Abstract
OBJECTIVE
Glycemia is a major risk factor for the development of long-term complications in type 1 diabetes; however, no specific genetic loci have been identified for glycemic control in individuals with type 1 diabetes. To identify such loci in type 1 diabetes, we analyzed longitudinal repeated measures of A1C from the Diabetes Control and Complications Trial.
RESEARCH DESIGN AND METHODS
We performed a genome-wide association study using the mean of quarterly A1C values measured over 6.5 years, separately in the conventional (n = 667) and intensive (n = 637) treatment groups of the DCCT. At loci of interest, linear mixed models were used to take advantage of all the repeated measures. We then assessed the association of these loci with capillary glucose and repeated measures of multiple complications of diabetes.
RESULTS
We identified a major locus for A1C levels in the conventional treatment group near SORCS1 (10q25.1, P = 7 × 10−10), which was also associated with mean glucose (P = 2 × 10−5). This was confirmed using A1C in the intensive treatment group (P = 0.01). Other loci achieved evidence close to genome-wide significance: 14q32.13 (GSC) and 9p22 (BNC2) in the combined treatment groups and 15q21.3 (WDR72) in the intensive group. Further, these loci gave evidence for association with diabetic complications, specifically SORCS1 with hypoglycemia and BNC2 with renal and retinal complications. We replicated the SORCS1 association in Genetics of Diabetes in Kidneys (GoKinD) study control subjects (P = 0.01) and the BNC2 association with A1C in nondiabetic individuals.
CONCLUSIONS
A major locus for A1C and glucose in individuals with diabetes is near SORCS1. This may influence the design and analysis of genetic studies attempting to identify risk factors for long-term diabetic complications.
Elevation in plasma glucose, as measured by A1C, is a major risk factor for long-term diabetic complications (1–13). Twin and family studies have shown that A1C levels are heritable in nondiabetic individuals (14–16). In addition, significant correlation in A1C between monozygotic twins both concordant and discordant for type 1 diabetes (14,17), as well as in siblings with type 1 diabetes (18,55), suggests that some genetic factors influence A1C in both diabetic and nondiabetic individuals. However, no study of dizygous twins has been performed. These observations have motivated genome-wide linkage studies of measures of glycemia (typically single fasting plasma glucose or A1C), predominantly in nondiabetic individuals, but have not led to the identification of novel genes possibly because of small effect sizes of individual loci and underpowered studies (15,19,20). More recently, association studies of fasting glucose in nondiabetic individuals have identified multiple loci that meet genome-wide significance criteria (P < 5 × 10−8, supplementary Table 1, available in an online appendix at http://diabetes.diabetesjournals.org/cgi/content/full/db09-0653/DC1) (20–29).
In individuals with type 1 diabetes, there is considerable variability in glycemic control both between and within individuals. Intra-individual variation is likely caused by changes in diabetes management (e.g., pump versus multiple daily injections [30]), diet, activity, weight control, and insulin dosing in response to glucose and A1C measures. Other factors that may influence A1C are erythrocyte turnover, hemoglobinopathies, defects of glucose transport into erythrocytes, mechanisms of glycation and de-glycation, and alterations in intracellular glucose metabolism. Specifically, in the conventional treatment arm of the Diabetes Control and Complications Trial (DCCT), the intra-class correlation between consecutive quarterly A1C values was 0.79, falling to 0.42 for values measured 3 years apart. In the face of such high variability over time, analyzing longitudinally repeated measures via mixed models can increase power to identify risk factors. Alternatively, taking the mean of the values over time may be sufficient to initially screen for important genetic effects. We hypothesized that common genetic variants associated with glycemic control in subjects with type 1 diabetes could be identified using repeated measures of A1C from subjects in the DCCT. Because glycemic control was the major intervention in DCCT, we performed genetic analysis separately in each treatment group, with the expectation that genetic effects may be larger in the conventional treatment group in which there was no attempt to alter A1C (apart from preventing very high values [i.e., >13.11%]).
RESEARCH DESIGN AND METHODS
DCCT subjects.
A total of 1,441 patients with type 1 diabetes were recruited for the DCCT (1,31), which was designed to determine if intensive diabetes management, with the goal of normalizing glycemic levels, would prevent or delay the development and/or progression of long-term diabetic complications. At screening for eligibility, A1C had to be >6.6%, and subjects with hemaglobinopathies were excluded (31). Subjects were randomized to either conventional therapy (CON) or intensive therapy (INT) (Table 1). CON consisted of one or two daily injections of mixed intermediate and short-acting insulins, daily self-monitoring of urine initially or later blood glucose, and education about diet and exercise (31,32), but did not usually include daily adjustments in the insulin dosage. INT included the administration of insulin three or more times daily by injection or treatment with an external pump. Insulin doses were adjusted according to the results of self-monitoring of blood glucose performed four or more times per day, dietary intake, and anticipated exercise. The goals of INT included preprandial blood glucose 70–120 mg/dl, postprandial <180 mg/dl, a weekly 3:00 a.m. measurement >65 mg/dl, and monthly A1C within the normal range (<6.05%). Women who became pregnant or were planning a pregnancy received INT until the time of delivery. The DCCT was terminated prematurely in 1993, when it was conclusively shown that intensive management delayed the development and progression of retinopathy, nephropathy, and neuropathy (1). DNA was collected (33), and participants gave written informed consent for its use. These studies were approved by the institutional review boards of all participating institutions.
TABLE 1.
Descriptive information of 1,304 white DCCT subjects, separately by treatment group
| Conventional | Intensive | |
|---|---|---|
| Sex (n) | ||
| Male | 363 | 332 |
| Female | 304 | 305 |
| Cohort (n) | ||
| Primary prevention | 344 | 307 |
| Secondary intervention | 323 | 330 |
| Age at DCCT baseline (years) | 26.5 ± 7.1 | 27.2 ± 7.1 |
| Duration in the DCCT (years) | 6.2 ± 1.6 | 6.3 ± 1.7 |
| Eligibility A1C (%) | 9.00 ± 1.61 | 9.07 ± 1.58 |
| A1C measures obtained* | 26 ± 7 (5–40) | 63 ± 21 (3–103) |
| Mean A1C (%)* | 9.06 ± 1.24 | 7.22 ± 0.93 |
| Mean daily glucose from seven-point capillary profile (mg/dl)* | 231 ± 80 | 156 ± 50 |
| Stimulated C-peptide at DCCT baseline (pmol/ml) | 0.117 ± 0.119 | 0.111 ± 0.119 |
Data are means ±SD and means ±SD (range) unless otherwise indicated. There were 667 individuals in the conventional group and 637 in the intensive group with genotype data.
*For the intensive group, values used were from DCCT year 1 onward, whereas in the conventional group, all DCCT values were used, and for A1C in the conventional group, quarterly values were used, while in the intensive group, monthly values were used.
A1C, glucose, and C-peptide measurements.
DCCT subjects had A1C measured centrally using a high-performance liquid chromatography method once during eligibility screening and quarterly (CON) or monthly (INT) during DCCT, for between 3 and 9 years (mean 6.5 years, Table 1) (34). The stability of this assay over time has been described (34). The mean quarterly A1C level in each CON participant during DCCT was 9.06% (SD 1.24), based on an average of 26 measures (Table 1, supplementary Fig. 1). The mean monthly A1C measures in INT from DCCT year 1 onward was 7.22% (SD 0.96, supplementary Fig. 7). A centrally measured capillary blood glucose daily profile was obtained quarterly, consisting of seven samples, obtained before and 90 min after each meal, and one before bedtime, from which a daily mean was calculated. Stimulated C-peptide was measured at screening for DCCT eligibility (35). CON subjects were masked to their centrally measured A1C, glucose, and C-peptide results (31). Further, at 82% of their visits, they were not adjusting their insulin dose in response to self-monitoring of blood glucose (30).
Genotyping and quality control.
We performed genome-wide genotyping in DCCT subjects using the Illumina 1M beadchip assay (www.illumina.com, San Diego, CA). Genotypes were called using BeadStudio using all individuals at once. Data from three probands was excluded because of discrepancies between reported sex and genotype data. No data were removed because of low genotype call rate (minimum call rate threshold was 0.988). A total of 58 probands were removed because of disagreements between genotypes of single-nucleotide polymorphisms (SNPs) from an earlier study (33). Genotypes from 24 duplicate samples had an agreement rate of 99.9995%. Sample contamination was assessed by calculation of the mean heterozygosity across the genome for each individual, and using a range of 0.25–0.32, none were removed. Consistency of genotypes with Mendelian inheritance was observed in 28 trios with an observed rate of 99.71%. To detect cryptically related individuals and/or sample mix-ups, identical-by-state estimates between all pairs of individuals were performed (36), and two probands were removed. A total of 841,342 SNPs with a minor allele frequency >1% were subsequently analyzed statistically.
