Abstract
Immunohistochemical (IHC) staining of formalin-fixed and paraffin-embedded tissues (FFPE) is widely used in diagnostic surgical pathology. All anatomical and surgical pathologists use IHC to confirm cancer cell type and possible origin of metastatic cancer of unknown primary site. What kinds of improvements in IHC are needed to boost and strengthen the use of IHC in future diagnostic pathology practice? The aim of this perspective is to suggest that continuing reliance on immunohistochemistry in cancer diagnosis, search and validation of biomarkers for predictive and prognostic studies and utility in cancer treatment selection means that minimum IHC data sets including “normalization methods” for IHC scoring, use of relative protein expression levels, use of protein functional pathways and modifications and protein cell type specificity may be needed when markers are proposed for use in diagnostic pathology. Furthermore evidence based methods (EBM), minimum criteria for diagnostic accuracy (STARD), will help in selecting antibodies for use in diagnostic pathology. In the near future, quantitative methods of proteomics, quantitative real-time polymerase chain reaction (qRT-PCR) and the use of high-throughput genomics for diagnosis and predictive decisions may become preferred tools in medicine.
Keywords: Immunoperoxidase, protein lifecycle, surgical pathology, proteomics, evidence based methods, normalization
Introduction
Immunohistochemical methods in diagnostic pathology has a long history [1, 2]. Immunohistochemical staining methods include use of fluorophore-labeled (immunofluorescence) and enzyme-labeled (immunoperoxidase) antibodies to identify proteins and other molecules in cells. In diagnostic surgical pathology, immunoperoxidase methods (usually single anti-gen-antibody and less commonly double anti-body-antigen combinations) (Figure1) are widely used to extract additional information that is not available by hematoxylin and eosin staining and light microscopy or by transmission electron-microscopy. The advantage is that the molecules are identified in-situ in the cell. Immunohistochemistry is now used in surgical pathology to determine cancer cell types, cancer subtype classifications and possible cell-of –origin in metastatic cancer of unknown or undetermined primary site. In all instances, accepted and standardized morphologic criteria are used in addition to immunohistochemical staining of the tissue. The morphologic criteria for cancer diagnosis do not encompass the proposed biologic hallmarks of cancer [3].
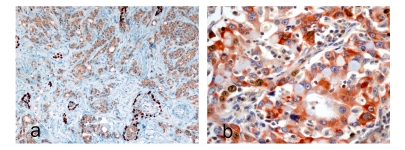
Figure 1.
a. Human prostate core biopsy with double immunohistochemical staining for high molecular weight cytokeratin (K903) and AMCAR (alpha-methyl-CoA-racemase). The dark brown stain (K903) highlights the basal epithelial cells and the light brown cytoplasmic stain AMCAR in prostate cancer cells including dysplastic cells in high-grade prostatic intraepithelial neoplasia (HGPIN). The differential localization and distribution are useful in confirming areas of invasive carcinoma (×40) in addition to conventional criteria for malignancy. b. Bcl-2 (anti-apoptotic protein) and Ki-67 in human lung carcinoma (courtesy Epitomics,Inc). This also highlights differential localization of the two proteins; Bcl-2 to cytoplasm and Ki-67 nuclear and also suggests that Ki-67 staining cells are different from Bcl-2 staining cells and the transcription cycle of the proteins.
This perspective is to review and promote the inclusion of some information to improve the interpretation of immunohistochemical data such as protein life-span and signaling, evidence-based methods and quantitative data and normalization.
