Abstract
The enzyme sirtuin 1 (SIRT1) is a critical regulator of many cellular functions, including energy metabolism. However, the precise mechanisms that modulate SIRT1 activity remain unknown. As SIRT1 activity in vitro was recently found to be negatively regulated by interaction with the deleted in breast cancer–1 (DBC1) protein, we set out to investigate whether DBC1 regulates SIRT1 activity in vivo. We found that DBC1 and SIRT1 colocalized and interacted, and that DBC1 modulated SIRT1 activity, in multiple cell lines and tissues. In mouse liver, increased SIRT1 activity, concomitant with decreased DBC1-SIRT1 interaction, was detected after 24 hours of starvation, whereas decreased SIRT1 activity and increased interaction with DBC1 was observed with high-fat diet (HFD) feeding. Consistent with the hypothesis that DBC1 is crucial for HFD-induced inhibition of SIRT1 and for the development of experimental liver steatosis, genetic deletion of Dbc1 in mice led to increased SIRT1 activity in several tissues, including liver. Furthermore, DBC1-deficient mice were protected from HFD-induced liver steatosis and inflammation, despite the development of obesity. These observations define what we believe to be a new role for DBC1 as an in vivo regulator of SIRT1 activity and liver steatosis. We therefore propose that the DBC1-SIRT1 interaction may serve as a new target for therapies aimed at nonalcoholic liver steatosis.
Introduction
Sirtuin 1 (SIRT1) protein, the mammalian homolog for yeast silent information regulator 2 (SIR2), is a NAD+-dependent deacetylase that has emerged as a key regulator of energy metabolism (1). SIRT1 and its orthologs have been implicated in the regulation of longevity in different organisms, including yeast, Caenorhabditis elegans, Drosophila melanogaster, and mice (2, 3). In mammals, SIRT1 activation promotes improvement in diseases of aging and in features of metabolic syndrome, including the development of liver steatosis (4). Moreover, liver SIRT1 activity is decreased during experimental liver steatosis (5), and SIRT1 activation prevents the development of both nonalcoholic and alcoholic-induced liver steatosis in mice (4–7).
SIRT1 deacetylates and regulates multiple substrates, including PGC-1α (8), p53 (9), forkhead transcription factors (10), NF-κB (11), Ku70 (12), MyoD (13), and histones (14, 15). Furthermore, SIRT1 activation influences gene silencing, apoptosis, stress resistance, senescence, circadian cycles, and fat and glucose metabolism (3, 8, 16–26). Although the cellular functions of SIRT1 have been extensively investigated, less is known about the mechanisms that regulate SIRT1 activity.
SIRT1 activity can be modulated by the metabolic state of the organism, with an increase in activity during starvation and a decrease with high-caloric diets (3, 4, 19, 20, 22, 25, 27). However, how changes in SIRT1 activity are achieved during different metabolic conditions remains unclear. Several studies suggest that SIRT1 may be regulated at the transcriptional level by FOXO3a (28), p53 (28), HIC1 (29), or E2F1 (30). SIRT1 was also shown to be regulated posttranscriptionally by mRNA stabilization (31), and posttranslationally by SUMOylation (32) and phosphorylation (33). Finally, SIRT1 activity may also be influenced by changes in cellular NAD+ levels (8, 34), which are controlled by the enzymes nicotinamide phosphoribosyltransferase (NAMPT) and CD38 (35–37). Recently, we and others have demonstrated that SIRT1 activity is also modulated by protein-protein interaction (38–40) through the association of SIRT1 with the deleted in breast cancer–1 (DBC1) protein.