Autosomal SNPs showing significant association with sex (P < 10−8) or deviating from the Hardy-Weinberg equilibrium (P < 10−8) were excluded from the analysis. To reduce the possibility of population stratification, we limited the analysis to individuals who self-identified as white and excluded individuals who were determined to be admixed between Caucasian and other ethnic groups through population genetic approaches (37), seeding with genotype data from the three major populations genotyped in HapMap Phase II (38). For this latter analysis, we first removed SNPs from two regions known to have strong linkage disequilibrium (major histocompatibility complex, the polymorphic chromosome 8 inversion) and then selected independent SNPs, required to have r2 < 0.2 over a shifting window of 500 kb, yielding ∼98 K SNPs. Principal components were selected based on a scree plot. In addition, cluster plots of called genotypes by allele intensities for SNPs showing association with outcomes were visually examined to ensure appropriate genotype calling. Imputation of an additional 1,677,236 ungenotyped autosomal SNPs was performed using release 22 Phase II CEU HapMap data (MACH v 1.0.16) and were analyzed using genotype probabilities and an additive genetic model (MACH2QTL v 1.04) (38,39). Genomic control lambda for mean A1C separately in CON, INT, and combined (COMB) treatment groups were all 1.01.
Statistical analysis.
We performed a five-stage analysis (supplementary Fig. 6). Given the level of multiple hypothesis testing, we required P values <5 × 10−8 for genome-wide significance in either the CON, INT, or COMB groups (40) and one-sided P < 0.05 for confirmation in the other treatment group with the same direction (41).
Stage 1 consisted of a genome-wide association study of the intra-individual mean A1C during DCCT, undertaken separately in each treatment group (667 CON individuals, 637 INT subjects) as well as in COMB including treatment as a covariate (Table 1). For this initial screen, we calculated the mean A1C from values obtained at eligibility screening, baseline, and up to 38 quarterly visits during DCCT for the conventional group. For INT and COMB, we excluded data obtained during the introduction of intensive therapy in the first year of DCCT. These means were converted to normal scores and tested for association with genotypes at each SNP using a 2df model, which does not assume a particular genetic model. Imputed SNPs were analyzed in a linear regression framework using the dosage of the imputed genotypes.
In stage 2, to identify loci that did not meet genome-wide significance using the mean A1C, but did so using the repeated measures, we tested the top SNPs from stage 1 (P < 10−4) using linear regression mixed models including all of the available longitudinal A1C measures. In the intensive group, we used monthly values. We analyzed the natural log transformation of the A1C values with genotypes as a categorical variable and the time from DCCT eligibility as a continuous covariate, specifying random intercept and slope, and assumed an autoregressive moving average (1,1) covariance structure (because it had the lowest Akaikes information criterion [AIC]). Association of A1C with covariates and principal components from the Eigenstrat analysis was tested by linear regression, with results based on type 3 sum of squares (SAS PROCs REG and MIXED v9.1.3, Cary, NC).
In stage 3, SNPs achieving, or close to, genome-wide significance in stage 2 were tested for association with the repeated mean daily glucose, as well as C-peptide measured at eligibility screening for DCCT.
In stage 4, we analyzed the same SNPs with COMB A1C repeated measures, including treatment group as a covariate, as well as testing for interaction between each SNP and treatment.
Finally, since glycemia is a major risk factor for many diabetes complications, in stage 5, we tested the top A1C-associated SNPs for association with the following: the prevalence of coronary calcium, confirmed clinical neuropathy, hypoglycemia, as well as the time to event from DCCT baseline for three retinal and two renal outcomes (supplementary Table 2). This was done separately in each treatment group.
GoKinD.
The Genetics of Diabetes in Kidneys (GoKinD) study was used to replicate associations with A1C and nephropathy. GoKinD is a case-extreme control study of renal disease in type 1 diabetes (42). Renal case and control subjects were defined as described elsewhere (42). Because, as expected the single cross-sectional A1C was significantly higher in renal case subjects than control subjects, the associations with A1C were analyzed separately. In addition, patients with a pancreas transplant were excluded from the A1C analysis. Genotype data from the Affymetrix SNP 5.0 were used (43) and imputed to HapMap phase 2 (release 24). Analyses were restricted to subjects with Caucasian ancestry as determined by genetic analysis and in whom appropriate genotype quality was observed.
MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium).
We determined if any of the SNPs associated with A1C in DCCT were also associated with A1C in nondiabetic individuals. For this purpose, we obtained meta-analysis results for A1C from 23 genome-wide association studies including 27,589–36,585 (depending on the SNP studied) nondiabetic individuals of European descent (56) (see supplementary Methods).
RESULTS
To examine familial aggregation of A1C in type 1 diabetes, use of the A1C measured at eligibility for DCCT probands and A1C from sibs with type 1 diabetes (measured either during the DCCT family study [44] or the Epidemiology of Diabetes Interventions and Complications [EDIC] genetics study [33]) revealed significant familial correlation (Spearman rank correlation = 0.33, P = 1 × 10−3, n = 95 pairs, supplementary Fig. 14).
Stage 1: genome-wide association study of mean A1C.
To address potential population stratification bias, we tested for association of mean A1C with principal components from Eigenstrat. When tested either individually or together, none of the three principal components were associated with mean A1C.
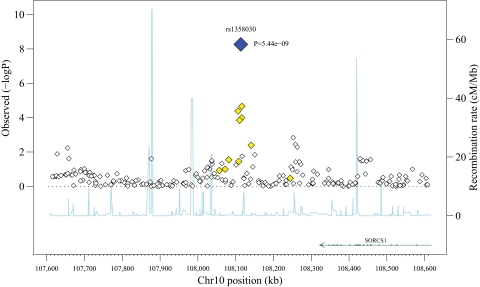
In the CON analysis, a single SNP (rs1358030, P = 5 × 10−9) on chromosome 10q25.1 met criteria for genome-wide significance (Table 2, supplementary Figs. 2 and 3), with five other SNPs within a 35-kb region having P < 10−2 (Fig. 1). SNPs in two other chromosomal regions also attained evidence for association close to the genome-wide significance criteria in CON: one on chromosome 9 (rs10810632, P = 4 × 10−×7), with eight other SNPs within a 20-kb interval (supplementary Fig. 11), and another on chromosome 18 (rs163061, P = 1 × 10−6), with no other associated SNPs. At each of the top SNPs in these three regions, the genotype-specific means were consistent with an additive model in which individuals carrying the rare allele had higher A1C (Table 2).
TABLE 2.
Results for the mean A1C in either the conventional, intensive, or combined treatment groups (stage 1) for any SNP with P < 10−6
| Chromosome | SNP | Position | Mean A1C P | Common homozygote |
Heterozygote |
Rare homozygote |
A1 | A2 | Minor allele frequency | Missing data* | Hardy-Weinberg equilibrium P | Location of SNP to the nearest gene | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| n | Mean ± SD | n | Mean ± SD | n | Mean ± SD | ||||||||||
| Conventional treatment group | |||||||||||||||
| 9 | rs10810632 | 16,779,024 | 3.7 × 10−7 | 576 | 8.96 ± 1.22 | 85 | 9.63 ± 1.22 | 5 | 10.63 ± 1.35 | C | T | 0.079 | 1 | 0.70 | Intron 1 BNC2 |
| 9 | rs6475082 | 16,779,436 | 7.4 × 10−7 | 575 | 8.96 ± 1.22 | 86 | 9.61 ± 1.23 | 5 | 10.63 ± 1.35 | G | A | 0.079 | 1 | 0.71 | Intron 1 BNC2 |
| 9 | rs4961760 | 16,779,878 | 7.1 × 10−7 | 575 | 8.96 ± 1.22 | 87 | 9.61 ± 1.22 | 5 | 10.63 ± 1.35 | C | T | 0.080 | 0 | 0.71 | Intron 1 BNC2 |
| 10 | rs1358030 | 108,113,589 | 5.4 × 10−9 | 268 | 8.71 ± 1.15 | 307 | 9.24 ± 1.25 | 90 | 9.44 ± 1.24 | C | T | 0.361 | 2 | 0.12 | 3′ SORCS1 |
| 18 | rs163061 | 22,727,740 | 9.7 × 10−7 | 350 | 8.83 ± 1.17 | 259 | 9.29 ± 1.26 | 35 | 9.62 ± 1.43 | G | C | 0.262 | 40 | 0.34 | 3′ C18orf16 |
| Intensive treatment group | |||||||||||||||
| 15 | rs493218 | 51,277,554 | 5.4 × 10−7 | 506 | 7.31 ± 0.96 | 120 | 7.01 ± 0.89 | 11 | 6.38 ± 0.69 | C | T | 0.109 | 2 | 0.20 | 3′ WDR72 |
| 15 | rs572221 | 51,291,924 | 6.8 × 10−7 | 504 | 7.31 ± 0.96 | 120 | 7.01 ± 0.89 | 11 | 6.38 ± 0.69 | A | G | 0.110 | 2 | 0.25 | 3′ WDR72 |
| 15 | rs690271 | 51,291,964 | 5.1 × 10−7 | 505 | 7.31 ± 0.96 | 121 | 7.01 ± 0.88 | 11 | 6.38 ± 0.69 | A | G | 0.110 | 0 | 0.26 | 3′ WDR72 |
| 15 | rs566369 | 51,295,884 | 5.1 × 10−7 | 505 | 7.31 ± 0.96 | 121 | 7.01 ± 0.88 | 11 | 6.38 ± 0.69 | A | G | 0.110 | 1 | 0.26 | 3′ WDR72 |
| 15 | rs482541 | 51,296,486 | 5.1 × 10−7 | 505 | 7.31 ± 0.96 | 121 | 7.01 ± 0.88 | 11 | 6.38 ± 0.69 | A | G | 0.110 | 1 | 0.26 | 3′ WDR72 |
| Combined treatment groups | |||||||||||||||
| 5 | rs286405 | 35,613,014 | 6.7 × 10−7 | 421 | 7.99 ± 1.32 | 631 | 8.18 ± 1.48 | 233 | 8.55 ± 1.68 | T | C | 0.427 | 19 | 0.95 | 5′ FLJ23577 |
| 10 | rs1358030 | 108,113,589 | 2.2 × 10−9 | 545 | 7.93 ± 1.32 | 574 | 8.35 ± 1.57 | 183 | 8.48 ± 1.53 | C | T | 0.361 | 2 | 0.12 | 3′ SORCS1 |
A1C values are %. Mean = mean A1C value, SD = standard deviation of A1C values. A1 = minor, A2 = major allele.