Protein structure, modifications, life-span and implications for Immunohistochemistry
Protein synthesis in the cell is highly regulated [4]. The proteins undergo many modifications before full maturation and functional activation. Life-span modifications in normal, stressed and cancer cells include summoylation and ubiquitination and subsequent degradation in the proteosome and probably rescued by de-ubiquitination, by chaperones and chaperonins [5-7] and the effects of microRNA [8]. A widely known functional modification is phosphorylation that occurs on serine and threonine amino-acids, and these changes may affect life-span [9]. There are numerous protein databases that are freely available that permit inquiry of protein structure, cellular and tissue distribution, developmental and evolutionary history, functional status, mutations and other relevant information [10]. Furthermore, since synthetic peptides are frequently used for generating antibodies (mono-and polyclonal), the functional significance and contribution of the peptide segment and structural information in relation to the function of the whole molecule should be taken into account when interpreting the immunohistochemical staining result. Phospho-specific antibodies are now available for immunohistochemical use to determine the functional status of the protein and their use may further improve the results of immunohistochemical staining [11]. The productive use of phospho-specific antibodies will rest heavily on further elucidation of the cellular phospho-proteome [12] and optimization of phospho-specific polyclonal and monoclonal antibodies and tissue processing [13]. The p53 Example (Figure 2 a-c): One of the most investigated proteins in cell biology and pathology is p53. As an example, p53 is altered in many human cancers (>18,000 mutations) and involved in cell death and survival, DNA damage response [14, 15] and affects the transcription of a large gene/protein set in the cell [16]. p53 undergoes many modifications as wild-type or mutant protein and influences its cytoplasmic or nuclear location [17-20], the function and life-span of p53 and cellular interactions with its known and unknown targets and their function [21]. There are now competing and continually improving methods of proteomics to quantify and determine presence of protein(s) in cells [22-25]. Proteomics is useful in searching for and defining biomarkers using high-throughput methods such as the whole cell proteome.
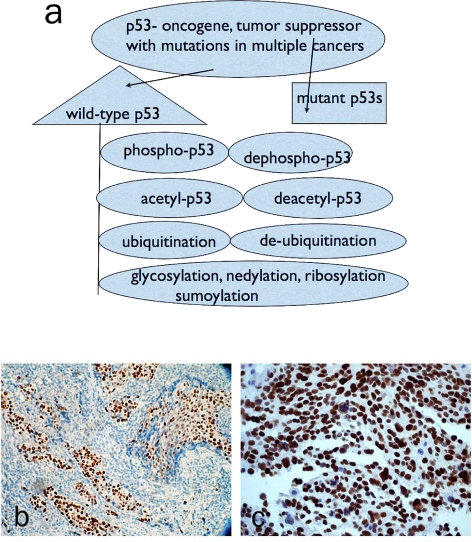
Figure 2.
a. Summarized p53 protein modifications b. immunohistochemical staining for wild-type p53 (Epitomics, Inc #1026-1) in human breast carcinoma. Note this antibody recognizes both wild-type and mutant p53. c. immunohistochemical staining for phospho-p53 (pS46) (Epitomics, Inc #2190) in human ovarian cancer tissue.
Diagnostic and predictive biomarkers
Biomarkers are currently proposed for various aspects of cancer such as early detection to selection of cancer patients for treatment. The biomarkers can be detected by immunohistochemical methods, quantitative proteomic methods and methods such as quantitative real-time polymerase chain reaction (qRT-PCR) [26]. The promotion of molecular and individualized medicine is based on the improvement and miniaturization of methods of proteomics and genomics in the search for bio-markers of disease onset, progression and treatment response [27]. A recent commentary also emphasizes the need to base the use of markers in diagnostic or predictive immunohistochemical staining o known biological pathways and underlying biology of the cancer or disease process [28]. Furthermore, there is growing interest in including defined biomarkers in clinical trials [29].
Evidence-based methods (EBM) and standards for reporting of diagnostic accuracy (STARD)
Another question is whether the rules of Evi-dence-based medicine (EBM) that are adopted in other sections of laboratory medicine (clinical chemistry) can be applied to immunohistochemical interpretation before adoption for routine use [30, 31]. The rules of EBM applied to laboratory values include agreement statistics (raw, kappa, expected and odds ratio), confidence intervals, sensitivity, specificity, positive and negative predictive values, likelihood ratios, pre-and post-test probabilities; all of these datasets are useful for estimating diagnostic accuracy and are used in other diagnostic settings [32]. The STARD criteria include 24 item check list. Including EBM and STARD criteria may reduce bias in their use in diagnosis or treatment selection. The proposal for minimum datasets in immunohistochemical publications (MISFISHIE) is encouraging, although quantitative analysis and protein structure data are not now included. Many publications provide immunohistochemical analysis as percentage of cases and control that are positive and negative for the anti-gen/protein under study. A recent study on immunohistochemical markers for mesothelioma listed the markers and percentage positive in the cases used but no sensitivity or specificity information [33]. Few studies provide sensitivity and specificity analysis; one recent study on lymphatic markers provided these analyses [34]. A recent study on use of markers for defining the possible primary site of metastatic adenocarcinomas had some markers with variable sensitivities and specificities [35]. The drawbacks of immunohistochemical staining that include inadequate antibody validation and many technical issues with a host of suggestions have been highlighted [36]. Some drawbacks of conventional immunohisto-chemical staining include lack of multiplexing, limited dynamic range and lack of correlation with functional protein and treatment response [36, 37]. The use of rabbit monoclonal versus mouse monoclonal antibody to estrogen receptor (ER) changed the level of positivity in breast cancer [38] (see Figure 3).
