Abstract
Patients with ulcerative colitis (UC) have an increased risk for developing colorectal cancer. Because UC tumorigenesis is associated with genomic field defects that can extend throughout the entire colon, including the non-dysplastic mucosa; we hypothesized that the same field defect will include abnormally expressed proteins. Here we applied proteomics to study the protein expression of UC neoplastic progression. The protein profiles of colonic epithelium were compared from 1) UC patients without dysplasia (non-progressors); 2) none-dysplastic colonic tissue from UC patient with high-grade dysplasia or cancer (progressors); 3) high-grade dysplastic tissue from UC progressors and 4) normal colon. We identified protein differential expression associated with UC neoplastic progression. Proteins relating to mitochondria, oxidative activity, calcium-binding proteins were some of interesting classes of these proteins. Network analysis discovered that Sp1 and c-myc proteins may play roles in UC early and late stages of neoplastic progression, respectively. Two over-expressed proteins in the non-dysplastic tissue of UC progressors, CPS1 and S100P, were further confirmed by IHC analysis. Our study provides insight into the molecular events associated with UC neoplastic progression, which could be exploited for the development of protein biomarkers in fields of non-dysplastic mucosa that identify a patient’s risk for UC dysplasia.
Keywords: biomarker, cancer, dysplasia, neoplastic progression, ulcerative colitis
1 Introduction
Ulcerative colitis (UC) patients have an elevated rate of colon cancer and the colonoscopic surveillance of these patients is expensive, time consuming, and invasive. Moreover, evaluation of UC neoplasia is a subjective interpretation; even the highly regarded, experienced GI (gastrointestinal) pathologists who developed the histologic standards for UC neoplasia could only agree on the diagnosis of indefinite for dysplasia half of the time[1]. Obviously, a biomarker of dysplasia would aid the clinical management of cancer risk in UC patients. Of even greater utility would be biomarkers that are present in large fields surrounding UC dysplasia, allowing their detection without the need for the very large numbers of biopsies now needed to safely identify focal dysplasia[2].
Neoplastic progression in UC occurs in a stepwise manner of histological changes from negative → indefinite for dysplasia →low-grade dysplasia →high-grade dysplasia[1]. UC cancer is preceded by and co-exists with dysplastic precursors, thus it is possible to analyze the cancer precursors to investigate biomarkers that could be exploited to improve cancer surveillance. Previous investigations have identified several markers of dysplasia including sialosyl-Tn, sphingomyelin, p53 abnormalities, changes in cyclin D1, abnormalities in Bcl-2 expression, and aneuploidy; unfortunately many of these markers are not sufficiently wide-spread and/or not sensitive enough for regular use as an ideal biomarker[3–5]. Our laboratories and others have previously showed that genetic factors (chromosomal instability, genomic instability and microsatellite instability) and epigenetic factors (methylation) play a role in UC neoplastic progression[6–9]. These genetic biomarkers are still under evaluation for their potential as diagnostic biomarker for UC dysplasia.
While it is clear that the histologically normal appearing mucosa of UC patients with cancer/pre-cancer is abnormal at the genome/DNA level, little exploration has taken place to examine whether protein expression is also abnormal. The study of various grades of UC dysplasia at the protein level may discern previously unrevealed events in UC tumorigenesis, and this knowledge could, in turn, be exploited for the development of biomarkers that predict UC dysplasia. The emerging field of proteomics provides systematic investigation of proteome expression within a complex biological system. While a large number of proteomic investigations have studied a variety of diseases and developed biomarkers for early cancer diagnosis, only a few have focused on IBD (inflammatory bowel disease)[10]. Ulcerative colitis is one of the two major forms of inflammatory bowel disease. Two reports using two-dimensional electrophoresis (2DE) and mass spectrometry were able to identify proteins associated with IBD [11, 12], but a global quantitative proteomic study of UC neoplastic progression has not been explored.
To date, one of the most effective and versatile quantitative proteomics approaches is the combination of high-resolution tandem mass spectrometry and stable isotope labeling[13]. The introduction of the stable isotope labeling technique allows for proteome identification and the direct comparison of 2 or more samples in a single measurement. We selected the iTRAQ labeling technique[14] which allows simultaneous detection of up to eight samples for quantitative proteomic comparison at a global scale, thus it is ideal for comparing multiple UC biopsies of varying degrees of dysplasia. Since both the identification and the quantification of peptides is achieved at the MS/MS level, the iTRAQ approach provides the ability for a multiplex comparison without complicating the MS spectrum, thereby minimizing ion suppression, and ultimately, enhancing the peptide identification and quantification.
UC tumorigenesis involves genomic abnormalities throughout the whole colon, including the non-dysplastic mucosa. This is referred as molecular field defect or genomic field defect [6–9]. Because of this genomic field defect in the colonic mucosa, we hypothesized that the same field defect would be associated with abnormally expressed proteins. In this report, we carried out an exploratory/discovery study to test this hypothesis. We used quantitative proteomics with iTRAQ labeling to compare the proteomes of both dysplastic and non-dysplastic mucosa from UC progressors (UC patients with high-grade dysplasia or cancer) to the colonic mucosa of UC non-progressors (UC patients who aredysplasia-free) and normal non-UC colon. We identified differentially expressed proteins that occur during the process of UC tumorigenesis, as the non-dysplastic mucosa turns into high-grade dysplasia. These differentially expressed proteins were further investigated to reveal the associated biological pathways which might underlie tumorigenesis. Finally, immunohistochemistry (IHC) was used to confirm two over-expressed proteins that are present at the earliest stages of neoplastic progression and which may serve as future candidate biomarkers to identify UC progressors.
2 Materials and methods
2.1 Patients and tissue specimen
Tissue specimens from patients with ulcerative colitis and normal non-UC colons were collected in accordance with approved Human Subject’s guidelines at the University of Washington and the Cleveland Clinic via institutional internal tissue banks. Once procured, all specimens are assigned with study IDs and specimen IDs. The specimens obtained at the time colonoscopy or from surgical resections were placed in frozen media containing Minimal Essential Medium with 10% DMSO, and kept frozen at −70°C until use. For proteomic analysis, colonic epithelial samples from 5 subjects (patients) from each of the following categories were used: 1) normal non-UC colon. 2) UC non-progressors 3) non-dysplastic biopsies from UC progressors: (UC progressor NEG). These specimens were from UC patients who had high-grade dysplasia (n=3) or colon cancer (n=2), but the particular specimens tested were histologically negative for dysplasia; 4) High-grade dysplasia (HGD) biopsies from UC progressors: (UC progressor HGD). The histological grades for each specimen were made by co-author (MPB) who has extensive experience in evaluating the pathology of IBD samples. The rational for selecting 5 subjects from each disease category was based on the notion that over 80% of the markers present in only 30% of patients can be identified when 5 cases are pooled for discovery study.
The clinical characteristics of these patients are listed in Supplemental Table 1. Efforts were made to match the progressor and non-progressor specimens for patient age, duration of disease, and inflammatory status of the samples.
