leviton A. Limitations of the World Health Organization classification of childhood supratentorial astrocytic tumors. Cancer 2000;88:1477–1483 [PubMed] [Google Scholar]
- 4.Prayson RA, Agamanolis DP, Cohen ML. Interobserver reproducibility among neuropathologists and surgical pathologists in fibrillary astrocytoma grading. J Neurol Sci 2000;175:33–39 [DOI] [PubMed] [Google Scholar]
- 5.Coons SW, Johnson PC, Scheithauer BW, et al. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer 1997;79:1381–1393 [DOI] [PubMed] [Google Scholar]
- 6.Jackson A, Kassner A, Annesley-Williams D. Abnormalities in the recirculation phase of contrast agent bolus passage in cerebral gliomas: comparison with relative blood volume and tumor grade. AJNR Am J Neuroradiol 2002;23:7–14 [PMC free article] [PubMed] [Google Scholar]
- 7.Law M, Oh S, Johnson G, et al. Perfusion magnetic resonance imaging predict patient outcome as an adjunct to histopathology. Neurosurgery 2006;58:1099–1107 [DOI] [PubMed] [Google Scholar]
- 8.Cha S. Update on brain tumor imaging: from anatomy to physiology. AJNR Am J Neuroradiol 2006;27:475–487 [PMC free article] [PubMed] [Google Scholar]
- 9.Lev MH, Rosen BR. Clinical applications of intracranial perfusion MR imaging. Neuroimaging Clin N Am 1999;9:309–331 [PubMed] [Google Scholar]
- 10.Henry RG, Vigneron DB, Fischbein NJ, et al. Comparison of relative cerebral blood volume and proton spectroscopy in patients with treated gliomas. AJNR Am J Neuroradiol 2000;21:357–366 [PMC free article] [PubMed] [Google Scholar]
- 11.Sugahara T, Korogi Y, Kochi M, et al. Correlation of MR imaging-determined cerebral blood volume maps with histologic and angiographic determination of vascularity of gliomas. AJR Am J Roentgenol 1998;171:1479–1486 [DOI] [PubMed] [Google Scholar]
- 12.Lupo JM, Cha S, Chang SM, et al. Dynamic susceptibility-weighted perfusion imaging of high-grade gliomas: characterization of spatial heterogeneity. AJNR Am J Neuroradiol 2005;26:1446–1454 [PMC free article] [PubMed] [Google Scholar]
- 13.Cha S, Lu S, Johnson G, et al. Dynamic susceptibility contrast MR imaging: correlation of signal intensity changes with cerebral blood volume measurements. J Magn Reson Imaging 2000;11:114–119 [DOI] [PubMed] [Google Scholar]
- 14.Cha S, Lupo JM, Chen MH, et al. Differentiation of glioblastoma multiforme and single brain metastasis by peak height and percentage of signal intensity recovery derived from dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol 2007;28:1078–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barajas RF, Chang JS, Sneed PK, Segal MR, McDermott MW, Cha S. Distinguishing recurrent brain metastasis from radiation necrosis following gamma knife radiosurgery using dynamic susceptibility weighted contrast-enhanced perfusion MR imaging. AJNR Am J Neuroradiol 2009;30:367–372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barajas RF, Jr, Chang JS, Segal MR, et al. Differentiation of recurrent glioblastoma multiforme from radiation necrosis after external beam radiation therapy with dynamic susceptibility-weighted contrast-enhanced perfusion MR imaging. Radiology 2009;253:486–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadeghi N, D’Haene N, Decaestecker C, et al. Apparent diffusion coefficient and cerebral blood volume in brain gliomas: relation to tumor cell density and tumor microvessel density based on stereotactic biopsies. AJNR Am J Neuroradiol 2008;29:476–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aronen HJ, Gazit IE, Louis DN, et al. Cerebral blood volume maps of gliomas: comparison with tumor grade and histologic findings. Radiology 1994;191:41–51 [DOI] [PubMed] [Google Scholar]
- 19.Cha S, Johnson G, Wadghiri YZ, et al. Dynamic, contrast-enhanced perfusion MRI in mouse gliomas: correlation with histopathology. Magn Reson Med 2003;49:848–855 [DOI] [PubMed] [Google Scholar]
- 20.Maia AC, Jr, Malheiros SM, da Rocha AJ, et al. MR cerebral blood volume maps correlated with vascular endothelial growth factor expression and tumor grade in nonenhancing gliomas. AJNR Am J Neuroradiol 2005;26:777–783 [PMC free article] [PubMed] [Google Scholar]
- 21.Rowley HA, Grant PE, Roberts TP. Diffusion MR imaging: theory and applications. Neuroimaging Clin N Am 1999;9:343–361 [PubMed] [Google Scholar]
- 22.Le Bihan D, Turner R, Douek P, Patronas N. Diffusion MR imaging: clinical applications. AJR Am J Roentgenol 1992;159:591–599 [DOI] [PubMed] [Google Scholar]
- 23.Kotsenas AL, Roth TC, Manness WK, Faerber EN. Abnormal diffusion-weighted MRI in medulloblastoma: does it reflect small cell histology?. Pediatr Radiol 1999;29:524–526 [DOI] [PubMed] [Google Scholar]
- 24.Kitis O, Altay H, Calli C, Yunten N, Akalin T, Yurtseven T. Minimum apparent diffusion coefficients in the evaluation of brain tumors. Eur J Radiol 2005;55:393–400 [DOI] [PubMed] [Google Scholar]