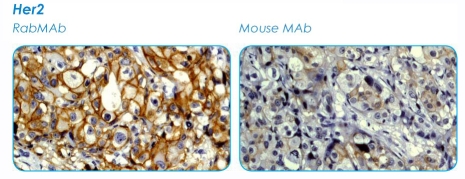
Figure 3.
Comparison of rabbit and mouse monoclonal antibodies in immunohistochemical detection of Her-2 (courtesy of Epitomics, Inc). The differences in intensities and percentage of cells stained is notable.
Normalizing IHC scores
How do we determine protein content in tissues? There are no reliable methods to quantify tissue protein content by immunohistochemical methods. Many authors use different methods to estimate protein / antigen level in the literature [39] including intensity levels (0-3) or percent of cells that stain or a combination of the two scoring methods and attempts at cutoff values. Many suggestions relating to quantitative methods in immunohistochemistry relate to its impact in high-throughput methods such as tissue microarray (TMA) [39, 40]. In a recent study, simulated mRNA levels were related to possible levels of protein detected by immunohistochemistry [41]; these methods will be difficult on an individual case based on protein life-span, modifications and mutations, tissue retrieval and fixation and other limitations. A study of Her-2 in breast cancer cell lines and tissue highlights the contribution of the primary antibody dilution on the level of Her-2 protein detected by immunohistochemical methods [40] especially as Her-2 belongs to a protein family with complex interacting networks (42). Unlike routine diagnostic immunohistochemical methods, high-throughput tissue microarrays, protein and DNA microarrays generate a lot of data. The methods of data analysis and presentation proposed for DNA and protein microarrays- including methods to remove noise in the data such as normalization, false and negative discovery rates [43-47] are designed to improve interpretation, and utility of the information. Can normalization be used in immunohistochemical evaluation and what methods can be used for normalization? One can use endogenous proteins for normalization as is used in northern blots for messenger ribonucleic acid (mRNA) levels (48) and for Western blotting and quantitative real-time polymerase chain reaction (qRT-PCR). A relative protein expression level can then be used. What are the minimum methods to quantify protein/antigen levels in the cell? Some investigators used bioinformatics tools to determine such cut-offs [49] and these attempts created different estimates for HER2 that are different from the standardized criteria for HER2 [50]. Image analysis computer programs that can be used easily are needed [51, 52] and [53] and are coming on-stream [54, 55]. A recent overview of quantitative image analysis software for immunostaining lists several commercial sources though costs may be a limitation to adoption of specific software [56]. Furthermore, as the interest in computer-assisted image analysis grows within the surgical pathology community an awareness of the multiple methods of image analysis, noise removal, image quality, and their effects on the results should be noted [57, 58]. The DAB-stained slides can be analyzed by spectral imaging [59], color deconvolution [54, 55, 60, 61], Hue-Saturation-Intensity [61], normalized RGB [62] and CMYK [63 and other methods. In a recent study of predictive biomarkers in breast cancer, automated image analysis was necessary to use 42 antibodies in the asseesment of marker utility [64]. The growth of many image analysis methods for the popular DAB-stained tissues needs internal normalization as done for RT-PCR and western blotting to truly compare the results.