2.2 Isolation of epithelial cells and protein lysis
Colonic epithelial cells were isolated from specimens by EDTA shake-off, which provides over 90% purity of epithelial cells as previously described as previously described[9]. Protein lysates were obtained by lysing the epithelial cells in T-Per (Pierce, Rockford, IL) or CHAPS (Millipore, Billerica, MA) with 1× Protease Inhibitor Cocktail (Pierce, Rockford, IL).
2.3 iTRAQ labeling and mass spectrometry
Four sample categories were compared using iTRAQ method, including: normal non-UC colon, UC non-progressors, UC progressors NEG, and UC progressors HGD. For each sample category, a pooled sample of 5 was generated using 50 ug of protein lysates from each sample. The final 250 μg of protein lysate from each sample category was resuspended in iTRAQ (Applied Biosystems, Foster City, CA) dissolution buffer (0.5 M triethylammonium bicarbonate). The cysteine groups of the proteins were reduced with 50 mM tris-(2 carboxyethyl)phosphine (TCEP) and blocked with 200 mM methyl methanethiosulfonate (MMTS). The proteins were then digested with trypsin (trypsin to protein ratio: 1/50) overnight (16–18 hours) and labeled with one channel of iTRAQ reagents per category according to the manufacturer’s instructions. The samples were then combined after labeling. The combined samples were separated with SCX into 12 fractions for LC MS/MS analysis. The samples were analyzed in triplicate.
Chromatographic separation was performed on a 100 mm × 75μm nanoAcquity 1.7μm BEH C18 analytical column (Waters, Milford, MA) using a nanoAcquity Ultra Performance Liquid Chromatography system (Waters, Milford MA.) The column temperature was maintained at 35°C and eluted with a gradient of 2–25% B over 50 minutes and 25–35% B over 10 minutes, where A = D.I. H2O with 0.1% formic acid and B = acetonitrile with 0.1% formic acid, for a total gradient duration of 60 minutes. The flow rate used was 300nL/min. Two μL of each sample was injected. Samples were analyzed on a Waters Q-TOF Premier mass spectrometer (Micromass, Manchester, UK) by data dependent acquisition using a standard Waters nano-ESI source operating in the positive mode. The capillary and cone voltages were 3.5kV and 36V, respectively. The source temperature was set to 90°C with a nanoflow gas pressure of 40,000 pascals.
2.4 Proteomics Data Processing
The MS/MS data were exported in .RAW format from Waters Q-TOF instrument using MassLynx software (v4.1.) and processed using Trans-Proteomic Pipeline (TPP: http://tools.proteomecenter.org/software.php). The MS/MS data was first converted into mzXML format using massWolf software provided with TPP and then searched against IPI human protein database (version 3.38, 70,856 protein entries, European Bioinformatics Institute) using the SEQUEST algorithm (version 27)[15]. The searches were performed using the following parameters: a mass tolerance of 3.0 Da, full trypsin specificity, static modification of iTRAQ labeling on N-terminus and lysine (+144.10), and blocking on cysteine using methyl methanethiosulfonate (+45.99). All the peptide identifications were validated using PeptideProphet[16]. PeptideProphet uses various SEQUEST scores and a number of other parameters to estimate the accuracy of peptide assignments to MS/MS spectra made by database search algorithms. A cut-off probability score of 0.9 was used for this study. A target/decoy search was performed and it revealed a false positive rate of less than 1% based on a PeptideProphet probability score cutoff at 0.9[17]. The false positive rate is defined as the percentage of the number of false positive identifications in the forward database to the total number of peptides identified with a probability score of 0.9 or higher.
The iTRAQ quantification was achieved using LIBRA program[18]. The threshold for the peptides used for quantification was a probability of 0.9. Protein quantitation is derived from the group of peptides associated with the protein. The integrated intensity of each peptide was normalized by the sum of its channel intensities. All of the normalized peptides of a protein were then averaged and the standard deviation of the mean was determined for each protein. Channel 114 was designated as a reference normalization channel, the protein quantitation of other channels (115, 116, and 117) was normalized with respect to this channel. Furthermore, the channel 115, 116,117 ratios were normalized by summing all iTRAQ ratios across the entire dataset, calculating an overall ratio (0.86 for 115, 1.00 for 116 and 1.02 for 117), and dividing the ratios for each protein by these values. This effectively accounts for experimental error and potential errors introduced by isotopic overlap between the iTRAQ reagent fragments [19]. More information about Trans-Proteomic Pipeline, PeptideProphet, LIBRA and other programs can be obtained from the Seattle Proteome Center (http://tools.proteomecenter.org).
2.5 Networks Analysis Using MetaCore
MetaCore (GeneGo, St. Joseph, MI) was used to map the differentially expressed proteins into biological networks. MetaCore is an integrated software suite for functional analysis of experimental data. It is based on a proprietary, manually-curated database of human protein-protein, protein-DNA and protein-compound interactions, metabolic and signaling pathways, and the effects of bioactive molecules. The differentially expressed proteins were converted into Entrez protein identifiers and uploaded into the MetaCore platform for analysis. The biological process enrichment was analyzed based on Gene Ontology processes. The Analyze Network algorithm was used to map protein pathways within two-step interactions. This algorithm generates sub-networks of a large network, where interactions are expanded from each gene (from our list), until the sub-networks intersect. Enrichment analysis was performed using MetaCore, in addition to the Database for Annotation, Visualization and Integrated Discovery (DAVID)[20] tool from the National Institute of Allergy and Infectious Diseases (NIAID), NIH.
2.6 IHC analysis
Paraffin-embedded, lightly paraformaldehyde-fixed slides were processed using a modification of previous protocols[21]. The deparaffinized sections were processed for antigen retrieval using Heat Induced Epitope Retrieval (HIER) Techniques in citrate buffer (0.1 M, pH 6.0), followed by cooling to room temperature and then primary antibody incubation. Dilution of the primary antibody CPS1 (Abcam Cambridge, MA) and S100P (R&D Systems, Minneapolis, MN) were 1:250 and 1:500, respectively. Dilution of the secondary antibodies for CPS1 and S100P were 1:500 and 1:1000 respectively. IHC staining was graded as 0–4+, from negative to intensely positive.
3 Results
3.1 Identification of differentially expressed proteins in UC and associated dysplasia
The overall workflow for the quantitative proteomics analysis of UC colonic epithelium is presented in Figure 1. UC colons have various degrees of leukocytes (inflammatory cells) in the epithelium depending on the degree of inflammation. However, these inflammatory cells usually stay in the lamina propria and submucosa; thus after epithelial cells isolation procedure, they are no longer in the epithelial cell fraction. The tryptic digestion of the samples was labeled with the appropriate iTRAQ reagents: normal non-UC colon with iTRAQ 114, UC non-progressors with iTRAQ 115, UC progressors NEG with iTRAQ 116, and UC progressors HGD with iTRAQ 117, respectively. The MS/MS spectra were searched against the IPI human protein database using the SEQUEST algorithm and peptide identifications were validated using PeptideProphet with a cutoff probability score of 0.9. At this cutoff, a false positive rate of 0.65% was estimated based on a target/decoy database search. The final, filtered dataset contains 4035 non-redundant peptides leading to the identification of 1107 proteins/protein groups including 269 single-peptide hits.
Figure 1.