*Number of individuals with missing genotype data. Asymptotic P values from the 2df normal score test. Position is the nucleotide location from build 36.3.
FIG. 1.
Association results for mean A1C levels in the conventional treatment group at a 500-kb region surrounding rs1358030 (SORCS1). On the left y-axis is the −log10 (P value) for each SNP genotyped. On the right y-axis and the line is the recombination rate estimated from our data. The annotated genes in the region are indicated along the bottom of the figure. SNPs are colored based on their linkage disequilibrium with the most significant SNP, i.e., rs1358030, where black indicates the index SNP, light grey indicates 0.2 < r2 < 0.5, and white indicates r2 ≤ 0.2 (see the online version of the figure for colors where blue indicates the index SNP and yellow indicates 0.2 < r2 < 0.5).
In the INT analysis, five SNPs on chromosome 15 had P values of 5 × 10−7 (Table 2, supplementary Figs. 8 and 13). The COMB analysis detected an additional SNP on chromosome 5, as well as the top CON chromosome 10 SNP (Table 2, supplementary Fig. 10). Analysis of imputed ungenotyped SNPs did not reveal any additional loci that met genome-wide significance using the mean A1C. Results for all SNPs are available from http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000086.v1.p1.
Stage 2: repeated A1C measures.
We selected the top SNPs from stage 1 according to a P value <10−4 and analyzed each of these 233 SNPs by mixed models using all the repeated measures. In CON, chromosome 9 and 10 SNPs from stage 1 became more significant (Table 3). In the INT group, however, there was little improvement for chromosome 15 SNPs over the analysis of the mean value (Table 3). In COMB (Table 3), three additional SNPs in a region on chromosome 14 approached genome-wide significance (P = 3 × 10−7, supplementary Fig. 12). Analysis of imputed SNPs did not provide evidence for additional loci meeting genome-wide significance (Table 3). Visual inspection of cluster plots for the SNPs in Tables 2 and 3 confirmed appropriate genotype calling (for rs1358030, see supplementary Fig. 15). Of the loci from the literature that were associated with glycemia in nondiabetic individuals, only rs13266634 (SLC30A8) was marginally associated with A1C in the DCCT, this being in the CON group (P = 0.03). This effect is in the same direction as the effect on A1C in nondiabetic individuals (28) and the effect on risk for type 2 diabetes (45) (N.B., supplementary Table 1).
TABLE 3.
Loci with P ≤ 10−7 in the conventional, intensive, or combined treatment groups using repeated A1C values (stage 2)
| Chromosome | SNP | Position | Repeated A1C P value | Mean A1C P value | Mean A1C by genotype from repeated analysis |
A1 | A2 | Minor allele frequency | Missing data* | Hardy-Weinberg equilibrium | Location of SNP to nearest gene | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C homo (SE) | Het (SE) | R homo (SE) | |||||||||||
| Conventional treatment group | |||||||||||||
| 9 | rs10810632 | 16,779,024 | 9.0 × 10−8 | 3.7 × 10−7 | 8.83 (0.05) | 9.50 (0.13) | 10.50 (0.63) | C | T | 0.08 | 1 | 0.70 | Intron 1 BNC2 |
| 9 | rs6475082 | 16,779,436 | 1.8 × 10−7 | 7.4 × 10−7 | 8.83 (0.05) | 9.48 (0.13) | 10.50 (0.63) | G | A | 0.08 | 1 | 0.71 | Intron 1 BNC2 |
| 9 | rs4961760 | 16,779,878 | 1.6 × 10−7 | 7.1 × 10−7 | 8.83 (0.05) | 9.48 (0.13) | 10.50 (0.63) | C | T | 0.08 | 0 | 0.71 | Intron 1 BNC2 |
| 9 | rs2254193 | 16,791,850 | 2.0 × 10−7 | 1.1 × 10−6 | 8.84 (0.05) | 9.57 (0.15) | 10.47 (0.70) | C | A | 0.07 | 8 | 0.67 | Intron 1 BNC2 |
| 10 | rs1358030 | 108,113,589 | 6.9 × 10−10 | 5.4 × 10−9 | 8.58 (0.07) | 9.12 (0.07) | 9.32 (0.13) | C | T | 0.36 | 2 | 0.12 | 3′ SORCS1 |
| Intensive treatment group | |||||||||||||
| 15 | rs493218 | 51,277,554 | 5.2 × 10−7 | 5.4 × 10−7 | 7.26 (0.04) | 6.99 (0.07) | 6.42 (0.22) | C | T | 0.11 | 2 | 0.20 | 3′ WDR72 |
| 15 | rs572221 | 51,291,924 | 6.6 × 10−7 | 6.8 × 10−7 | 7.26 (0.04) | 6.99 (0.07) | 6.42 (0.22) | A | G | 0.11 | 2 | 0.25 | 3′ WDR72 |
| 15 | rs690271 | 51,291,964 | 5.0 × 10−7 | 5.1 × 10−7 | 7.26 (0.04) | 6.99 (0.07) | 6.42 (0.22) | A | G | 0.11 | 0 | 0.26 | 3′ WDR72 |
| 15 | rs566369 | 51,295,884 | 5.0 × 10−7 | 5.1 × 10−7 | 7.26 (0.04) | 6.99 (0.07) | 6.42 (0.22) | A | G | 0.11 | 1 | 0.26 | 3′ WDR72 |
| 15 | rs482541 | 51,296,486 | 5.0 × 10−7 | 5.1 × 10−7 | 7.26 (0.04) | 6.99 (0.07) | 6.42 (0.22) | A | G | 0.11 | 1 | 0.26 | 3′ WDR72 |
| Combined treatment groups | |||||||||||||
| 10 | rs1358030 | 108,113,589 | 3.8 × 10−10 | 2.2 × 10−9 | 7.84 (0.04) | 8.16 (0.04) | 8.36 (0.08) | C | T | 0.36 | 2 | 0.12 | 3′ SORCS1 |
| 14 | rs11624318 | 94,375,765 | 2.7 × 10−7 | 7.6 × 10−6 | 8.07 (0.04) | 8.11 (0.05) | 7.43 (0.12) | A | C | 0.21 | 1 | 0.28 | 5′ GSC |
| 14 | rs11160219 | 94,405,244 | 4.3 × 10−7 | 3.4 × 10−5 | 8.10 (0.04) | 8.08 (0.05) | 7.47 (0.11) | A | G | 0.24 | 1 | 0.49 | 5′ GSC |
| 14 | rs8007115 | 94,407,241 | 5.4 × 10−7 | 4.1 × 10−5 | 8.10 (0.04) | 8.07 (0.05) | 7.47 (0.11) | T | C | 0.23 | 3 | 0.59 | 5′ GSC |
Mean A1C values are results from stage 1. Longitudinal repeated A1C P values are the results from stage 2. C homo, Het, and R homo are least square means of A1C in the common homozygote, heterozygote and rare homozygote genotype groups, respectively, with SEs in parentheses, back-transformed from analysis of repeated lnA1C. NB. The statistical results for the repeated A1C were based on normal scores. A1 = minor, A2 = major allele.
*Number of individuals with missing genotype data. Position is the nucleotide location from build 36. Location relative to gene is from dbSNP build 129.
Stage 3: repeated mean daily glucose measures and C-peptide.
Analysis of the 13 SNPs in Table 3, with repeated mean daily glucose measures from all quarterly measures during DCCT in the CON group, revealed associations with SNPs in the chromosome 9 and 10 regions, consistent in direction with their effect on A1C (Table 4). There was also evidence for association of the chromosome 10 SNP with mean daily glucose in the INT group (Table 4). This argues that SNP variation at these loci influences A1C through effects on glucose. None of the SNPs from Table 3 were associated with C-peptide measured at eligibility screening for DCCT (P > 0.05).
TABLE 4.