Alternative methods for biomarker identification and selection
The role of IHC data sets and analysis in diagnostic pathology are being challenged by other quantitative methods such as DNA microarray and qRT-PCR in cancer detection, classification and predicting cancer treatment response. Recently proposed molecular classifications of cancers and their use in cancer treatment planning are based on DNA microarray methods that have well-defined methods and analysis [65-68] and in some cases new entities unknown by light microscopic methods have emerged. The DNA microarray methods have been used to separate primary and secondary cancers in lung [69] and separate colonic from ovarian cancer origins [70] and to determine cancer of unknown primary sites [71]. The future of a needle core biopsy of suspicious mass may be (a) routine hematoxylin and eosin, immunohistochemistry including normalization and analysis, EBM and STARD (b) isolation of protein content for 2dimensional gel electrophoresis and western blotting, protein and antibody arrays and mass spectrometry (c) isolation of total messenger ribonucleic acids(mRNA), cDNA synthesis, microarray expression studies, single nucleotide polymorphism(SNP) and array comparative genomic hybridization (array CGH) and copy number variation (CNV) of genes (Table 1). The continued use and dependence on immunohistochemical staining in diagnostic surgical pathology will need the use of EBM and STARD methods and minimum datasets and integration of protein networks and function, and image analysis with normalization or definable cut-offs.
Table 1.
Summarized Comparison of Immunohistochemical Method and Liquid Chromatography-Mass Spectrometry (LC-MS) in Tissue Proteomics
| Method of Protein Detection | Advantages | Drawbacks |
|---|---|---|
| Immunohistochemistry | Protein location and distribution seen Detectable in small and large tissue biopsies and fixed tissues Validation of other highthroughput studies ( DNA microarray) | Limited ability to quantitate protein content Problems with antibody types, limited ability to detect protein modifications Limited or lack of Evidence based Criteria Single or dual detection ability Variable scoring methods and reproducibility No normalization methods Limited throughput Limited capacity for clinical biomarker profiling( only with tissue microarrays) |
| Other Proteomics (i.e LC-MS) | 100's to 1000's of peptides and proteins detected Can peruse databases for protein function and Gene ontology Robust Bioinformatics High throughput High level quantitation Can detect modified proteins Great potential for use in detecting clinical biomarkers | Cannot locate identified peptides to cell type(s) Need fresh or frozen tissue samples |
Acknowledgments
I am grateful to Epitomics, Inc (www.epitomics.com) for permission to use their figures.
References
- 1.Robinson G, Dawson I. Immunochemical studies of the endocrine cells of the gastrointestinal tract II An immunoperoxidase technique for the localization of secretin-containing cells in human duodenum. J Clin Patho. 1975;28:631–635. doi: 10.1136/jcp.28.8.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakane P, Pierce G. Enzyme-labeled antibodies; preparation and application for the localization of antigens. J Histochem and Cytochem. 1966;14:929–931. doi: 10.1177/14.12.929. [DOI] [PubMed] [Google Scholar]
- 3.Hanahan D, Weinberg R. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 4.Lewin B. GENES VIII. Upper Saddle River, NJ 07458: Pearson/Prentice Hall; 2004. Protein Synthesis; pp. 135–166. [Google Scholar]