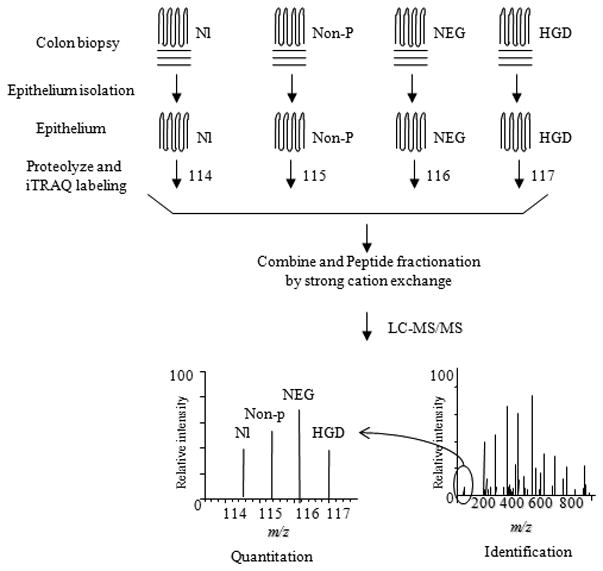
Work flow for quantitative proteomics analysis of colon epithelium. Colonic epithelium was first isolated from colon specimens. The protein lysates from colon epithelium were proteolyzed and iTRAQ labeled with different iTRAQ channel reagents. The labeled samples were then combined, fractionated, purified and analyzed by tandem mass spectrometer. The proteins were identified by database search of the tandem mass spectrum for matching peptides. The relative abundance was determined by the ratio of signal intensities of the signature peaks of the target sample compared to normal colon (Nl). Non-p = non-progressors.
The iTRAQ quantification was achieved using the LIBRA program. We performed technical replicates for the experiment. There is a strong correlation in the protein ratios between the replicates: r2 = 0.94 (P<0.0001). This level of variation (6%) is consistent with the results reported in the literature[22]. For each iTRAQ channel, the data was normalized to reduce the systematic error and a Z-score was calculated for each protein based on the normalized iTRAQ ratio. Using a 2.0 fold change in protein expression as cutoff value, there were 60 differentially expressed proteins in UC non-progressors compared to normal non-UC colon, comprising 45 down-regulated and 15 up-regulated proteins. The numbers of differentially expressed proteins in the UC progressor samples (compared to normal non-UC colon) were increased to 98 and 99 for UC progressors NEG and HGD, respectively. While the number of down-regulated proteins in UC progressors NEG (n= 73) was much higher than in UC non-progressors (n= 45); the number of under-expressed proteins in HGD (n= 49) was close to that of UC non-progressors. In contrast, the number of over-expressed proteins increased as the UC colon was becoming more dysplastic: non-progressor (n= 15 proteins) → NEG (n= 25 proteins) → HGD (n= 50 proteins) (Supplemental Figure 1A). A subset of over-expressed proteins in UC progressor with at least 2-fold expression change and at least two-peptide quantification is presented in Table 1. The protein identification information is provided in Supplemental Table 2. As shown in Supplemental Figure 1B, forty-four proteins were differentially expressed in all UC samples compared to normal non-UC colon. These included 8 up-regulated and 36 down-regulated proteins. There were 6, 35, and 37 differentially expressed proteins in UC non-progressors, UC progressor NEG and HGD respectively.
Table 1.
Over-expressed proteins in UC progressor by at least two-fold (proteins with at least two peptides quantification were included here
| Database IDa | protein_description | gene symbol | NP/NLb (115:114) | SEMc | NEG/NLb (116:114) | SEMc | HGD/NLb (117:114) | SEMc | peptides for quantification |
|---|---|---|---|---|---|---|---|---|---|
| IPI00385149 | Putative uncharacterized protein | PTMA | 1.2 | 0.2 | 1.0 | 0.1 | 2.2 | 0.4 | 9 |
| IPI00258804 | ATPase inhibitory factor 1 isoform 3 precursor | ATPIF1 | 1.3 | 0.5 | 1.1 | 0.5 | 2.1 | 1.2 | 10 |
| IPI00004962 | Golgi integral membrane protein 4 | GOLIM4 | 2.7 | 4.0 | 1.1 | 1.0 | 2.6 | 3.3 | 5 |
| IPI00003734 | Putative S100 calcium-binding protein H_NH0456N16.1 | LOC347701 | 1.0 | 0.2 | 1.2 | 0.3 | 2.0 | 0.6 | 14 |
| IPI00027412 | Carcinoembryonic antigen-related cell adhesion molecule 6 | CEACAM6 | 1.4 | 0.1 | 1.3 | 0.2 | 2.4 | 0.3 | 5 |
| IPI00019872 | Estradiol 17-beta-dehydrogenase 2 | HSD17B2 | 1.3 | 0.3 | 1.3 | 0.1 | 2.6 | 0.5 | 12 |
| IPI00793199 | annexin IV | ANXA4 | 1.2 | 0.1 | 1.4 | 0.3 | 2.1 | 0.5 | 78 |
| IPI00001539 | 3-ketoacyl-CoA thiolase, mitochondrial | ACAA2 | 1.6 | 0.3 | 1.4 | 0.3 | 2.0 | 0.5 | 118 |
| IPI00026328 | Thioredoxin domain-containing protein 12 precursor | TXNDC12 | 1.4 | 0.2 | 1.5 | 0.3 | 2.1 | 0.4 | 11 |
| IPI00030122 | Isoform 1 of Urotensin-2 precursor | UTS2 | 1.7 | 0.0 | 1.5 | 0.0 | 2.1 | 0.1 | 2 |
| IPI00020599 | Calreticulin precursor | CALR | 1.5 | 0.2 | 1.6 | 0.3 | 2.3 | 0.6 | 139 |
| IPI00386755 | ERO1-like protein alpha precursor | ERO1L | 1.7 | 0.3 | 1.6 | 0.5 | 4.0 | 1.4 | 16 |
| IPI00009904 | Protein disulfide-isomerase A4 precursor | PDIA4 | 1.5 | 0.3 | 1.6 | 0.4 | 2.3 | 0.7 | 152 |
| IPI00171421 | Uncharacterized protein C8orf55 precursor | C8orf55 | 1.5 | 0.1 | 1.7 | 0.1 | 2.5 | 0.4 | 2 |
| IPI00013895 | Protein S100-A11 | S100A11 | 1.1 | 0.1 | 1.7 | 0.3 | 2.9 | 0.5 | 22 |
| IPI00024993 | Enoyl-CoA hydratase, mitochondrial precursor | ECHS1 | 1.7 | 0.4 | 1.8 | 0.5 | 2.0 | 0.6 | 99 |
| IPI00008934 | Hydroxymethylglutaryl-CoA synthase, mitochondrial | HMGCS2 | 1.7 | 0.4 | 1.8 | 0.5 | 2.0 | 0.7 | 180 |
| IPI00025252 | Protein disulfide-isomerase A3 precursor | PDIA3 | 1.6 | 0.3 | 1.8 | 0.4 | 2.4 | 0.6 | 282 |
| IPI00010491 | Ras-related protein Rab-27B | RAB27B | 0.9 | 0.0 | 1.8 | 0.4 | 2.0 | 0.1 | 2 |
| IPI00007427 | AGR2 | AGR2 | 1.2 | 0.5 | 1.8 | 0.6 | 2.3 | 1.0 | 117 |
| IPI00010796 | Protein disulfide-isomerase precursor | P4HB | 1.9 | 0.4 | 1.8 | 0.4 | 2.7 | 0.7 | 292 |
| IPI00021828 | Cystatin-B | CSTB | 1.5 | 0.6 | 1.8 | 0.9 | 2.0 | 1.0 | 3 |
| IPI00025062 | Isoform 1 of Cathepsin E precursor | CTSE | 0.7 | 0.1 | 1.8 | 0.3 | 3.7 | 0.7 | 5 |
| IPI00220362 | 10 kDa heat shock protein, mitochondrial | HSPE1 | 1.6 | 0.2 | 1.8 | 0.3 | 2.1 | 0.4 | 110 |