Loci for which there is evidence for association using repeated A1C values (stages 3 and 4)
| Chromosome | SNP | Position | Stage 2: repeated A1C P |
Stage 3: repeated daily glucose P |
Stage 3: mean glucose by genotype (mg/dl) |
Stage 4: repeated A1C P value (COMB = CON +INT) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CON | INT | CON | INT | COMB | C homo (SE) | Het (SE) | R homo (SE) | SNP P | SNP treatment interaction | |||
| 9 | rs10810632 | 16,779,024 | 9.0 × 10−8 | 0.44 | 9.7 × 10−3 | 0.62 | 0.52 | 216 (2)* | 226 (5)* | 259 (25)* | 9.2 × 10−3 | 7.9 × 10−4 |
| 9 | rs6475082 | 16,779,436 | 1.8 × 10−7 | 0.44 | 0.011 | 0.62 | 0.52 | 216 (2)* | 225 (5)* | 259 (25)* | 0.011 | 1.1 × 10−3 |
| 9 | rs4961760 | 16,779,878 | 1.6 × 10−7 | 0.44 | 0.012 | 0.62 | 0.53 | 216 (2)* | 225 (5)* | 259 (25)* | 0.010 | 1.1 × 10−3 |
| 9 | rs2254193 | 16,791,850 | 2.0 × 10−7 | 0.67 | 8.3 × 10−3 | 0.50 | 0.34 | 216 (2)* | 229 (6)* | 253 (27)* | 0.011 | 3.1 × 10−3 |
| 10 | rs1358030 | 108,113,589 | 6.9 × 10−10 | 0.012 | 2.7 × 10−5 | 5.8 × 10−3 | 1.5 × 10−6 | 208 (3)* | 223 (3)* | 228 (5)* | 3.8 × 10−10 | 0.013 |
| 14 | rs11624318 | 94,375,765 | 9.4 × 10−4 | 2.9 × 10−3 | 0.40 | 0.035 | 0.025 | 180 (1)‡ | 182 (2)‡ | 170 (4)‡ | 2.7 × 10−7 | 0.95 |
| 14 | rs11160219 | 94,405,244 | 1.1 × 10−5 | 0.023 | 0.37 | 0.58 | 0.49 | 181 (1)‡ | 180 (2)‡ | 176 (4)‡ | 4.3 × 10−7 | 0.25 |
| 14 | rs8007115 | 94,407,241 | 2.2 × 10−5 | 0.017 | 0.38 | 0.78 | 0.49 | 181 (1)‡ | 180 (2)‡ | 176 (4)‡ | 5.4 × 10−7 | 0.37 |
| 15 | rs493218 | 51,277,554 | 0.77 | 5.2 × 10−7 | 0.32 | 0.10 | 0.074 | 149 (1)† | 145 (2)† | 142 (7)† | 8.9 × 10−3 | 2.4 × 10−4 |
| 15 | rs572221 | 51,291,924 | 0.83 | 6.6 × 10−7 | 0.26 | 0.088 | 0.056 | 149 (1)† | 145 (2)† | 142 (7)† | 8.3 × 10−3 | 3.6 × 10−4 |
| 15 | rs690271 | 51,291,964 | 0.83 | 5.0 × 10−7 | 0.26 | 0.085 | 0.050 | 149 (1)† | 145 (2)† | 142 (7)† | 7.6 × 10−3 | 3.4 × 10−4 |
| 15 | rs566369 | 51,295,884 | 0.82 | 5.0 × 10−7 | 0.26 | 0.085 | 0.050 | 149 (1)† | 145 (2)† | 142 (7)† | 7.6 × 10−3 | 3.4 × 10−4 |
| 15 | rs482541 | 51,296,486 | 0.82 | 5.0 × 10−7 | 0.26 | 0.085 | 0.050 | 149 (1)† | 145 (2)† | 142 (7)† | 7.6 × 10−3 | 3.4 × 10−4 |
*Data are presented only for SNPs that were close to or met genome-wide significance (P < 5 × 10−8) from stage 2 using the longitudinal repeated measures in the conventional treatment group and either nominal significance in stage 3 for the intensive group, or reach genome-wide significance when both treatment groups were combined. For the intensive group only analysis, monthly measures were used. For the CON + INT analysis, only quarterly measures were used for the intensive group, so that they would be comparable to the conventional group. C homo, Het, and R homo are least square means of daily mean of the back-transformed Ln(glucose) in the common homozygote, heterozygote, and rare homozygote genotype groups, respectively, in one of the following groups: *CON,
†INT,
‡COMB. NB. The statistical analysis of the glucose measures was performed using normal scores.
Stage 4: repeated A1C measures in both treatment groups with SNP treatment interaction.
Testing for SNP by treatment interaction provided strong evidence for differences in the SNP associations between CON and INT at the regions on chromosomes 9 (P = 1 × 10−3) and 15 (P = 3 × 10−4), weaker on chromosome 10 (P = 0.01), but no evidence on chromosome 14 (P = 0.25, Table 4).
Stage 5: association of the 13 SNPs associated with A1C with complications.
We then tested the A1C-associated SNPs (Table 3) for association with complications in the same subjects and found several biologically consistent associations of SNPs on chromosomes 9 and 10. The chromosome 10 signal, rs1358030, was associated with both time to persistent microalbuminuria (P = 0.05) and severe nephropathy (P = 0.03) in CON (Table 5 and supplementary Table 3). In COMB, rs1358030 was associated with the prevalence of coronary calcium (P = 4 × 10−3, supplementary Table 4). Even more striking, was that in CON, this SNP was associated with hypoglycemia either requiring medical assistance (P = 1 × 10−4) or resulting in coma or seizure (P = 1 × 10−3; Table 5 and supplementary Table 5). Inclusion of the mean DCCT A1C as a covariate in these later models reduced the association of the SNP with hypoglycemia (P = 0.03 and 0.02, respectively), consistent with an effect being mediated through glycemic control (supplementary Table 5). Similarly, in CON, SNPs in the chromosome 9 locus were associated with the following: time to persistent microalbuminuria (P = 5 × 10−4), severe nephropathy (P = 2 × 10−3, Table 6 and supplementary Table 3), mild retinopathy (P = 2 × 10−4), clinically significant macular edema (P = 1 × 10−3), and severe retinopathy (P = 2 × 10−3, Table 6 and supplementary Table 6) as well as hypoglycemia requiring assistance (P = 4 × 10−3, Table 6 and supplementary Table 5). More modest results were obtained for SNPs in the chromosome 14 and 15 regions for time to mild retinopathy (P = 0.01 and 0.02, respectively, supplementary Table 6). The directions of these associations at these three regions were consistent with their effects on A1C, i.e., the genotype with higher A1C had higher risk of renal, retinal, cardiac, and neuropathic complications, while having lower risk of hypoglycemia (Tables 5 and 6).
TABLE 5.
Association of rs1358030 (SORCS1) with complications in the DCCT/EDIC
| Outcome | Conventional |
Intensive |
||
|---|---|---|---|---|
| OR or HR (95% CI) | P | OR or HR (95% CI) | P | |
| Hypoglycemia | ||||
| Requiring medical assistance | 0.63 (0.50–0.80) | 0.0001 | 0.89 (0.72–1.10) | 0.28 |
| Resulting in coma/seizure | 0.60 (0.44–0.81) | 0.001 | 1.05 (0.84–1.31) | 0.64 |
| Retinopathy | ||||
| Mild nonproliferative retinopathy | 1.15 (0.99–1.34) | 0.07 | 1.16 (0.99–1.35) | 0.07 |
| Severe nonproliferative retinopathy | 1.30 (1.04–1.63) | 0.02 | 1.27 (0.92–1.77) | 0.15 |
| Clinically significant macular edema | 1.16 (0.92–1.46) | 0.22 | 1.10 (0.8–1.5) | 0.57 |
| Nephropathy | ||||
| Persistent microalbuminuria | 1.25 (1.01–1.56) | 0.04 | 1.25 (0.93–1.67) | 0.14 |
| Severe nephropathy | 1.39 (1.04–1.87) | 0.03 | 1.00 (0.61–1.64) | 0.99 |
| Coronary calcium (β ± SE) | 0.83 ± 0.36 | 0.05 | 0.68 ± 0.35 | 0.02 |
| Confirmed clinical neuropathy | 1.32 (1.01–1.73) | 0.05 | 1.08 (0.81–1.43) | 0.19 |
See supplementary Table 2 for definition of complications outcomes. Analysis was performed using additive coding of genotype, with the direction expressed as the number of copies of the C alleles (compared with the T allele). HR, hazard ratio; OR, odds ratio.
TABLE 6.