- 5.Krappmann D, Scheidereit C. A pervasive role of ubiquitin conjugation in activation and termination of IkB kinase pathways. EMBO Reports. 2005;6(4):321–326. doi: 10.1038/sj.embor.7400380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Seet B, Dikic I, Zhou M-M, Pawson T. Reading protein modifications with interaction domains. Nat Rev Mol Cell Biol. 2006;7:473–483. doi: 10.1038/nrm1960. [DOI] [PubMed] [Google Scholar]
- 7.Wilson VG, Rosas-Acosta G. Wrestling with SUMO in a New Arena. Science STKE. 2005 doi: 10.1126/stke.2902005pe32. (pe32) [DOI] [PubMed] [Google Scholar]
- 8.Baek D, Villen J, Shin C, Camargo F, Gygi S, Bartel D. The impact of microRNAs on protein output. Nature. 2008 doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hunter T. The Age of Crosstalk: Phosphorylation, Ubiquitination, and Beyond. Molecular Cell. 2007;28:730–738. doi: 10.1016/j.molcel.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Peri J, Navarro J, Amanchy R, Kristiasen T, Jonnalagada C, Surendranath Vea. Development of Human Protein Reference Database as an Initial Platform for Approaching Systems Biology in Humans. Genome Res. 2003;13:2363–2371. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mandell J. Phosphorylation State-Specific Antibodies Applications in Investigative and Diagnostic Pathology. Am J Pathol. 2003;163:1687–1698. doi: 10.1016/S0002-9440(10)63525-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lim Y. Mining the Tumor Phosphoproteome for Cancer Markers. Clin Cancer Res. 2005;11(9):3163–3169. doi: 10.1158/1078-0432.CCR-04-2243. [DOI] [PubMed] [Google Scholar]
- 13.Baker A, Dragovich T, Ihle N, Williams R, Fenoglio-Preiser C, Powls G. Stability of Phosphoprotein as a Biological Marker of Tumor Signaling. Clin Cancer Res. 2005;11(12):4338–4340. doi: 10.1158/1078-0432.CCR-05-0422. [DOI] [PubMed] [Google Scholar]
- 14.Jin S, Levine A. The p53 functional circuit. Journal of Cell Science. 2001;114:4139–4140. doi: 10.1242/jcs.114.23.4139. [DOI] [PubMed] [Google Scholar]
- 15.Haupt S, Berger M, Goldberg Z, Haupt Y. Apoptosis- the p53 network. J Cell Sci. 2003;116:4077–4085. doi: 10.1242/jcs.00739. [DOI] [PubMed] [Google Scholar]
- 16.Wei C-L, Wu Q, Vega V, Chiu K, Ng P, Zhang T, et al. A Global Map of p53 Transcription-Factor Binding Sites in the Human Genome. Cell. 2006;124:207–219. doi: 10.1016/j.cell.2005.10.043. [DOI] [PubMed] [Google Scholar]
- 17.Bode A, Dong Z. Post-translational modification of p53 in tumorigenesis. Nat Rev Cancer. 2004;4:793–805. doi: 10.1038/nrc1455. [DOI] [PubMed] [Google Scholar]
- 18.Bourdon J-C, Fernandes K, Murray-Zmijeweski F, Liu G, Diot A, Xirodimas D, et al. p53 isoforms can regulate p53 transcriptional activity. Genes Dev. 2005;19:2122–2137. doi: 10.1101/gad.1339905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vousden K, Prives C. P53 and Prognosis: New Insights and Further Complexity. Cell. 2005;120:7–10. doi: 10.1016/j.cell.2004.12.027. [DOI] [PubMed] [Google Scholar]
- 20.Schmitt C, Fridman J, Yang M, Baranov E, Hoffman R. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1:289–298. doi: 10.1016/s1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 21.Trigiante G, Lu X. ASPPs and cancer. Nat Rev Cancer. 2006;6:217–226. doi: 10.1038/nrc1818. [DOI] [PubMed] [Google Scholar]
- 22.Petricoin E, Zoon K, Kohn E, Liotta L. Clinical Proteomics: Translating Benchside Promise Into Bedside Reality. Nat Rev Drug Discov. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 23.Celis J, Gromov P, Gromova I, Moreira J, Cabezon T, Ambartsumian Nea. Integrating Proteomic and Functional Genomic Technologies in Discovery-driven Translational Breast Cancer Research. Mol Cell Proteomics. 2003;2:369–377. doi: 10.1074/mcp.R300007-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Graham D, Elliott S, Van Eyk J. Broad -based proteomic strategies: a practical guide to proteomics and functional screening. J Physiol. 2004;563(1):1–9. doi: 10.1113/jphysiol.2004.080341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sanchez-Carbayo M, Socci N, Lozano J, Haab B, Cordon-Cardo C. Profiling Bladder Cancer Using Targeted Antibody Microarrays. Am J Pathol. 2006;168:93–103. doi: 10.2353/ajpath.2006.050601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ludwig J, Weinstein J. Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat Rev Cancer. 2005;5:845–856. doi: 10.1038/nrc1739. [DOI] [PubMed] [Google Scholar]