| IPI00008840 | POU domain, class 3, transcription factor 1 | POU3F1 | 1.1 | 0.1 | 1.9 | 0.1 | 2.3 | 0.2 | 14 |
| IPI00010290 | FABP1 protein (Fragment) | FABP1 | 2.4 | 1.0 | 1.9 | 0.9 | 2.0 | 0.9 | 157 |
| IPI00216071 | Eosinophil lysophospholipase | CLC | 1.6 | 0.0 | 2.0 | 0.0 | 1.5 | 0.0 | 2 |
| IPI00008367 | Isoform 1 of Inactive carboxylesterase 4 precursor | CES4 | 1.5 | 0.2 | 2.0 | 0.2 | 1.4 | 0.2 | 4 |
| IPI00005668 | Aldo-keto reductase family 1 member C2 | AKR1C2 | 1.4 | 0.3 | 2.0 | 0.4 | 1.2 | 0.2 | 4 |
| IPI00006663 | Aldehyde dehydrogenase, mitochondrial precursor | ALDH2 | 2.0 | 0.4 | 2.0 | 0.5 | 2.0 | 0.5 | 234 |
| IPI00027028 | Paraneoplastic antigen Ma2 | PNMA2 | 2.1 | 0.0 | 2.0 | 0.0 | 1.7 | 0.0 | 2 |
| IPI00167072 | Isoform 5 of Abhydrolase domain-containing protein 11 | ABHD11 | 1.5 | 0.3 | 2.0 | 1.0 | 1.8 | 0.9 | 7 |
| IPI00011603 | 26S proteasome non-ATPase regulatory subunit 3 | PSMD3 | 3.1 | 3.0 | 2.0 | 1.9 | 2.6 | 2.5 | 2 |
| IPI00027463 | Protein S100-A6 | S100A6 | 1.1 | 0.1 | 2.1 | 0.5 | 2.9 | 0.8 | 45 |
| IPI00103436 | Intelectin-2 precursor | ITLN2 | 0.7 | 0.1 | 2.1 | 0.4 | 1.5 | 0.2 | 8 |
| IPI00216932 | Isoform 1 of Acetyl-coenzyme A synthetase 2-like, | ACSS1 | 1.5 | 0.4 | 2.1 | 0.7 | 1.7 | 0.4 | 5 |
| IPI00027701 | Short-chain specific acyl-CoA dehydrogenase, | ACADS | 1.7 | 0.5 | 2.2 | 0.8 | 1.7 | 0.4 | 62 |
| IPI00291737 | Intelectin-1 precursor | ITLN1 | 0.9 | 0.2 | 2.3 | 0.5 | 1.7 | 0.4 | 28 |
| IPI00018909 | trefoil factor 3 precursor | TFF3 | 1.4 | 0.4 | 2.4 | 0.9 | 1.7 | 0.7 | 17 |
| IPI00242956 | IgGFc-binding protein precursor | FCGBP | 1.4 | 0.3 | 2.5 | 0.8 | 2.0 | 0.6 | 258 |
| IPI00008339 | 78 kDa protein | 2.1 | 0.5 | 2.5 | 0.3 | 2.6 | 1.3 | 2 | |
| IPI00791006 | Mucin 4 | MUC4 | 4.6 | 2.4 | 3.3 | 1.9 | 2.8 | 1.8 | 3 |
| IPI00020716 | Isoform 2 of Cathepsin E precursor | CTSE | 1.3 | 0.2 | 5.0 | 0.5 | 11.9 | 1.2 | 2 |
| IPI00011062 | Isoform 1 of Carbamoyl-phosphate synthase, mitochondrial | CPS1 | 1.1 | 0.3 | 5.3 | 4.1 | 3.6 | 2.5 | 43 |
| IPI00017526 | Protein S100-P | S100P | 1.3 | 0.3 | 6.4 | 3.8 | 6.5 | 3.9 | 18 |
when there are multiple matches in database, only the first IPI ID is listed here.
NP: non-progressor; NL: normal; NEG: negative fro dysplasia from progressor patietn; HGD: high grade dysplasia from progression patient.
SEM: standard error of the mean here
We do not rule out the possibility that some of the differentially expressed proteins may be due to biological heterogeneity. As we discussed in the following sections, many of the proteins in Table 1 are found to be functionally associated with neoplastic progression. It is also interesting to note that several keratins were identified as underexpressed in the UC diseased samples compared to the normal non-UC colon. Keratins can be possible contaminations in proteomics analysis. However, in this particular study, we used isolated colonic epithelial tissue, which is particularly rich in keratin content, for proteomics analysis. Thus, it is not surprised to observe the presence of keratins in our samples. In addition, it has been previously observed that defects in keratin result in a variety of tissue diseases that can be reflected by the changes in the keratin expression profile.
3.2 Functional analysis of differentially expressed proteins in UC non-progressors and progressors
To functionally annotate the differentially expressed proteins identified in this study, these proteins (≥2.0-fold change) were entered into GeneGo’s MetaCore, and DAVID databases for enrichment and network analysis.
Common biological processes in UC
First we examined the common biologic processes that were represented by the shared differentially expressed proteins present in all UC samples. This was accomplished using GeneGo’s Compare Experiments Workflow from the MetaCore platform. The top five common network processes were: 1) interphase of mitotic cell cycle and cell cycle; 2) mitotic spindle organization and biogenesis and microtubule-based movement; 3) localization of cell, cell motility, and actin cytoskeleton organization and biogenesis; 4) structure, organ and system development; 5) cell differentiation and cellular developmental process. These common processes reflect the underlining biological events in UC condition: active cellular turn-over, cellular regeneration and tissue repair.
Unique biological processes in UC neoplastic progression
We then analyzed the biologic processes for the differentially expressed proteins unique for each stage of UC neoplastic progression using the MetaCore platform mentioned above. The most relevant unique processes for the UC non-progressors (by degree of significance) were nuclear transport, recombination, spindle organization and biogenesis, and mRNA processing, indicating the very active cellular organelle activity in UC conditions. The most relevant unique processes for UC progressors NEG were triacylglycerol mobilization, cellular component organization and biogenesis, cell development, apoptosis, and programmed cell death. Finally, in UC progressors HGD, the unique processes included immune response, multi-cellular organismal process, system development, and multi-cellular organismal development.