DBC1, localized in the cell nucleus, was initially described as being absent in certain human breast cancers. DBC1 binds directly to the catalytic domain of SIRT1, preventing substrate binding to SIRT1 and inhibiting SIRT1 activity (39, 40). However, how the DBC1-SIRT1 interaction is modulated and whether DBC1 is a regulator of SIRT1 at the physiological level in animals has not yet been determined. In the present study, we investigated the role of DBC1 as a regulator of SIRT1 during different metabolic conditions. We showed that DBC1-SIRT1 interaction was regulated during high-fat diet (HFD) feeding and during starvation, which suggests that changes in SIRT1 activity observed during different metabolic conditions could be explained by changes in DBC1-SIRT1 association. Furthermore, using our recently developed Dbc1 KO mouse model, we investigated the role of DBC1 in hepatic metabolism and the development of experimental liver steatosis induced by HFD. We found that Dbc1 KO led to both activation of hepatic SIRT1 and protection against the development of experimental liver steatosis and inflammation. Furthermore, we observed that the beneficial effect of deletion of DBC1 on the development of cellular lipid accumulation was mediated by a SIRT1-dependent mechanism. These data indicate that disruption of the DBC1-SIRT interaction may serve as a new target for the development of therapies against liver steatosis and other components of the metabolic syndrome.
Results
SIRT1 protein and NAD+ levels do not change in the liver during starvation and HFD.
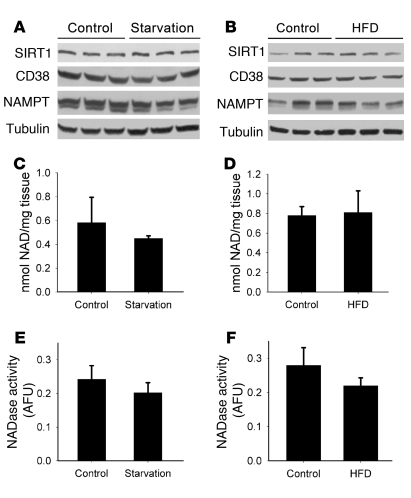
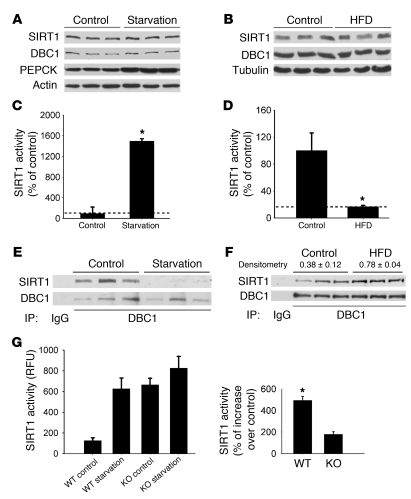
Several researchers have proposed that changes in SIRT1 activity observed during different metabolic conditions are primarily regulated by its own expression or by changes in NAD+ concentration inside the cell (8, 16, 34, 41). However, at least in the liver, the published data regarding these changes are contradictory (8, 27, 34). We therefore analyzed the levels of these molecules in mouse liver under different metabolic conditions. Levels of SIRT1 protein (Figure 1, A and B) and NAD+ did not vary either after 24 hours of starvation (Figure 1C, P = 0.37) or after 4 weeks of HFD (Figure 1D, P = 0.42). To further characterize NAD+ metabolism in our samples, we analyzed the expression level of 2 of the main enzymes involved in the synthesis and degradation of NAD+ in cells, namely, NAMPT and CD38 (35–37, 42, 43). Similar to our observations for NAD+, CD38 and NAMPT protein levels remained unchanged under starvation and after 4 weeks of HFD (Figure 1, A and B). Furthermore, hepatic CD38 (NADase) activity did not vary between these 2 conditions (starvation, P = 0.32; HFD, P = 0.37; Figure 1, E and F). These results demonstrate that under our experimental conditions, levels of NAD+, SIRT1, and the 2 main NAD+ metabolizing enzymes, NAMPT and CD38, remained unchanged under different metabolic conditions. These findings suggest that although NAD+ is essential for SIRT1 activity, it may not be the only factor that controls SIRT1 activity in the liver.