- 27.Dalton W, Friend S. Cancer Biomarkers- An Invitation to the Table. Science. 2006;312:1165–1168. doi: 10.1126/science.1125948. [DOI] [PubMed] [Google Scholar]
- 28.Natkunam Y, Mason D. Prognostic immunohistologic markers in human tumors: why are so few used in clinical practice? Lab Invest. 2006;86:742–747. doi: 10.1038/labinvest.3700447. [DOI] [PubMed] [Google Scholar]
- 29.Weil R. Incorporating Molecular Tools into Early- Stage Clinical Trials. PLos Med. 2008;5:e21. doi: 10.1371/journal.pmed.0050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hawkins R. The Evidence Based Medicine Approach to Diagnostic Testing: practicalities and limitations. Clinical Biochem Review. 2005:7–18. 26(May) [PMC free article] [PubMed] [Google Scholar]
- 31.McQueen M. Overview of Evidence -based Medicine: Challenges for Evidence-based Laboratory Medicine. Clin Chem. 2001;47(8):1536–1546. [PubMed] [Google Scholar]
- 32.Mallet S, Deeks J, Halligan S, Hopewell S, Cornelius V, Altman D. Systematic reviews of diagnostic tests in cancer: review of methods and reporting. BMJ. 2006;333:413. doi: 10.1136/bmj.38895.467130.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ordonez N. The diagnostic utility of immunohistochemistry in distinguishing between epitheloid mesotheliomas and squamous cell carcinomas of the lung: a comparative study. Mod Pathol. 2006;19:417–429. doi: 10.1038/modpathol.3800544. [DOI] [PubMed] [Google Scholar]
- 34.Evangelou E, Kyzas P, Trikalinos T. Comparison of the diagnostic accuracy of lymphatic endothelium markers: Bayesian approach. Mod Pathol. 2005;18(11):1490–1497. doi: 10.1038/modpathol.3800457. [DOI] [PubMed] [Google Scholar]
- 35.Dennis J, Hvidsen T, Wit E, Komorowski J, Bell A, Downie I, et al. Markers of Adenocarcinoma Characteristic of the site of OrigDevelopment of a Diagnostic Algorithm. Clin Cancer Res. 2005;11(10):3766–3772. doi: 10.1158/1078-0432.CCR-04-2236. [DOI] [PubMed] [Google Scholar]
- 36.Bast R, Lilja H, Urban N, Rimm D, Fritsche H, Gray J, et al. Translational Crossroads for Biomarkers. Clin Cancer Res. 2005;11(17):6103–6108. doi: 10.1158/1078-0432.CCR-04-2213. [DOI] [PubMed] [Google Scholar]
- 37.Rimm D. What brown cannot do for you. Nat Biotechnol. 2006;24(8):914–916. doi: 10.1038/nbt0806-914. [DOI] [PubMed] [Google Scholar]
- 38.Cheang M, Treaba D, Speers C, Olivoto I, Badjik C, Chia S, et al. Immunohistochemical Detection Using the New Rabbit Monoclonal Antibody SP1 of Estrogen Receptor in Breast Cancer Is Superior to Mouse Monoclonal Antibody 1D5 in Predicting Survival. J Clin Oncol. 2006;24(36):5637–5644. doi: 10.1200/JCO.2005.05.4155. [DOI] [PubMed] [Google Scholar]
- 39.True L, Feng Z. Immunohistochemical Validation of Expression Microarray Results. J Mol Diagn. 2005;7(2):149–151. doi: 10.1016/S1525-1578(10)60540-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McCabe A, Dolled-Filhart M, Camp R, Rimm D. Automated Quantitative Analysis (AQUA) of In situ Protein Expression, Antibody Concentration, and Prognosis. J Natl Cancer Inst. 2005;97:1808–1815. doi: 10.1093/jnci/dji427. [DOI] [PubMed] [Google Scholar]