Mitochondrial proteins were frequently differentially expressed
Mitochondrial proteins were among the most differentially expressed proteins in UC non-progressors and even more in the UC progressors: 6, 8 and 12 mitochondrial proteins were differentially expressed by at least twofold in UC non-progressors, UC progressors NEG and UC progressors HGD, respectively. The enrichment of mitochondrial proteins among the differentially expressed proteins were all statistically significant (p=0.049, 0.05, and 0.0005 for UC non-progressor, UC progressor NEG, and HGD respectively.)
Oxidative activity
Ulcerative colitis is associated with increased level of reactive oxygen species and oxidative stress. The number of differentially expressed proteins in oxido-reductase activity increased with UC neoplastic progression: 5 in UC non-progressors, 7 in UC progressors NEG, and 13 in UC progressors HGD (p=0.072, 0.059, and 0.0017 for UC non-progressor, UC progressor NEG, and HGD respectively).
3.3 Unique differentially expressed networks associated with UC neoplastic progression
To analyze the broader interacting network among the differentially expressed proteins in UC progressors, we used GeneGo’s network algorithms from the MetaCore platform using the Analyze Network option, with a 50 node limit. The top sub-network for UC progressors NEG brought together 12 differentially expressed proteins (≥2.0 fold) with the most prominent network protein being transcriptional factor Sp1. Sp1 displays direct interactions with half of the targets in the UC network, 12 of the 25 were the differentially expressed proteins in UC progressors NEG. The top network for UC progressors HGD brought together 8 differentially expressed proteins with ≥2.0 fold changes. In contrast to the central role of Sp1 in UC progressor NEG, the transcriptional factor c-myc played a role in the HGD protein network. Among the 8 differentially expressed proteins in the network, c-myc directly interacted with 6 of them, and had two steps (e.g. indirect) interactions with the other two proteins.
3.4 Progressive S100 protein dysregulation associated with UC neoplastic progression
The S100 proteins are a family of calcium-binding proteins that are involved in a broad range of intra- and extra-cellular functions. Several members of this S100 family were differentially expressed in UC neoplastic progression. Four S100 proteins, including S100P, S100A6, S100A11, and S100H, displayed increased expression in the UC neoplastic progression, e.g., normal colon < UC non-progressors < UC progressors NEG < UC progressors HGD, with HGD samples having the highest expression (Figure 2). Four other proteins, S100A8, S100A9, S100A10 and S100A4 displayed various degrees of decreased expression in UC progressor NEG and HGD.
Figure 2.
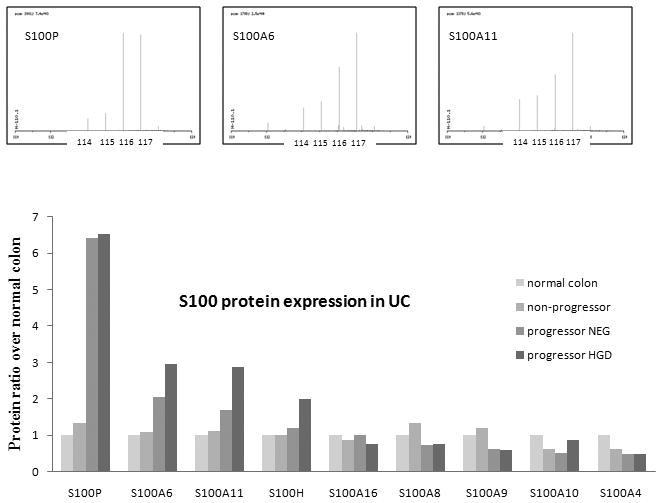
S100 protein expression in UC. The upper panel shows the ion chromatogram of the signature peaks for fragments 114, 115, 116, and 117 which represent normal colon, UC non-progressors, UC progressors NEG, and UC progressors HGD, respectively. The lower panel displays the ratio of S100 abundance in the 4 sample types.
3.5 Over-expression of CPS1 and S100P in UC progressors: IHC confirmation
The two proteins with highest degree of over-expression in UC progressor, CPS1 and S100P were selected for further IHC analysis.
CPS1
Proteomic analysis revealed that carbamoyl-phosphate synthase 1 (CPS1) was over-expressed in UC progressors (5.3 fold in NEG, and 3.6 fold in HGD) and its expression was normal in UC non-progressors (Table 1). IHC staining was performed to confirm this result. As showed in Figure 3B, colonic epithelium from UC progressors NEG exhibited strong staining, while the UC non-progressor mucosa stained mildly positive (Figure 3A). The mean IHC staining scores in UC progressors is significantly higher that of UC non-progressors (p=0.0001, Figure 4). All but one of the 17 specimens from UC progressor displayed IHC score of 3+ or 4+. If the IHC score of 3+ or higher was used as a cutoff, CPS1 could achieve 80% specificity and 94% sensitivity in distinguishing UC progressor from UC non-progressors.
Figure 3.
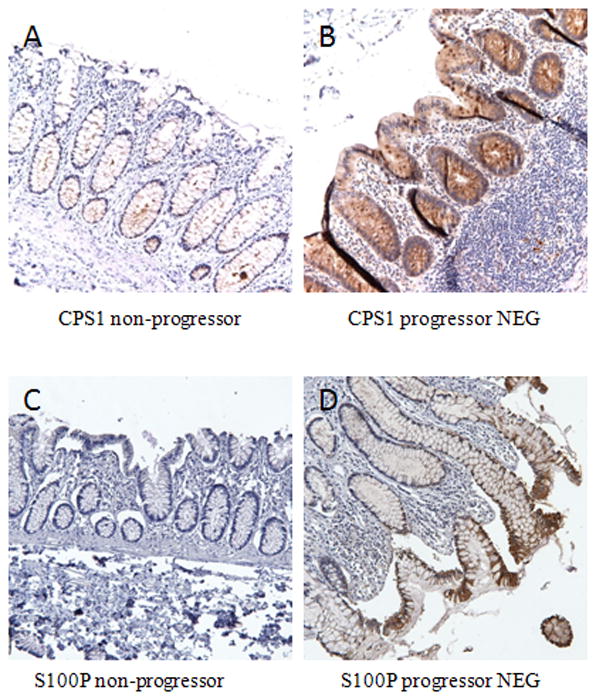
IHC analysis of CPS1 and S100P protein expression in UC non-progressors and UC progressors. (A–B): CPS1 is mildly positive in the epithelium of non-progressor (A); in contrast, overexpression of CPS1 was apparent in the epithelium of UC progressors NEG (B). (C–D): Note the overexpression of S100P in the epithelium of UC progressors NEG while UC non-progressor colon has absence of staining.
Figure 4.