Association of rs10810632 (BNC2) with complications in the DCCT/EDIC
| Outcome | Conventional |
Intensive |
||
|---|---|---|---|---|
| OR or HR (95% CI) | P | OR or HR (95% CI) | P | |
| Hypoglycemia | ||||
| Requiring medical assistance | 0.49 (0.30–0.80) | 0.004 | 1.02 (0.70–1.47) | 0.94 |
| Resulting in coma/seizure | 0.60 (0.32–1.10) | 0.10 | 0.89 (0.60–1.33) | 0.58 |
| Retinopathy | ||||
| Mild nonproliferative retinopathy | 1.73 (1.31–2.27) | 0.0002 | 1.02 (0.77–1.35) | 0.90 |
| Severe nonproliferative retinopathy | 1.83 (1.29–2.62) | 0.002 | 0.61 (0.3–1.25) | 0.15 |
| Clinically significant macular edema | 1.87 (1.31–2.67) | 0.001 | 0.97 (0.56–1.69) | 0.92 |
| Nephropathy | ||||
| Persistent microalbuminuria | 1.84 (1.31–2.59) | 0.001 | 0.80 (0.46–1.41) | 0.43 |
| Severe nephropathy | 1.85 (1.19–2.87) | 0.01 | 0.32 (0.08–1.31) | 0.06 |
| Coronary calcium (β ± SE) | 0.64 ± 0.62 | 0.30 | 1.34 ± 0.57 | 0.02 |
| Confirmed clinical neuropathy | 1.64 (1.00–2.68) | 0.05 | 0.69 (0.40–1.20) | 0.6 |
See legend to Table 5. Additive coding of genotype, with the direction expressed as the number of copies of the C alleles (compared with the T allele).
Confirmation of rs1358030 associated with A1C in GoKinD.
We then tested the top 13 SNPs that were identified from the four regions for A1C in DCCT for association with A1C in GoKinD separately by case-control status. The mean ± SD A1C in case subjects without pancreas transplant was 8.29 ± 1.56 (n = 531), while in control subjects, it was 7.47 ± 1.15 (n = 851). There was nominal evidence for association of rs1358030 with A1C in control subjects, but not case subjects (P = 0.01 and P = 0.8, respectively). In the control subjects, the direction of the effect at rs1358030 is consistent with that in DCCT, with higher A1C values in individuals with more copies of the rare allele (supplementary Table 7). Testing association of the same 13 SNPs with renal disease, separately in 795 case subjects and 856 control subjects, revealed no significant evidence for association at any of the 13 SNPs (P ≥ 0.1, supplementary Table 8).
Confirmation of BNC2 association with A1C in nondiabetic individuals.
To determine if there was evidence for association of the SNPs identified in DCCT with A1C in nondiabetic individuals, we obtained summary statistics from an analysis of A1C in >27.5 K nondiabetic individuals, carried out by the MAGIC investigators. There was nominal evidence consistent with association at SNPs in the chromosome 9 region (e.g., rs10810632, Table 7), which is in the same direction as the association observed with A1C in the DCCT CON group (Tables 2 and 3).
TABLE 7.
Association results of top 13 SNPs from DCCT with A1C in the MAGIC study of nondiabetic individuals
| Chromosome | Position | SNP | Allele 1 | Allele 2 | Effect | SE | P | Sample size |
|---|---|---|---|---|---|---|---|---|
| BNC2 gene region | ||||||||
| 9 | 16,779,024 | rs10810632 | t | c | −0.0220 | 0.0065 | 7.1 × 10−4 | 36,446 |
| 9 | 16,779,436 | rs6475082 | a | g | −0.0235 | 0.0064 | 2.7 × 10−4 | 36,557 |
| 9 | 16,779,878 | rs4961760 | t | c | −0.0194 | 0.0065 | 0.0030 | 35,837 |
| 9 | 16,791,850 | rs2254193 | a | c | −0.0235 | 0.0066 | 3.5 × 10−4 | 36,458 |
| SORCS1 gene region | ||||||||
| 10 | 108,113,589 | rs1358030 | a | g | 0.0014 | 0.0036 | 0.70 | 35,304 |
| GSC gene region | ||||||||
| 14 | 94,375,765 | rs11624318 | a | c | 0.0016 | 0.0042 | 0.71 | 35,801 |
| 14 | 94,405,244 | rs11160219 | a | g | −0.0018 | 0.0044 | 0.68 | 32,898 |
| 14 | 94,407,241 | rs8007115 | t | c | −0.0012 | 0.0045 | 0.79 | 27,589 |
| WDR72 gene region | ||||||||
| 15 | 51,277,554 | rs493218 | t | c | −0.0015 | 0.0057 | 0.79 | 36,530 |
| 15 | 51,291,924 | rs572221 | a | g | 0.0015 | 0.0057 | 0.79 | 36,561 |
| 15 | 51,291,964 | rs690271 | a | g | 0.0014 | 0.0057 | 0.81 | 36,585 |
| 15 | 51,295,884 | rs566369 | a | g | 0.0012 | 0.0057 | 0.83 | 36,575 |
| 15 | 51,296,486 | rs482541 | a | g | 0.0010 | 0.0057 | 0.86 | 36,577 |
| 18 | 22,727,740 | rs163061 | c | g | −0.0048 | 0.0041 | 0.25 | 33,297 |
Allele 1 indicates the effect allele compared with allele 2 as the reference. Alleles are aligned to HapMap forward strand, but are not aligned in terms of minor or major allele. The A1C values were untransformed % [National Glycohemoglobin Standardization Program transformation: %A1C = 91.48 × (A1C/Hb) + 2.152], adjusted for age and sex.
DISCUSSION
Using repeated longitudinal measures of A1C in the DCCT and a multistage analysis, we identified a novel locus (rs1358030) with genome-wide significant association in the conventional treatment group. This locus is also associated with mean glucose during DCCT, arguing against a specific effect on glycation of hemoglobin or red cell survival. Analysis of repeated A1C in the intensive group supported this locus at P = 0.012, with additional supportive evidence in renal control subjects from the GoKinD study (P = 0.01).
None of the SNPs that were associated A1C were found to be associated with C-peptide at DCCT baseline. Important caveats to these negative results, however, are important to remember, since the DCCT was not an inception cohort of newly diagnosed type 1 diabetes, C-peptide levels were an inclusion criteria, and it has been shown that glycemic control influences persistence of C-peptide (1,35,46).
The SNP with the most significant association (rs1358030) is ∼200 kb 3′ from SORCS1 (sortilin-related vacuolar protein sorting 10 domain containing receptor 1), the nearest annotated gene. Because there are prior data supporting the involvement of this gene in glycemic traits, we focus discussion below on SORCS1. A previous study mapped a major locus for fasting plasma insulin levels to a 242-kb interval containing only the promoter, first exon, and most of the first intron of Sorcs1, using a conditioned cross between mice carrying the “obese” Leptin mutation. Between the two strains, 46 sequence differences were identified in Sorcs1, including three nonsynonymous changes, making it difficult to ascribe phenotypic differences to a specific nucleotide change (47). Differences in Sorcs1 expression in islets between the two strains were documented (47). Independent data supporting SORCS1 as a locus for glycemic traits comes from a mapping study that determined that Sorcs1 was the only gene in a major locus for post-intraperitoneal glucose tolerance tests in a rat model of diabetes (48). There was modest evidence for association of SNPs in SORCS1, with a single measure of fasting insulin in 574 nondiabetic Mexican Americans from 102 families (P = 4 × 10−3). Further, in the Framingham Heart Study, there was nominal association of rs1416406, which is ∼120 kb 5′ of SORCS1, with fasting insulin, insulin sensitivity index, and insulin resistance (P = 0.02–0.005) (49). However, this SNP was not statistically significantly associated (P > 0.4) with type 2 diabetes in four replication studies. Importantly, neither of the SNPs from these two studies (49,50) are in strong linkage disequilibrium with rs1358030. Further, rs1358030 is not in strong linkage disequilibrium with any other SNP genotyped in HapMap phase 2 or 3 (r2 < 0.5, supplementary Fig. 4) (38), arguing that it is either the etiological variant or is in strong linkage disequilibrium with a causal variant not genotyped in HapMap.
The regions associated with A1C on chromosomes 9 (BNC2), 14 (WDR72), and 15 (GSC) have not been previously shown to be associated with glycemic traits in humans or animals. It is interesting to note that there is evidence for confirmation of the BNC2 region in a large meta-analysis of A1C from nondiabetic individuals, suggesting that some of the loci influencing glycemic control in type 1 diabetes also influence glycemia in nondiabetic individuals, consistent with twin studies (14). The associated SNPs in BNC2 are close to LOC648570, a hypothetical gene.
Because of the larger “environmental” effect of INT during DCCT, mean A1C was ∼1.8% lower than in the CON group (Table 1), so INT cannot be considered a straightforward replication group for loci identified in CON (Table 4). Despite the concern that in the INT, subjects adjusted insulin dose to achieve treatment aims, we specifically show at the four loci that inclusion of insulin dose as a covariate does not diminish the SNP association with A1C (supplementary Table 9). If a genetic association depends on the level of A1C, then evidence for association may differ between the two groups. Moreover, overestimation of the SNP effects in one treatment group, due to the Winner's curse (51), could explain a SNP-by-treatment interaction (Table 4). Nevertheless, the findings on chromosome 10 in repeated measures analysis of the intensive group (P = 0.012) argue for an effect in both treatment groups, as well as GoKinD control subjects. The nonsignificant results in the GoKinD case subjects may be due to lower power (estimated to be 43%).
Importance of repeated A1C measures.