- 41.Betensky R, Nutt C, Batchelor T, Louis D. Statistical Considerations for Immunohistochemical Panel Development after Gene Expression Profiling of Human Cancers. J Mol Diagn. 2005;7(2):276–282. doi: 10.1016/S1525-1578(10)60555-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones R, Gordus A, Krail J, MacBeath G. A quantitative protein interaction network for the ErbB receptors using protein microarrays. Nature. 2006;439:168–174. doi: 10.1038/nature04177. [DOI] [PubMed] [Google Scholar]
- 43.Norris A, Kahn C. Analysis of gene expression in pathophysiological states: Balancing false discovery and false negative rates. Proc Natl Acad Sci U S A. 2006;103:649–653. doi: 10.1073/pnas.0510115103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simon R. Development and Validation of Therapeutically Relevant Multi-Gene Biomarker Classifiers. JNCI. 2005;97(12):866–867. doi: 10.1093/jnci/dji168. [DOI] [PubMed] [Google Scholar]
- 45.Simon R, Radmacher M, Dobbin K, McShane L. Pitfalls in the Use of DNA Microarray Data for Diagnostic and Prognostic Classification. JNCI. 2003;95(1):14–18. doi: 10.1093/jnci/95.1.14. [DOI] [PubMed] [Google Scholar]
- 46.Tinker A, Boussioutas A, Bowtell D. The challenge of gene expression microarrays for the study of human cancer. Cancer Cell. 2006;9(5):333–339. doi: 10.1016/j.ccr.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 47.Storey J, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.de Kok J, Roelofs R, Giesendorf B, Pennings J, Waas E, Feuth T, et al. Normalization of gene expression measurements in tumor tissues: comparison of 13 endogenous control genes. Lab Invest. 2005;85:154–159. doi: 10.1038/labinvest.3700208. [DOI] [PubMed] [Google Scholar]
- 49.Camp R, Dolled-Filhart M, Rimm D. X-Tile: A New Bio-Informatics Tool for Biomarker Assessment and Outcome-Based Cut-Point Optimization. Clin Cancer Res. 2004;10:7252–7259. doi: 10.1158/1078-0432.CCR-04-0713. (Nov1) [DOI] [PubMed] [Google Scholar]
- 50.Dowsett M, Hanna W, Kockx M, Pennault-Llorca F, Ruschoff J, Gutjahr T, et al. Standardization of HER2 testing: results of an international proficiency testing ring study. Mod Pathol. 2007;20:584–591. doi: 10.1038/modpathol.3800774. [DOI] [PubMed] [Google Scholar]
- 51.Matkowskyj K, Cox R, Jensen R, Benya R. Quantitative Immunohistochemistry by Measuring Cumulative Signal Strength Accurately Measures Receptor Number. J Histochem Cytochem. 2003;51(2):205–214. doi: 10.1177/002215540305100209. [DOI] [PubMed] [Google Scholar]
- 52.Hu J-J, Ambrus A, Fossum T, Miller M, Humphrey J, Wilson E. Time Courses of Growth and Remodeling of Porcine Aortic Media During Hypertension: A Quantitative Immunohistochemical Examination. J Histochem Cytochem. 2008;56(4):359–370. doi: 10.1369/jhc.7A7324.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wester K, Wahlund E, Sundstrom C, Ranefall P, bengtsson E, Russell P, et al. Parrafin Section Storage and Immunohistochemistry. Appl Immunohistochem Mol Morphol. 2000;8(1):61–70. [PubMed] [Google Scholar]
- 54.Halushka M, Cornish T, Lu J, Selvin S, Selvin E. Creation, validation, and quantitative analysis of protein expression in vascular tissue microarrays. Cardiovasc Pathol. 2009 doi: 10.1016/j.carpath.2008.12.007. doi: 10.1016/j.carpath.2008.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Halushka M, Selvin E, Macgregor A, Cornish T. The use of vascular tissue microarrays for measurement of advanced glycation end products. J Histochem Cytochem. 2009 doi: 10.1369/jhc.2009.953273. DOI: 10.1369/jhc.2009.953273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cregger M, Berger A, Rimm D. Immunohistochemistry and Quantitative Analysis of Protein Expression. Arch Pathol Lab Med. 2006;130:1026–1030. doi: 10.5858/2006-130-1026-IAQAOP. [DOI] [PubMed] [Google Scholar]