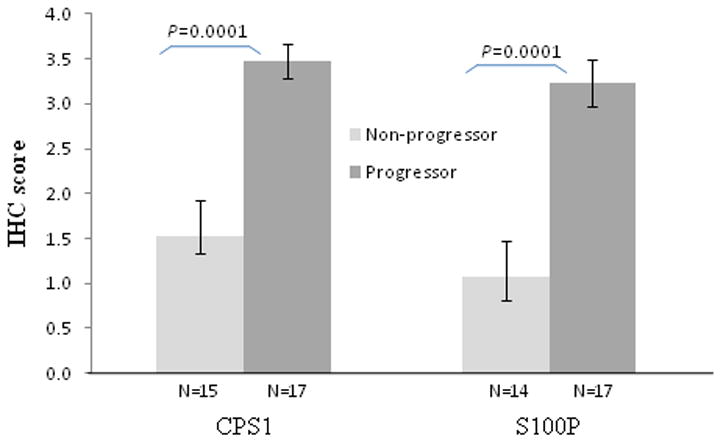
IHC scores of CPS1 and S100P. The mean IHC score for progressor is significantly higher than non-progressor for both CPS1 and S100P. IHC scores ranges from 0–4+, 0 being negative and 4+ being strongest. N, number of specimens tested.
S100P
S100P was over-expressed in UC progressors NEG (6.4 fold) and HGD (6.5 fold) by proteomic analysis. IHC staining revealed that majority of non-progressor samples had negative or mild (1+) staining of S100P (Figure 3C). In contrast, the majority of UC progressors showed strong epithelial staining (Figure 3D). The mean IHC staining scores in UC progressors is significantly higher than that of UC non-progressors (p=0.0001, Figure 4). All but three of the 17 specimens from UC progressor displayed IHC score of 3+ or 4+. If the IHC score of 3+ or higher was used as a cutoff, S100P could achieve 79% specificity and 82% sensitivity in distinguishing UC progressor from UC non-progressors.
4 Discussion
In this study, we investigated the proteome that underlies UC neoplastic progression. The protein profiles of colonic epithelium from UC non-progressors versus UC progressors (both the non-dysplastic and dysplastic mucosa) were compared to normal colonic epithelium from non-UC colons. Not surprisingly, the colonic mucosa of UC progressors displayed more differentially expressed proteins than the UC non-progressors. What was surprising was the discovery that the proteome of the non-dysplastic mucosa from the progressors was also abnormal --that it was more akin to the proteome of high-grade dysplasia, than it was to the non-dysplastic mucosa of non-progressors. These findings suggest that there are changes in protein expression early in the neoplastic progression, before the histologic changes become evident in the epithelial cells. IHC studies provided confirmation of the overexpression of two proteins in UC progressors (in both the dysplastic tissue and non-dysplastic tissue) compared to absent or minimal expression in UC non-progressors and normal colon. The data from this proteomic analysis may help shed light on the process of UC tumorigenesis, as well as provide candidates for future biomarkers. In considering the changes in protein expression between UC non-progressor epithelium and the UC progressors, it is theoretically possible that some of the changes could be due to the inflammatory cells retained in the isolated epithelial cell fractions used for proteomics analysis. However, we used the following strategies to minimize the chance of identifying biomarkers from inflammation: a) our epithelial cell isolation protocol usually obtains over 90% purity of epithelial cell; 2) the degree of inflammation varies between patients and biopsies; we use inflammation matching for each group of pooled specimens used in this study including some matched UC specimens that had minimal to no inflammation.
4.1 Differentially expressed Mitochondrial Proteins
One of the enriched classes of differentially expressed proteins in both UC non-progressors and UC progressors was the mitochondrial proteins (8% and 12% of all differentially expressed proteins, respectively). As the main source of energy and endogenous oxidative stress in the cell, mitochondria require continuous turnover and regeneration. Inability to maintain mitochondrial function and integrity is associated with degenerative diseases, cancer, and aging. We have previously identified ulcerative colitis as a disease of premature aging of the colon, with the colon of a young person with 8 years of disease sharing the same colonic telomere lengths and DNA damage markers as those of a person who is 60 years old [21]. In ulcerative colitis, the colon epithelium undergoes repeated cycles of inflammation and tissue repair, resulting in oxidative stress and accumulation of reactive oxidative species (ROS). Mitochondrial DNA is more susceptible than nuclear DNA to damage by ROS because there are no adequate repair mechanisms for mtDNA. Mutations in mitochondrial DNA have been found in UC colonic epithelium, and animal models of colitis suggest that these mutations may be preventable with the use of selenium [23]. It is well known that mitochondrial dysfunction and mitochondrial mutations are associated with and accumulate in cancer cells, although the precise role of impaired mitochondria in tumorigenesis is still unclear. One mechanism in which the mitochondria could influence tumorigenesis stems from observations that impaired mitochondria can lead to premature senescence [24]. Senescent cells have recently been recognized as having a potential role in promoting neoplastic progression, for example, by promoting a pro-inflammatory micro-environment that promotes tumor evolution [25]. Thus, increasingly impaired mitochondria may help drive the process of UC tumorigenesis. In one study the transfer of mitochondrial DNA from cells that have a high metastatic potential into cells that have a low metastatic potential leads a change in the functional behavior of the cells. The cells take on the phenotype that is associated with the mitochondria in this case the low metastatic cells became highly metastatic. Interestingly, the process was reversed by introduction of anti-oxidants[26]. The premature aging in UC colons, and the increased cancer risk, is not just due to injury of nuclear DNA but perhaps chronic injury to the mitochondria as well.
4.2 Sp1 in the earliest stages of UC progression
Sp1 is a ubiquitously expressed transcription factor which can both positively and negatively regulate genes. In addition to the major targeted genes that encode proteins for intermediary metabolism, Sp1 target genes are key players in cell proliferation and oncogenesis[27], as well as the ulcerative colitis susceptibility gene of interferon regulatory factor 5 (TRF5)[28]. A polymorphism in the TRF5 gene provides additional binding sites for Sp1, increasing the transcription of the TRF5 and likely causing an increase in production of pro-inflammatory cytokinesand the perpetuation of inflammation [28]. In our study, GeneGo network analysis revealed that Sp1 played a central role in the most significant network of the differentially expressed proteins in UC progressor NEG. This result suggests that Sp1 may play a role not only in UC susceptibility by increasing transcription of TRF5, but it also appears to be important in early UC neoplastic progression.
4.3 c-myc may regulate advanced UC dysplasia
The transcription factor c-myc is a key regulator of cell proliferation, cell growth, differentiation, and apoptosis. Our previous studies have identified c-myc as a common prominent regulatory protein in pancreatic cancer and chronic pancreatitis [29]. In the current study, in UC progressors HGD, the most significant network of differentially expressed proteins merged through c-myc regulation. This finding is supported by previous studies showing enhanced c-myc expression, as measured by IHC and c-myc mRNA, in UC and particularly in UC dysplasia [30]. Together these findings suggest that c-myc plays an important role in late UC neoplastic progression (HGD). Recent in vitro studies in human colon cancer cell lines revealed that high doses of mesalamine, an anti-inflammatory agent used to treat UC patients, can significantly reduce expression of c-myc mRNA and protein and increase apoptosis in a dose-dependent manner [31]. Chemoprevention of UC dysplasia using mesalamine to reduce of inflammation and c-myc overexpression would certainly be an area of future interest [32].