Figure 1. SIRT1 protein levels and NAD+ do not change in the mouse liver in response to starvation or HFD.
Mice were put under 24 hours of starvation (A, C, and E) or fed HFD for 4 weeks (B, D, and F). Control groups consisted of mice fed a normal chow diet ad libitum (n = 3). (A and B) Western blots were performed for liver SIRT1, CD38, NAMPT, and tubulin. Each lane represents 1 different mouse liver. (C and D) Total NAD+ was isolated and measured from liver extracts after starvation (P = 0.37) and HFD (P = 0.42). (E and F) CD38 NADase activity was measured from liver extracts from starved (P = 0.32) and HFD-fed mice (P = 0.37). All experiments were performed with at least 6 mice in each group. Data are mean ± SD.
DBC1 expression regulates SIRT1 activity.
Recently, the protein DBC1 was found to bind to and inhibit SIRT1 activity (39, 44). To further examine the role of DBC1 in SIRT1 activity, we investigated whether DBC1 regulates SIRT1 in vivo and whether the interaction between these 2 proteins is regulated by different metabolic conditions.
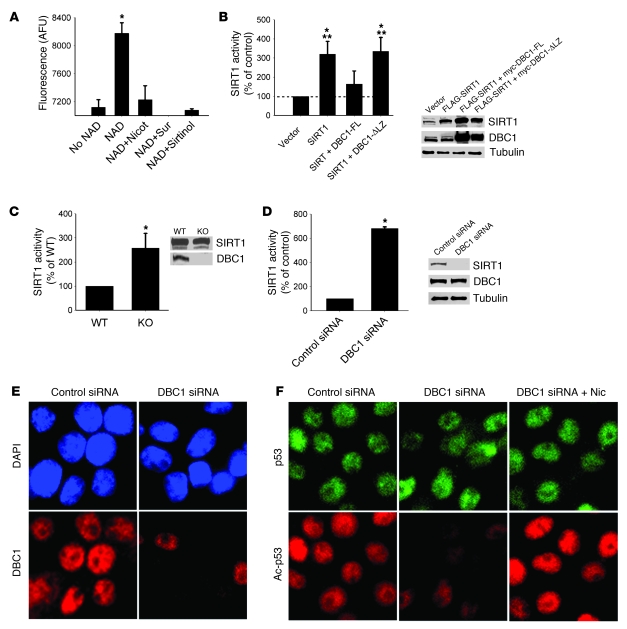
When SIRT1 was overexpressed in 293T cells, we observed a robust increase in SIRT1 protein levels that correlated with an increase in NAD+-dependent deacetylase activity compared with nontransfected cells (Figure 2B). In contrast, when SIRT1 was coexpressed with DBC1, SIRT1 activity decreased (Figure 2B), which confirmed that DBC1 inhibits SIRT1 deacetylase activity. SIRT1 activity was measured by a specific fluorometric assay extensively characterized in our laboratory (42).
Figure 2. DBC1 regulates NAD-dependent SIRT1 activity.
(A) Isolated rat liver nuclei extracts were incubated in the absence or presence of 200 μM NAD+, or with NAD+ plus nicotinamide (2 mM), suramin (100 μM), or sirtinol (100 μM), and SIRT1 activity was measured. (B) Flag-SIRT1, full-length DBC1 (myc-DBC1-FL), and DBC1 with the leucine zipper deleted (myc-DBC1-ΔLZ) were overexpressed in 293T cells. At 24 hours after transfection, cell lysates were obtained, and SIRT1 activity was measured. NAD-dependent SIRT1 activity was expressed as a percentage of control (empty vector) activity. Protein overexpression was confirmed by Western blot. *P < 0.05 versus control; **P < 0.05 versus myc-DBC1-ΔLZ, t test. (C) MEFs derived from WT or Dbc1 KO mice were analyzed for NAD-dependent SIRT1 activity. Expression levels of SIRT1 and DBC1 were analyzed by Western blot. (D) DBC1 was knocked down in INS cells by siRNA transfection. SIRT1 activity was measured from cell lysates 72 hours after the first transfection. DBC1 knockdown was confirmed by Western blot. (E) INS cells were transfected with DBC1 siRNA, fixed after 72 hours, and stained for DBC1. Nuclei were stained with DAPI. Original magnification, ×600. (F) Endogenous p53 acetylation was analyzed by immunofluorescence after DBC1 siRNA transfection. Cells were treated with control siRNA, DBC1 siRNA, or DBC1 siRNA plus 5 mM nicotinamide (added 16 hours before fixation). Original magnification, ×600. Data are mean ± SD (n = 3). *P < 0.05, t test.