- 57.Paizis M, Engelhardt J, Siklos L. Quantitative assessment of relative changes of immunohistochemical staining by light microscopy in specified anatomical regions. J Microsc. 2009;234(1):103–112. doi: 10.1111/j.1365-2818.2009.03146.x. [DOI] [PubMed] [Google Scholar]
- 58.Kayser K, Gorter J, Metze K, Goldmann T, Vollmer E, Mireskandari M, et al. How to measure image quality in tissue-based diagnosis (diagnostic surgical pathology) BMC Diagn Pathol. 2008;3(Supl 1):S11. doi: 10.1186/1746-1596-3-S1-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van der Loos C. Multiple Immunoenzyme Staining: Methods and Visualizations for the Observation With Spectral Imaging. J Histochem Cytochem. 2008;56(4):313–328. doi: 10.1369/jhc.2007.950170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ruifrok A, Johnson D. Quantification of histochemical staining by color deconvolution. Anal Quant Cytol Histol. 2001;23:291–299. [PubMed] [Google Scholar]
- 61.Ruifrok A, Katz R, Johnson D. Comparison of Quantification of Histochemical Staining By Hue-Saturation-Intensity (HSI) Transformation and Color-Deconvolution. Appl Immunohistochem Mol Morphol. 2003;11(1):85–91. doi: 10.1097/00129039-200303000-00014. [DOI] [PubMed] [Google Scholar]
- 62.Brey E, Lalani Z, Johnston C, Wong M, McIntire L, Duke P, et al. Automated Selection of DABlabeled Tissue for Immunohistochemical Quantification. J Histochem Cytochem. 2003;51(5):575–584. doi: 10.1177/002215540305100503. [DOI] [PubMed] [Google Scholar]
- 63.Pham N-A, Morrison A, Schwock J, Aviel-Ronen S, Iakolev V, Tsao M-S, et al. Quantitative image analysis of immunohistochemical stains using a CMYK color model. BMC Diagn Pathol. 2007;2:8. doi: 10.1186/1746-1596-2-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Charpin C, Secq V, Giusiano S, Carpenter S, Andrac L, Lavaut M-N, et al. A signature predictive of disease outcome in breast carcinomas, identified by quantitative immunocytochemical assays. Int J Cancer. 2009;124:2124–2134. doi: 10.1002/ijc.24177. [DOI] [PubMed] [Google Scholar]
- 65.Su A, Welsh J, Sapinoso L, Kern S, Dimitrov P, Lapp H, et al. Molecular classification of Human Carcinomas by Use of Gene Expression Signatures. Cancer Res. 2001;61:7388–7393. [PubMed] [Google Scholar]
- 66.Glinsky GV, Higashiyama T, Glinski AB. Classification of Human Breast Cancer Using Gene Expression Profiling as a Component of the Survival Predictor Algorithm. Clin Cancer Res. 2004;10:2272–2283. doi: 10.1158/1078-0432.ccr-03-0522. [DOI] [PubMed] [Google Scholar]
- 67.Rhodes D, Chinnayan A. Integrative analysis of the cancer transcriptome. Nat Genet Supplement. 2005;37:S31–37. doi: 10.1038/ng1570. [DOI] [PubMed] [Google Scholar]
- 68.Pinkel D, Albertson D. Array comparative genomic hybridization and its applications in cancer. Nature Genetics Supplement. 2005;37:S11–17. doi: 10.1038/ng1569. [DOI] [PubMed] [Google Scholar]
- 69.Talbot S, Estilo C, Maghami E, Sarkaria I, Pham D, O-Charoenrat P, et al. Gene Expression Profiling Allows Distinction between Primary and Metastatic Squamous Cell Carcinomas in the Lung. Cancer Research. 2005;65(8):3063–3071. doi: 10.1158/0008-5472.CAN-04-1985. [DOI] [PubMed] [Google Scholar]
- 70.Nishizuka S, Chen S-T, Gwadry F, Alexander J, Major S, Scher U, et al. Diagnostic Markers That Distinguish Colon and Ovarian Adenocarcinomas: Identification by Genomic, Proteomic, and Tissue Array Profiling. Cancer Research. 2003;63:5243–5250. (Sep 1) [PubMed] [Google Scholar]
- 71.Veradhachary G, Talantov D, Raber M, Meng C, Hess K, Jatkoe T, et al. Molecular Profiling of Carcinoma of Unknown Primary and Correlation with Clinical Evaluation. J Clin Oncology. 2008;26(27):4442–4448. doi: 10.1200/JCO.2007.14.4378. [DOI] [PubMed] [Google Scholar]