4.4 Dysregulation of S100 proteins
The S100 proteins are a multi-gene calcium-binding family comprising 20 known human members[33]. They have a broad range of intracellular and extracellular functions, including regulation of protein phosphorylation and enzyme activity, calcium homeostasis, regulation of cytoskeletal components and regulation of transcriptional factors [33]. Some S100 proteins are located on the mitochondrial membrane and some play a role in apoptosis by inducing a rapid decrease in the mitochondrial membrane potential [34, 35]. Several members of S100 proteins were differentially expressed in the colonic epithelium of UC progressors compared to UC non-progressors in our study, suggesting their importance in UC tumorigenesis.
S100P is frequently overexpressed in several epithelial tumor types including pancreas [36] and colon [37]. A recent study suggests that S100P may play an important role in sporadic colon cancer by stimulating cell growth, migration, and signaling pathways that affect a range of pro-inflammatory molecules [37]. Up-regulation of S100P is an early molecular event in the development of pancreatic cancer and it is expressed at high levels in both precursor lesions and invasive cancer [38, 39]. In a similar vein, we detected over-expression of S100P in UC-associated HGD and colon cancer and in the non-dysplastic mucosa of UC progressors. Furthermore, IHC analysis verified that S100P protein expression was markedly higher in the non-dysplastic colonic tissue in UC progressors than in normal or UC non-progressor colonic mucosa. Further studies are warranted to investigate whether S100P could be a potential biomarker for predicting colon cancer development in the setting of UC.
In addition to dysregulation of S100P, there were three other S100 proteins displayed increased expression in UC progressors (S100A6, S100A11 and S100H [Putative S100 calcium-binding protein H], and two other S100 proteins displayed decreased expression. Given the substantial changes in expression of S100 proteins in UC progressors, it would be worthwhile to explore further the role of these calcium-binding proteins in UC tumorigenesis.
4.5 Carbamoyl phosphate synthetase 1 (CPS1)
CPS1 is a mitochondrial enzyme involved in the urea cycle. The protein, which is expressed mainly in intestinal epithelial and liver cells, detoxifies ammonia and, together with other enzymes of the urea cycle, is the de novo source of arginine. Variations in the supply of arginine, due to alterations in urea cycle function, affect the production of nitric oxide [40]. Nitric oxide, in turn, can cause DNA damage, laxity in DNA repair, and is associated with inflammation-associated cancers [41]. The rate of CPS1 gene amplification is elevated 50- to 100-fold in human cell lines deficient in mismatch repair (due to MLH1 or MSH6), as compared with mismatch repair-proficient control cells. Previous studies have showed increased expression of CPS1 in gastric cancer [42], and down-regulation in human hepatocellular carcinoma[43]. Our study identifies over-expression of CPS1 in UC progressors, in both histologically normal and dysplastic tissues. CPS1 is an abundant protein that constitutes 20% to 30% of mitochondrial matrix protein. There is usually an excess amount of CPS1 that is actually required for its function. Further study is needed to investigate if increased CPS1 protein expression level reflects an increase in its functional activity.
4.6 Summary
In this study, we applied quantitative proteomics to investigate differentially expressed proteins associated with UC neoplastic progression. Proteins relating to mitochondrion, oxidative activity, and calcium ion binding were some of the interesting classes of these differentially expressed proteins. Further network analysis discovered that Sp1 and c-myc may play roles in the early and late events of UC tumorigenesis, respectively. Finally, two up-regulated proteins in both the dysplastic and non-dysplastic mucosa of UC progressors, CPS1 and S100P, were confirmed by IHC analysis. The differentially expressed proteins identified in this study, especially CPS1 and S100P, could be exploited for the future development of biomarkers that predict UC dysplasia.
Supplementary Material
Acknowledgments
We thank Rosa Ana Rrisques, Josephine Maurer, Allyn Stevens, Jimmy Eng, and Ashley Eastham for technical support and study coordination. This work was supported by grants from Crohn’s & Colitis Foundation (RC), NIH NCI R01CA068124 (TB) and National Heart, Lung and Blood Institute NIH, under contract # NOI-HV-28179 (RA).
Abbreviations
- HGD
high-grade dysplasia
- NEG
negative for dysplasia
- UC
ulcerative colitis
Footnotes
Conflict of interest statement. The authors declare that there is no conflict of interest.
Reference List
- 1.Riddell RH, Goldman H, Ransohoff DF, Appelman HD, et al. Dysplasia in inflammatory bowel disease: standardized classification with provisional clinical applications. Hum Pathol. 1983;14:931–968. doi: 10.1016/s0046-8177(83)80175-0. [DOI] [PubMed] [Google Scholar]
- 2.Rubin CE, Haggitt RC, Burmer GC, Brentnall TA, et al. DNA aneuploidy in colonic biopsies predicts future development of dysplasia in ulcerative colitis. Gastroenterology. 1992;103:1611–1620. doi: 10.1016/0016-5085(92)91185-7. [DOI] [PubMed] [Google Scholar]
- 3.Agoff SN, Brentnall TA, Crispin DA, Taylor SL, et al. The role of cyclooxygenase 2 in ulcerative colitis-associated neoplasia. Am J Pathol. 2000;157:737–745. doi: 10.1016/S0002-9440(10)64587-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brentnall TA, Crispin DA, Rabinovitch PS, Haggitt RC, et al. Mutations in the p53 gene: an early marker of neoplastic progression in ulcerative colitis. Gastroenterology. 1994;107:369–378. doi: 10.1016/0016-5085(94)90161-9. [DOI] [PubMed] [Google Scholar]
- 5.Willenbucher RF, Zelman SJ, Ferrell LD, Moore DH, Waldman FM. Chromosomal alterations in ulcerative colitis-related neoplastic progression. Gastroenterology. 1997;113:791–801. doi: 10.1016/s0016-5085(97)70173-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen R, Rabinovitch PS, Crispin DA, Emond MJ, et al. DNA fingerprinting abnormalities can distinguish ulcerative colitis patients with dysplasia and cancer from those who are dysplasia/cancer-free. Am J Pathol. 2003;162:665–672. doi: 10.1016/S0002-9440(10)63860-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Issa JP, Ahuja N, Toyota M, Bronner MP, Brentnall TA. Accelerated age-related CpG island methylation in ulcerative colitis. Cancer Res. 2001;61:3573–3577. [PubMed] [Google Scholar]
- 8.O’Sullivan JN, Bronner MP, Brentnall TA, Finley JC, et al. Chromosomal instability in ulcerative colitis is related to telomere shortening. Nat Genet. 2002;32:280–284. doi: 10.1038/ng989. [DOI] [PubMed] [Google Scholar]
- 9.Rabinovitch PS, Dziadon S, Brentnall TA, Emond MJ, et al. Pancolonic chromosomal instability precedes dysplasia and cancer in ulcerative colitis. Cancer Res. 1999;59:5148–5153. [PubMed] [Google Scholar]