To emphasize the importance of longitudinal measures in this study, we compared the results from the stage 1 (mean A1C analysis) with analysis of a single A1C measure (obtained at DCCT eligibility screening) at the most significant CON SNP (rs1358030). The latter produced nominal significance (P = 0.027) and accounted for only 0.61% of the variance in 1,304 individuals. This compares to 5.2% (P = 5 × 10−9) of the variance in the former analysis (noting however that this latter estimate may be upwardly biased [51]). We calculated that 6,580 subjects (i.e., approximately five times larger than DCCT) would be needed to detect a comparable effect using a single A1C measure (α = 5 × 10−8, 1 − β = 0.8) (52). These calculations, however, assume 1) the mean A1C values from a contemporary study would be similar to values at DCCT eligibility (mean = 9.0%) and 2) the genetic effect is similar across the range of A1C values. Current clinical studies in type 1 diabetes report lower mean A1C levels than that observed at DCCT eligibility (53). Further, in the conventional group, the mean A1C dropped from 9.1 to 7.8% during follow-up in EDIC (12). In addition, rs1358030 accounted for only 1.7% of the variance in mean A1C in the intensive group (compared with 5.2% in the conventional group), consistent with a possible relationship between effect size and A1C levels. Therefore, the combination of these two factors could result in even larger sample sizes being required for the same power to detect a genetic effect using a single A1C measure in a contemporary setting. This clearly emphasizes the advantage of exploiting longitudinal measures of variable traits such as A1C, as has been described in other studies (54).
In conclusion, although a major locus for A1C levels in type 1 diabetes was identified, a number of outstanding questions remain. These include evaluation in other groups: nonwhite individuals, type 1 diabetic subjects who do not meet DCCT eligibility criteria, and type 2 diabetic subjects. SORCS1 is not a known susceptibility loci for either type 1 or type 2 diabetes, nor does it overlap with the loci identified for fasting glucose in nondiabetic individuals (Table 7) (20–23). Further, the etiological variants and mechanisms by which genetic variation influences glycemic control are unknown. Nonetheless, understanding of genetic factors that influence an individual's ability to control their A1C levels has potential application to clinical care and may provide insight into why patients on the same regimen have different A1C values. In the future, it may be possible to recommend different treatment or regimen approaches to subjects based on genotype to achieve similar targets. In the design and analysis of genetic studies, attempting to identify risk factors for long-term diabetic complications confounding by loci for glycemic control may be reduced by inclusion of longitudinal measures of A1C as covariates.
Supplementary Material
ACKNOWLEDGMENTS
The DCCT/EDIC Research Group is sponsored through research contracts from the National Institute of Diabetes, Endocrinology and Metabolic Diseases of the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) and the National Institutes of Health. S.B.B. held a Canadian Institutes of Health Research (CIHR) Senior Investigator award (2002–2007). A.D.P. holds a Canada Research Chair in the Genetics of Complex Diseases. This work received support from National Institute of Diabetes and Digestive and Kidney Diseases contract N01-DK-6-2204, National Institute of Diabetes and Digestive and Kidney Diseases Grants R01-DK-077510, R01-DK077489, and P60-DK20595 and support from the Canadian Network of Centres of Excellence in Mathematics and Genome Canada through the Ontario Genomics Institute. Clinical data and DNA from the DCCT/EDIC study will be made available through the National Institute of Diabetes and Digestive and Kidney Diseases repository at https://www.niddkrepository.org/niddk/home.do. The GoKinD study sample collection was supported by the Juvenile Diabetes Research Foundation in collaboration with the Joslin Diabetes Center and George Washington University and by the U.S. Centers for Disease Control and Prevention.
No potential conflicts of interest relevant to this article were reported.
The authors are grateful to the subjects in the DCCT/EDIC cohort for their long-term participation.
Footnotes
Clinical trial registry nos. NCT00360815 (DCCT) and NCT00360893 (EDIC), clinicaltrials.gov.
The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the National Institute of Diabetes and Digestive and Kidney Diseases or the National Institutes of Health.
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
See accompanying commentary, p. 332..
REFERENCES
- 1.The effect of intensive treatment of diabetes on the development and progression of long-term complications in insulin-dependent diabetes mellitus. The Diabetes Control and Complications Trial Research Group. N Engl J Med 1993; 329:977–986 [DOI] [PubMed] [Google Scholar]
- 2.The effect of intensive diabetes therapy on the development and progression of neuropathy. The Diabetes Control and Complications Trial Research Group. Ann Intern Med 1995; 122:561–568 [DOI] [PubMed] [Google Scholar]
- 3.Effect of intensive diabetes treatment on nerve conduction in the Diabetes Control and Complications Trial. Ann Neurol 1995; 38:869–880 [DOI] [PubMed] [Google Scholar]
- 4.Effect of intensive diabetes management on macrovascular events and risk factors in the Diabetes Control and Complications Trial. Am J Cardiol 1995; 75:894–903 [DOI] [PubMed] [Google Scholar]
- 5.Effect of intensive therapy on the development and progression of diabetic nephropathy in the Diabetes Control and Complications Trial. The Diabetes Control and Complications (DCCT) Research Group. Kidney Int 1995; 47:1703–1720 [DOI] [PubMed] [Google Scholar]
- 6.The effect of intensive diabetes therapy on measures of autonomic nervous system function in the Diabetes Control and Complications Trial (DCCT). Diabetologia 1998; 41:416–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Effect of intensive diabetes treatment on carotid artery wall thickness in the epidemiology of diabetes interventions and complications. Epidemiology of Diabetes Interventions and Complications (EDIC) Research Group. Diabetes 1999; 48:383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Retinopathy and nephropathy in patients with type 1 diabetes four years after a trial of intensive therapy. The Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. N Engl J Med 2000; 342:381–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Effect of intensive therapy on the microvascular complications of type 1 diabetes mellitus. JAMA 2002; 287:2563–2569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cleary PA, Orchard TJ, Genuth S, Wong ND, Detrano R, Backlund JY, Zinman B, Jacobson A, Sun W, Lachin JM, Nathan DMDCCT/EDIC Research Group. The effect of intensive glycemic treatment on coronary artery calcification in type 1 diabetic participants of the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study. Diabetes 2006; 55:3556–3565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Writing Team for the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Sustained effect of intensive treatment of type 1 diabetes mellitus on development and progression of diabetic nephropathy: the Epidemiology of Diabetes Interventions and Complications (EDIC) study. JAMA 2003; 290:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nathan DM, Cleary PA, Backlund JY, Genuth SM, Lachin JM, Orchard TJ, Raskin P, Zinman B.Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study Research Group. Intensive diabetes treatment and cardiovascular disease in patients with type 1 diabetes: the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications (DCCT/EDIC) Study Research Group. N Engl J Med 2005; 353:2643–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nathan DM, Lachin J, Cleary P, Orchard T, Brillon DJ, Backlund JY, O'Leary DH, Genuth S.Diabetes Control and Complications Trial, Epidemiology of Diabetes Interventions and Complications Research Group. Intensive diabetes therapy and carotid intima-media thickness in type 1 diabetes mellitus. N Engl J Med 2003; 348:2294–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Snieder H, Sawtell PA, Ross L, Walker J, Spector TD, Leslie RD: HbA(1c) levels are genetically determined even in type 1 diabetes: evidence from healthy and diabetic twins. Diabetes 2001;50:2858–2863 [DOI] [PubMed] [Google Scholar]