- 25.Le Bihan D, Breton E, Lallemand D, et al. MR imaging of intravoxel incoherent motions: application to diffusion and perfusion in neurologic disorders. Radiology 1986;161:401–407 [DOI] [PubMed] [Google Scholar]
- 26.Guo AC, Cummings TJ, Dash RC, Provenzale JM. Lymphomas and high-grade astrocytomas: comparison of water diffusibility and histologic characteristics. Radiology 2002;224:177–183 [DOI] [PubMed] [Google Scholar]
- 27.Stadnik TW, Chaskis C, Michotte A, et al. Diffusion-weighted MR imaging of intracerebral masses: comparison with conventional MR imaging and histologic findings. AJNR Am J Neuroradiol 2001;22:969–976 [PMC free article] [PubMed] [Google Scholar]
- 28.Sugahara T, Korogi Y, Kochi M, et al. Usefulness of diffusion-weighted MRI with echo-planar technique in the evaluation of cellularity in gliomas. J Magn Reson Imaging 1999;9:53–60 [DOI] [PubMed] [Google Scholar]
- 29.Gupta RK, Cloughesy TF, Sinha U, et al. Relationships between choline magnetic resonance spectroscopy, apparent diffusion coefficient and quantitative histopathology in human glioma. J Neurooncol 2000;50:215–226 [DOI] [PubMed] [Google Scholar]
- 30.Cha S, Johnson G, Yuz M, et al. The role of contrast-enhanced perfusion MR imaging in differentiating between recurrent tumor and radiation necrosis (abstr). Radiology 1999;213(P):188 [Google Scholar]
- 31.Cha S, Knopp EA, Johnson G, et al. Dynamic contrast-enhanced T2*-weighted MR imaging of recurrent malignant gliomas treated with thalidomide and carboplatin. AJNR Am J Neuroradiol 2000;21:881–890 [PMC free article] [PubMed] [Google Scholar]
- 32.Cha S, Pierce S, Knopp EA, et al. Dynamic contrast-enhanced T2*-weighted MR imaging of tumefactive demyelinating lesions. AJNR Am J Neuroradiol 2001;22:1109–1116 [PMC free article] [PubMed] [Google Scholar]
- 33.Sorensen AG, Buonanno FS, Gonzalez RG, et al. Hyperacute stroke: evaluation with combined multisection diffusion-weighted and hemodynamically weighted echo-planar MR imaging. Radiology 1996;199:391–401 [DOI] [PubMed] [Google Scholar]
- 34.Mischel PS, Cloughesy TF, Nelson SF. DNA-microarray analysis of brain cancer: molecular classification for therapy. Nat Rev Neurosci 2004;5:782–792 [DOI] [PubMed] [Google Scholar]
- 35.Mischel PS, Shai R, Shi T, et al. Identification of molecular subtypes of glioblastoma by gene expression profiling. Oncogene 2003;22:2361–2373 [DOI] [PubMed] [Google Scholar]
- 36.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell 2006;9:157–173 [DOI] [PubMed] [Google Scholar]
- 37.Diehn M, Nardini C, Wang DS, et al. Identification of noninvasive imaging surrogates for brain tumor gene-expression modules. Proc Natl Acad Sci U S A 2008;105:5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Van Meter T, Dumur C, Hafez N, et al. Microarray analysis of MRI-defined tissue samples in glioblastoma reveals differences in regional expression of therapeutic targets. Diagn Mol Pathol 2006;15:195–205 [DOI] [PubMed] [Google Scholar]
- 39.Lim DA, Cha S, Mayo MC, et al. Relationship of glioblastoma multiforme to neural stem cell regions predicts invasive and multifocal tumor phenotype. Neuro Oncol 2007;9:424–429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aroca F, Renaud W, Bartoli C, et al. Expression of PECAM-1/CD31 isoforms in human brain gliomas. J Neurooncol 1999;43:19–25 [DOI] [PubMed] [Google Scholar]
- 41.DeYoung BR, Swanson PE, Argenyi ZB, et al. Cd31 immunoreactivity in mesenchymal neoplasms of the skin and subcutis: report of 145 cases and review of putative immunohistologic markers of endothelial differentiation. J Cutan Pathol 1995;22:215–222 [DOI] [PubMed] [Google Scholar]
- 42.Moore A, Marecos E, Simonova M, et al. Novel gliosarcoma cell line expressing green fluorescent protein: a model for quantitative assessment of angiogenesis. Microvasc Res 1998;56:145–153 [DOI] [PubMed] [Google Scholar]
- 43.Takano S, Yoshii Y, Kondo S, et al. Concentration of vascular endothelial growth factor in the serum and tumor tissue of brain tumor patients. Cancer Res 1996;56:2185–2190 [PubMed] [Google Scholar]
- 44.Burger PC, Shibata T, Kleihues P. The use of the monoclonal antibody Ki-67 in the identification of proliferating cells: application to surgical neuropathology. Am J Surg Pathol 1986;10:611–617 [DOI] [PubMed] [Google Scholar]
- 45.Karamitopoulou E, Perentes E, Diamantis I. Ki-67 immunoreactivity in human central nervous system tumors: a study with MIB 1 monoclonal antibody on archival material. Acta Neuropathol 1994;87:47–54 [DOI] [PubMed] [Google Scholar]
- 46.RNA sample preparation, labeling, and array hybridizations protocols. UCSF Shared Microarray Core Facilities Web site. http://www.arrays.ucsf.edu. Accessed March 10, 2008.