- 10.Felley-Bosco E, Andre M. Proteomics and chronic inflammatory bowel diseases. Pathol Res Pract. 2004;200:129–133. doi: 10.1016/j.prp.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 11.Hsieh SY, Shih TC, Yeh CY, Lin CJ, et al. Comparative proteomic studies on the pathogenesis of human ulcerative colitis. Proteomics. 2006;6:5322–5331. doi: 10.1002/pmic.200500541. [DOI] [PubMed] [Google Scholar]
- 12.Shkoda A, Werner T, Daniel H, Gunckel M, et al. Differential protein expression profile in the intestinal epithelium from patients with inflammatory bowel disease. J Proteome Res. 2007;6:1114–1125. doi: 10.1021/pr060433m. [DOI] [PubMed] [Google Scholar]
- 13.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 14.Ross PL, Huang YN, Marchese JN, Williamson B, et al. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 15.Eng J, McCormack AL, Yates JR. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 16.Keller A, Nesvizhskii AI, Kolker E, Aebersold R. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Analytical Chemistry. 2002;74:5383–5392. doi: 10.1021/ac025747h. [DOI] [PubMed] [Google Scholar]
- 17.Elias JE, Haas W, Faherty BK, Gygi SP. Comparative evaluation of mass spectrometry platforms used in large-scale proteomics investigations. Nat Methods. 2005;2:667–675. doi: 10.1038/nmeth785. [DOI] [PubMed] [Google Scholar]
- 18.Deutsch EW, Lam H, Aebersold R. PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 2008;9:429–434. doi: 10.1038/embor.2008.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- 20.Dennis G, Jr, Sherman BT, Hosack DA, Yang J, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- 21.Risques RA, Lai LA, Brentnall TA, Li L, et al. Ulcerative colitis is a disease of accelerated colon aging: evidence from telomere attrition and DNA damage. Gastroenterology. 2008;135:410–418. doi: 10.1053/j.gastro.2008.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gan CS. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ) J Proteome Res. 2007;6:821–827. doi: 10.1021/pr060474i. [DOI] [PubMed] [Google Scholar]
- 23.Nishikawa M, Oshitani N, Matsumoto T, Nishigami T, et al. Accumulation of mitochondrial DNA mutation with colorectal carcinogenesis in ulcerative colitis. Br J Cancer. 2005;93:331–337. doi: 10.1038/sj.bjc.6602664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kujoth GC, Bradshaw PC, Haroon S, Prolla TA. The role of mitochondrial DNA mutations in mammalian aging. PLoS Genet. 2007;3:e24. doi: 10.1371/journal.pgen.0030024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Campisi J. Aging, tumor suppression and cancer: high wire-act! Mech Ageing Dev. 2005;126:51–58. doi: 10.1016/j.mad.2004.09.024. [DOI] [PubMed] [Google Scholar]
- 26.Ishikawa K, Takenaga K, Akimoto M, Koshikawa N, et al. ROS-generating mitochondrial DNA mutations can regulate tumor cell metastasis. Science. 2008;320:661–664. doi: 10.1126/science.1156906. [DOI] [PubMed] [Google Scholar]
- 27.Wierstra I. Sp1: emerging roles--beyond constitutive activation of TATA-less housekeeping genes. Biochem Biophys Res Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 28.Dideberg V, Kristjansdottir G, Milani L, Libioulle C, et al. An insertion-deletion polymorphism in the interferon regulatory Factor 5 (IRF5) gene confers risk of inflammatory bowel diseases. Hum Mol Genet. 2007;16:3008–3016. doi: 10.1093/hmg/ddm259. [DOI] [PubMed] [Google Scholar]
- 29.Chen R, Brentnall TA, Pan S, Cooke K, et al. Quantitative Proteomics Analysis Reveals That Proteins Differentially Expressed in Chronic Pancreatitis Are Also Frequently Involved in Pancreatic Cancer. Mol Cell Proteomics. 2007;6:1331–1342. doi: 10.1074/mcp.M700072-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Macpherson AJ, Chester KA, Robson L, Bjarnason I, et al. Increased expression of c-myc proto-oncogene in biopsies of ulcerative colitis and Crohn’s colitis. Gut. 1992;33:651–656. doi: 10.1136/gut.33.5.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pavelic ZP, Pavelic L, Kuvelkar R, Gapany SR. High c-myc protein expression in benign colorectal lesions correlates with the degree of dysplasia. Anticancer Res. 1992;12:171–175. [PubMed] [Google Scholar]
- 32.Chu EC, Chai J, Ahluwalia A, Tarnawski AS. Mesalazine downregulates c-Myc in human colon cancer cells. A key to its chemopreventive action? Aliment Pharmacol Ther. 2007;25:1443–1449. doi: 10.1111/j.1365-2036.2007.03336.x. [DOI] [PubMed] [Google Scholar]
- 33.Salama I, Malone PS, Mihaimeed F, Jones JL. A review of the S100 proteins in cancer. Eur J Surg Oncol. 2008;34:357–364. doi: 10.1016/j.ejso.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 34.Brezova A, Heizmann CW, Uhrik B. Immunocytochemical localization of S100A1 in mitochondria on cryosections of the rat heart. Gen Physiol Biophys. 2007;26:143–149. [PubMed] [Google Scholar]
- 35.Ghavami S, Kerkhoff C, Chazin WJ, Kadkhoda K, et al. S100A8/9 induces cell death via a novel, RAGE-independent pathway that involves selective release of Smac/DIABLO and Omi/HtrA2. Biochim Biophys Acta. 2008;1783:297–311. doi: 10.1016/j.bbamcr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 36.Dowen SE, Crnogorac-Jurcevic T, Gangeswaran R, Hansen M, et al. Expression of S100P and its novel binding partner S100PBPR in early pancreatic cancer. Am J Pathol. 2005;166:81–92. doi: 10.1016/S0002-9440(10)62234-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fuentes MK, Nigavekar SS, Arumugam T, Logsdon CD, et al. RAGE activation by S100P in colon cancer stimulates growth, migration, and cell signaling pathways. Dis Colon Rectum. 2007;50:1230–1240. doi: 10.1007/s10350-006-0850-5. [DOI] [PubMed] [Google Scholar]
- 38.Arumugam T, Simeone DM, Van Golen K, Logsdon CD. S100P promotes pancreatic cancer growth, survival, and invasion. Clin Cancer Res. 2005;11:5356–5364. doi: 10.1158/1078-0432.CCR-05-0092. [DOI] [PubMed] [Google Scholar]
- 39.Komatsu K, Andoh A, Ishiguro S, Suzuki N, et al. Increased expression of S100A6 (Calcyclin), a calcium-binding protein of the S100 family, in human colorectal adenocarcinomas. Clin Cancer Res. 2000;6:172–177. [PubMed] [Google Scholar]
- 40.Mitchell S, Ellingson C, Coyne T, Hall L, et al. Genetic variation in the urea cycle: a model resource for investigating key candidate genes for common diseases. Hum Mutat. 2008;30:56–60. doi: 10.1002/humu.20813. [DOI] [PubMed] [Google Scholar]
- 41.Wink DA, Ridnour LA, Hussain SP, Harris CC. The reemergence of nitric oxide and cancer. Nitric Oxide. 2008;19:65–67. doi: 10.1016/j.niox.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu TH, Li DC, Gu CF, Ye SF. Carbamyl phosphate synthetase I. A novel marker for gastric carcinoma. Chin Med J (Engl) 1989;102:630–638. [PubMed] [Google Scholar]
- 43.Kinoshita M, Miyata M. Underexpression of mRNA in human hepatocellular carcinoma focusing on eight loci. Hepatology. 2002;36:433–438. doi: 10.1053/jhep.2002.34851. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