- 15.Meigs JB, Panhuysen CI, Myers RH, Wilson PW, Cupples LA: A genome-wide scan for loci linked to plasma levels of glucose and HbA(1c) in a community-based sample of Caucasian pedigrees: The Framingham Offspring Study. Diabetes 2002;51:833–840 [DOI] [PubMed] [Google Scholar]
- 16.Simonis-Bik AM, Eekhoff EM, Diamant M, Boomsma DI, Heine RJ, Dekker JM, Willemsen G, van Leeuwen M, de Geus EJ: The heritability of HbA1c and fasting blood glucose in different measurement settings. Twin Res Hum Genet 2008;11:597–602 [DOI] [PubMed] [Google Scholar]
- 17.Dubrey SW, Reaveley DR, Seed M, Lane DA, Ireland H, O'Donnell M, O'Connor B, Noble MI, Leslie RD: Risk factors for cardiovascular disease in IDDM: a study of identical twins. Diabetes 1994;43:831–835 [DOI] [PubMed] [Google Scholar]
- 18.Borch-Johnsen K, Nørgaard K, Hommel E, Mathiesen ER, Jensen JS, Deckert T, Parving HH: Is diabetic nephropathy an inherited complication? Kidney Int 1992;41:719–722 [DOI] [PubMed] [Google Scholar]
- 19.An P, Freedman BI, Hanis CL, Chen YD, Weder AB, Schork NJ, Boerwinkle E, Province MA, Hsiung CA, Wu X, Quertermous T, Rao DC: Genome-wide linkage scans for fasting glucose, insulin, and insulin resistance in the National Heart, Lung, and Blood Institute Family Blood Pressure Program: evidence of linkages to chromosome 7q36 and 19q13 from meta-analysis. Diabetes 2005;54:909–914 [DOI] [PubMed] [Google Scholar]
- 20.Meigs JB, Manning AK, Fox CS, Florez JC, Liu C, Cupples LA, Dupuis J: Genome-wide association with diabetes-related traits in the Framingham Heart Study. BMC Med Genet 2007;8 (Suppl. 1):S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, Cavalcanti-Proença C, Marchand M, Hartikainen AL, Sovio U, De Graeve F, Rung J, Vaxillaire M, Tichet J, Marre M, Balkau B, Weill J, Elliott P, Jarvelin MR, Meyre D, Polychronakos C, Dina C, Sladek R, Froguel P: A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 2008; 320: 1085– 1088 [DOI] [PubMed] [Google Scholar]
- 22.Chen WM, Erdos MR, Jackson AU, Saxena R, Sanna S, Silver KD, Timpson NJ, Hansen T, Orrù M, Grazia Piras M, Bonnycastle LL, Willer CJ, Lyssenko V, Shen H, Kuusisto J, Ebrahim S, Sestu N, Duren WL, Spada MC, Stringham HM, Scott LJ, Olla N, Swift AJ, Najjar S, Mitchell BD, Lawlor DA, Smith GD, Ben-Shlomo Y, Andersen G, Borch-Johnsen K, Jørgensen T, Saramies J, Valle TT, Buchanan TA, Shuldiner AR, Lakatta E, Bergman RN, Uda M, Tuomilehto J, Pedersen O, Cao A, Groop L, Mohlke KL, Laakso M, Schlessinger D, Collins FS, Altshuler D, Abecasis GR, Boehnke M, Scuteri A, Watanabe RM: Variations in the G6PC2/ABCB11 genomic region are associated with fasting glucose levels. J Clin Invest 2008;118:2620–2628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, Roos C, Tewhey R, Rieder MJ, Hall J, Abecasis G, Tai ES, Welch C, Arnett DK, Lyssenko V, Lindholm E, Saxena R, de Bakker PI, Burtt N, Voight BF, Hirschhorn JN, Tucker KL, Hedner T, Tuomi T, Isomaa B, Eriksson KF, Taskinen MR, Wahlstrand B, Hughes TE, Parnell LD, Lai CQ, Berglund G, Peltonen L, Vartiainen E, Jousilahti P, Havulinna AS, Salomaa V, Nilsson P, Groop L, Altshuler D, Ordovas JM, Kathiresan S: Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 2008;57:3112–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bouatia-Naji N, Bonnefond A, Cavalcanti-Proença C, Sparsø T, Holmkvist J, Marchand M, Delplanque J, Lobbens S, Rocheleau G, Durand E, De Graeve F, Chèvre JC, Borch-Johnsen K, Hartikainen AL, Ruokonen A, Tichet J, Marre M, Weill J, Heude B, Tauber M, Lemaire K, Schuit F, Elliott P, Jørgensen T, Charpentier G, Hadjadj S, Cauchi S, Vaxillaire M, Sladek R, Visvikis-Siest S, Balkau B, Lévy-Marchal C, Pattou F, Meyre D, Blakemore AI, Jarvelin MR, Walley AJ, Hansen T, Dina C, Pedersen O, Froguel P: A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 2009;41:89–94 [DOI] [PubMed] [Google Scholar]
- 25.Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, Spégel P, Bugliani M, Saxena R, Fex M, Pulizzi N, Isomaa B, Tuomi T, Nilsson P, Kuusisto J, Tuomilehto J, Boehnke M, Altshuler D, Sundler F, Eriksson JG, Jackson AU, Laakso M, Marchetti P, Watanabe RM, Mulder H, Groop L: Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 2009;41:82–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, Thorleifsson G, Loos RJ, Manning AK, Jackson AU, Aulchenko Y, Potter SC, Erdos MR, Sanna S, Hottenga JJ, Wheeler E, Kaakinen M, Lyssenko V, Chen WM, Ahmadi K, Beckmann JS, Bergman RN, Bochud M, Bonnycastle LL, Buchanan TA, Cao A, Cervino A, Coin L, Collins FS, Crisponi L, de Geus EJ, Dehghan A, Deloukas P, Doney AS, Elliott P, Freimer N, Gateva V, Herder C, Hofman A, Hughes TE, Hunt S, Illig T, Inouye M, Isomaa B, Johnson T, Kong A, Krestyaninova M, Kuusisto J, Laakso M, Lim N, Lindblad U, Lindgren CM, McCann OT, Mohlke KL, Morris AD, Naitza S, Orrù M, Palmer CN, Pouta A, Randall J, Rathmann W, Saramies J, Scheet P, Scott LJ, Scuteri A, Sharp S, Sijbrands E, Smit JH, Song K, Steinthorsdottir V, Stringham HM, Tuomi T, Tuomilehto J, Uitterlinden AG, Voight BF, Waterworth D, Wichmann HE, Willemsen G, Witteman JC, Yuan X, Zhao JH, Zeggini E, Schlessinger D, Sandhu M, Boomsma DI, Uda M, Spector TD, Penninx BW, Altshuler D, Vollenweider P, Jarvelin MR, Lakatta E, Waeber G, Fox CS, Peltonen L, Groop LC, Mooser V, Cupples LA, Thorsteinsdottir U, Boehnke M, Barroso I, Van Duijn C, Dupuis J, Watanabe RM, Stefansson K, McCarthy MI, Wareham NJ, Meigs JB, Abecasis GR: Variants in MTNR1B influence fasting glucose levels. Nat Genet 2009;41:77–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sabatti C, Service SK, Hartikainen AL, Pouta A, Ripatti S, Brodsky J, Jones CG, Zaitlen NA, Varilo T, Kaakinen M, Sovio U, Ruokonen A, Laitinen J, Jakkula E, Coin L, Hoggart C, Collins A, Turunen H, Gabriel S, Elliot P, McCarthy MI, Daly MJ, Järvelin MR, Freimer NB, Peltonen L: Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet 2009;41:35–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pare G, Chasman DI, Parker AN, Nathan DM, Miletich JP, Zee RY, Ridker PM: Novel association of HK1 with glycated hemoglobin in a non-diabetic population: a genome-wide evaluation of 14,618 participants in the Women's Genome Health Study. PLoS Genet 2008;4:e1000312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnefond A, Vaxillaire M, Labrune Y, Lecoeur C, Chevre JC, Bouatia-Naji N, Cauchi S, Balkau B, Marre M, Tichet J, Riveline JP, Hadjadj S, Gallois Y, Czernichow S, Hercberg S, Kaakinen M, Wiesner S, Charpentier G, Levy-Marchal C, Elliott P, Jarvelin MR, Horber F, Dina C, Pedersen O, Sladek R, Meyre D, Froguel P: A genetic variant in HK1 is associated with proanemic state and A1C but not other glycemic control-related traits. Diabetes 2009;58:2687–2697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Implementation of treatment protocols in the Diabetes Control and Complications Trial. Diabetes Care 1995; 18:361–376 [DOI] [PubMed] [Google Scholar]
- 31.Diabetes Control and Complications Trial (DCCT). Design and methodologic considerations for the feasibility phase. The DCCT Research Group. Diabetes 1986; 35:530–545 [PubMed] [Google Scholar]
- 32.Diabetes Control and Complications Trial Research Group. Manual of Operations for the Diabetes Control and Complications Trial. Springfield, Virginia, U.S.Department of Commerce, National Technical Information Service, 1993 [Google Scholar]
- 33.Al-Kateb H, Boright AP, Mirea L, Xie X, Sutradhar R, Mowjoodi A, Bharaj B, Liu M, Bucksa JM, Arends VL, Steffes MW, Cleary PA, Sun W, Lachin JM, Thorner PS, Ho M, McKnight AJ, Maxwell AP, Savage DA, Kidd KK, Kidd JR, Speed WC, Orchard TJ, Miller RG, Sun L, Bull SB, Paterson AD.Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Research Group. Multiple superoxide dismutase 1/splicing factor serine alanine 15 variants are associated with the development and progression of diabetic nephropathy: the Diabetes Control and Complications Trial/Epidemiology of Diabetes Interventions and Complications Genetics study. Diabetes 2008; 57:218–228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steffes M, Cleary P, Goldstein D, Little R, Wiedmeyer HM, Rohlfing C, England J, Bucksa J, Nowicki M: Hemoglobin A1c measurements over nearly two decades: sustaining comparable values throughout the Diabetes Control and Complications Trial and the Epidemiology of Diabetes Interventions and Complications Study. Clin Chem 2005;51:753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Diabetes Control and Complications Trial Research Group. Effect of intensive therapy on residual beta-cell function in patients with type 1 diabetes in the Diabetes Control and Complications Trial: a randomized, controlled trial. The Diabetes Control and Complications Trial Research Group. Ann Intern Med 1998; 128:517–523 [DOI] [PubMed] [Google Scholar]