- 47.RNA sample preparation, labeling, and array hybridizations protocols. Agilent Technologies Web site. http://www.agilent.comedu. Accessed February 18, 2008.
- 48.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 2003;19:185–193 [DOI] [PubMed] [Google Scholar]
- 49.Gentleman RC, Carey VJ, Bates DM, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol 2004;5:R80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B 1995;57:289–300 [Google Scholar]
- 51.Lonnstedt I, Speed TP. Replicated microarray data. Statist Sinica 2002;2:31–46 [Google Scholar]
- 52.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat 1979;6:65–70 [Google Scholar]
- 53.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol 2004;3:Article 3. [DOI] [PubMed] [Google Scholar]
- 54.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 2009;4:44–57 [DOI] [PubMed] [Google Scholar]
- 55.Gene Ontology Annotation (GOA) Human Gene-Association Database. http://www.ebi.ac.uk/GOA. Accessed March 25, 2008.
- 56.Shweiki D, Neeman M, Itin A, et al. Induction of vascular endothelial growth factor expression by hypoxia and by glucose deficiency in multicell spheroids: implications for tumor angiogenesis. Proc Natl Acad Sci U S A 1995;92:768–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fischer I, Gagner JP, Law M, et al. Angiogenesis in gliomas: biology and molecular pathophysiology. Brain Pathol 2005;15:297–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Miyagami M, Katayama Y. Angiogenesis of glioma: evaluation of ultrastructural characteristics of microvessels and tubular bodies (Weibel–Palade) in endothelial cells and immunohistochemical findings with VEGF and p53 protein. Med Mol Morphol 2005;38:36–42 [DOI] [PubMed] [Google Scholar]
- 59.Deane BR, Lantos PL. The vasculature of experimental brain tumours. I. A sequential light and electron microscope study of angiogenesis. J Neurol Sci 1981;49:55–66 [DOI] [PubMed] [Google Scholar]
- 60.Deane BR, Lantos PL. The vasculature of experimental brain tumours. II. A quantitative assessment of morphological abnormalities. J Neurol Sci 1981;49:67–77 [DOI] [PubMed] [Google Scholar]
- 61.Kleihues P, Cavenee WK, eds. World Health Organization classification of tumors: pathology and genetics of tumors of the nervous system Lyon, France: IARC, 2000 [Google Scholar]
- 62.Knopp EA, Cha S, Johnson G, et al. Glial neoplasms: dynamic contrast-enhanced T2-weighted MR imaging. Radiology 1999;211:791–798 [DOI] [PubMed] [Google Scholar]
- 63.Brem S, Cotran R, Folkman J. Tumor angiogenesis: a quantitative method for histologic grading. J Natl Cancer Inst 1972;48:347–356 [PubMed] [Google Scholar]
- 64.Batra A, Tripathi RP, Singh AK. Perfusion magnetic resonance imaging and magnetic resonance spectroscopy of cerebral gliomas showing imperceptible contrast enhancement on conventional magnetic resonance imaging. Australas Radiol 2004;48:324–332 [DOI] [PubMed] [Google Scholar]
- 65.Ginsberg LE, Fuller GN, Hashmi M, et al. The significance of lack of MR contrast enhancement of supratentorial brain tumors in adults: histopathological evaluation of a series. Surg Neurol 1998;49:436–440 [DOI] [PubMed] [Google Scholar]
- 66.Law M, Yang S, Wang H, et al. Glioma grading: sensitivity, specificity, and predictive values of perfusion MR imaging and proton MR spectroscopic imaging compared with conventional MR imaging. AJNR Am J Neuroradiol 2003;24(10):1989–1998 [PMC free article] [PubMed] [Google Scholar]
- 67.Pope WB, Chen JH, Dong J, et al. Relationship between gene expression and enhancement in glioblastoma multiforme: exploratory DNA microarray analysis. Radiology 2008;249(1):268–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.