- 36.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, Sham PC: PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patterson N, Price AL, Reich D: Population structure and eigenanalysis. PLoS Genet 2006;2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.International HapMap Consortium. Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, Pasternak S, Wheeler DA, Willis TD, Yu F, Yang H, Zeng C, Gao Y, Hu H, Hu W, Li C, Lin W, Liu S, Pan H, Tang X, Wang J, Wang W, Yu J, Zhang B, Zhang Q, Zhao H, Zhao H, Zhou J, Gabriel SB, Barry R, Blumenstiel B, Camargo A, Defelice M, Faggart M, Goyette M, Gupta S, Moore J, Nguyen H, Onofrio RC, Parkin M, Roy J, Stahl E, Winchester E, Ziaugra L, Altshuler D, Shen Y, Yao Z, Huang W, Chu X, He Y, Jin L, Liu Y, Shen Y, Sun W, Wang H, Wang Y, Wang Y, Xiong X, Xu L, Waye MM, Tsui SK, Xue H, Wong JT, Galver LM, Fan JB, Gunderson K, Murray SS, Oliphant AR, Chee MS, Montpetit A, Chagnon F, Ferretti V, Leboeuf M, Olivier JF, Phillips MS, Roumy S, Sallée C, Verner A, Hudson TJ, Kwok PY, Cai D, Koboldt DC, Miller RD, Pawlikowska L, Taillon-Miller P, Xiao M, Tsui LC, Mak W, Song YQ, Tam PK, Nakamura Y, Kawaguchi T, Kitamoto T, Morizono T, Nagashima A, Ohnishi Y, Sekine A, Tanaka T, Tsunoda T, Deloukas P, Bird CP, Delgado M, Dermitzakis ET, Gwilliam R, Hunt S, Morrison J, Powell D, Stranger BE, Whittaker P, Bentley DR, Daly MJ, de Bakker PI, Barrett J, Chretien YR, Maller J, McCarroll S, Patterson N, Pe'er I, Price A, Purcell S, Richter DJ, Sabeti P, Saxena R, Schaffner SF, Sham PC, Varilly P, Altshuler D, Stein LD, Krishnan L, Smith AV, Tello-Ruiz MK, Thorisson GA, Chakravarti A, Chen PE, Cutler DJ, Kashuk CS, Lin S, Abecasis GR, Guan W, Li Y, Munro HM, Qin ZS, Thomas DJ, McVean G, Auton A, Bottolo L, Cardin N, Eyheramendy S, Freeman C, Marchini J, Myers S, Spencer C, Stephens M, Donnelly P, Cardon LR, Clarke G, Evans DM, Morris AP, Weir BS, Tsunoda T, Mullikin JC, Sherry ST, Feolo M, Skol A, Zhang H, Zeng C, Zhao H, Matsuda I, Fukushima Y, Macer DR, Suda E, Rotimi CN, Adebamowo CA, Ajayi I, Aniagwu T, Marshall PA, Nkwodimmah C, Royal CD, Leppert MF, Dixon M, Peiffer A, Qiu R, Kent A, Kato K, Niikawa N, Adewole IF, Knoppers BM, Foster MW, Clayton EW, Watkin J, Gibbs RA, Belmont JW, Muzny D, Nazareth L, Sodergren E, Weinstock GM, Wheeler DA, Yakub I, Gabriel SB, Onofrio RC, Richter DJ, Ziaugra L, Birren BW, Daly MJ, Altshuler D, Wilson RK, Fulton LL, Rogers J, Burton J, Carter NP, Clee CM, Griffiths M, Jones MC, McLay K, Plumb RW, Ross MT, Sims SK, Willey DL, Chen Z, Han H, Kang L, Godbout M, Wallenburg JC, L'Archevêque P, Bellemare G, Saeki K, Wang H, An D, Fu H, Li Q, Wang Z, Wang R, Holden AL, Brooks LD, McEwen JE, Guyer MS, Wang VO, Peterson JL, Shi M, Spiegel J, Sung LM, Zacharia LF, Collins FS, Kennedy K, Jamieson R, Stewart J: A second generation human haplotype map of over 3.1 million SNPs. Nature 2007;449:851–861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y, Abecasis G: Mach 1.0: Rapid haplotype reconstruction and missing genotype inference. Presented at the annual meeting of the American Society of Human Genetics, 9–13 October 2006,New Orleans, Louisiana (program number 2290) [Google Scholar]
- 40.Dudbridge F, Gusnanto A: Estimation of significance thresholds for genomewide association scans. Genet Epidemiol 2008;32:227–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.NCI-NHGRI Working Group on Replication in Association Studies. Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS: Replicating genotype-phenotype associations. Nature 2007;447:655–660 [DOI] [PubMed] [Google Scholar]
- 42.Mueller PW, Rogus JJ, Cleary PA, Zhao Y, Smiles AM, Steffes MW, Bucksa J, Gibson TB, Cordovado SK, Krolewski AS, Nierras CR, Warram JH: Genetics of Kidneys in Diabetes (GoKinD) study: a genetics collection available for identifying genetic susceptibility factors for diabetic nephropathy in type 1 diabetes. J Am Soc Nephrol 2006;17:1782–1790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pezzolesi MG, Poznik GD, Mychaleckyj JC, Paterson AD, Barati MT, Klein JB, Ng DP, Placha G, Canani LH, Bochenski J, Waggott D, Merchant ML, Krolewski B, Mirea L, Wanic K, Katavetin P, Kure M, Wolkow P, Dunn JS, Smiles A, Walker WH, Boright AP, Bull SB, DCCT/EDIC Research Group. Doria A, Rogus JJ, Rich SS, Warram JH, Krolewski AS: Genome-wide association scan for diabetic nephropathy susceptibility genes in type 1 diabetes. Diabetes 2009;58:1403–1410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clustering of long-term complications in families with diabetes in the Diabetes Control and Complications Trial. The Diabetes Control and Complications Trial Research Group. Diabetes 1997; 46:1829–1839 [PubMed] [Google Scholar]
- 45.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S, Balkau B, Heude B, Charpentier G, Hudson TJ, Montpetit A, Pshezhetsky AV, Prentki M, Posner BI, Balding DJ, Meyre D, Polychronakos C, Froguel P: A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature 2007;445:881–885 [DOI] [PubMed] [Google Scholar]
- 46.Palmer JP, Fleming GA, Greenbaum CJ, Herold KC, Jansa LD, Kolb H, Lachin JM, Polonsky KS, Pozzilli P, Skyler JS, Steffes MW: C-peptide is the appropriate outcome measure for type 1 diabetes clinical trials to preserve beta-cell function: report of an ADA workshop, 21–22 October 2001. Diabetes 2004;53:250–264 [DOI] [PubMed] [Google Scholar]
- 47.Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET, Stapleton DS, Gray-Keller MP, Young MB, Stoehr JP, Lan H, Boronenkov I, Raess PW, Flowers MT, Attie AD: Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat Genet 2006;38:688–693 [DOI] [PubMed] [Google Scholar]
- 48.Granhall C, Park HB, Fakhrai-Rad H, Luthman H: High-resolution quantitative trait locus analysis reveals multiple diabetes susceptibility loci mapped to intervals <800 kb in the species-conserved Niddm1i of the GK rat. Genetics 2006;174:1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Florez JC, Manning AK, Dupuis J, McAteer J, Irenze K, Gianniny L, Mirel DB, Fox CS, Cupples LA, Meigs JB: A 100K genome-wide association scan for diabetes and related traits in the Framingham Heart Study: replication and integration with other genome-wide datasets. Diabetes 2007;56:3063–3074 [DOI] [PubMed] [Google Scholar]
- 50.Goodarzi MO, Lehman DM, Taylor KD, Guo X, Cui J, Quiñones MJ, Clee SM, Yandell BS, Blangero J, Hsueh WA, Attie AD, Stern MP, Rotter JI: SORCS1: a novel human type 2 diabetes susceptibility gene suggested by the mouse. Diabetes 2007;56:1922–1929 [DOI] [PubMed] [Google Scholar]
- 51.Beavis WD: QTL analyses: power, precision, and accuracy. In Molecular Dissection of Complex Traits. Paterson AH. Ed.New York,CRC Press, 1998, p.145–162 [Google Scholar]
- 52.Gauderman WJ: Candidate gene association analysis for a quantitative trait, using parent-offspring trios. Genet Epidemiol 2003;25:327–338 [DOI] [PubMed] [Google Scholar]
- 53.Gerstl EM, Rabl W, Rosenbauer J, Gröbe H, Hofer SE, Krause U, Holl RW: Metabolic control as reflected by HbA1c in children, adolescents and young adults with type-1 diabetes mellitus: combined longitudinal analysis including 27,035 patients from 207 centers in Germany and Austria during the last decade. Eur J Pediatr 2008;167:447–453 [DOI] [PubMed] [Google Scholar]
- 54.Diggle PJ, Heagerty P, Liang KY, Zeger SL: Analysis of Longitudinal Data. Oxford, U.K., Oxford University Press,2002 [Google Scholar]
- 55.Hietala K, Forsblom C, Summanen P, Groop PH: FinnDiane Study Group. Heritability of proliferative diabetic retinopathy. Diabetes 2008; 57:2176–2180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sanna S, Soranzo N, Wheeler E, Gieger C, Radke D, Dupuis J, Stolerman E, Bouatia-Naji N, Largenberg C, Prokopenko I, Sandhu MS, Kao WHL, Wareham NJ, Florez JC, Uda M, Barroso I, Meigs JB: on behalf of MAGIC. Common variants at ten loci influence glycated hemoglobin levels via glycemic and non-glycemic pathways. American Society of Human Genetics annual meeting 2010 Program #1008, Honolulu, Hawaii, 21 October 2009 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.