Kim et al. have previously shown that SIRT1 and DBC1 interact directly and that this interaction is dependent on the leucine zipper (LZ) domain present on DBC1 (39). When we coexpressed SIRT1 and a mutant form of DBC1 with a deletion of the LZ domain, we found no inhibition of SIRT1 activity (Figure 2B), which confirmed that the DBC1 effect on SIRT1 activity is dependent on DBC1-SIRT1 interaction mediated by the DBC1 LZ domain.
Next, we examined the effect of DBC1 downregulation on endogenous SIRT1 deacetylase activity. Mouse embryonic fibroblasts (MEFs) from WT and Dbc1 KO mice were analyzed for SIRT1 deacetylase activity. DBC1 null MEFs showed a more than 2-fold increase in SIRT1 deacetylase activity, although SIRT1 protein levels remained unchanged (Figure 2C). To confirm these observations, we transfected pancreatic INS cells with DBC1 siRNA. Under these conditions, DBC1 protein expression was undetectable, and endogenous SIRT1-dependent deacetylase activity increased compared with cells transfected with control siRNA (Figure 2D). To further confirm that DBC1 regulates endogenous SIRT1 activity, we determined the levels of acetylation of endogenous p53 in INS cells transfected with DBC1 siRNA. Downregulation of DBC1 protein expression (Figure 2E) decreased endogenous p53 acetylation (Figure 2F), and this decrease was prevented by treating the cells with the SIRT1 inhibitor nicotinamide (Figure 2F). These results show that the decrease in acetylation levels observed were indeed SIRT1 dependent and clearly demonstrate that DBC1 regulates SIRT1 activity in multiple cell types.
DBC1 regulates glucose-dependent SIRT1 activity in human hepatocytes.
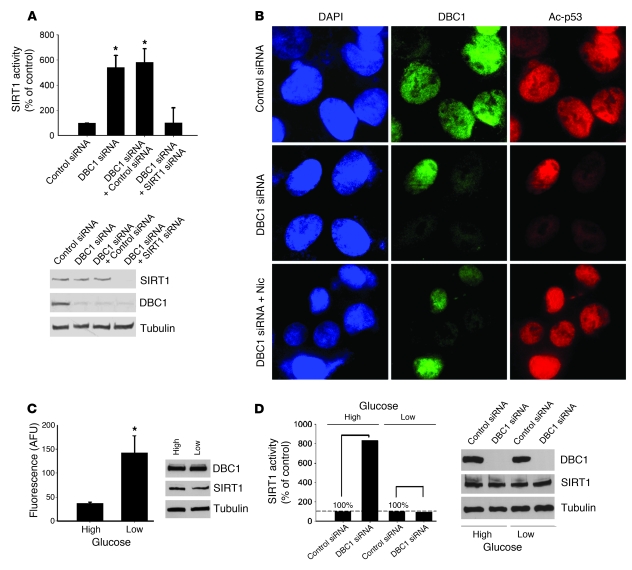
Hou et al. previously proposed that glucose levels regulate SIRT1 activity in the hepatocyte-derived cell line HEPG2 (24). Therefore, we investigated the possible role of DBC1 in the regulation of SIRT1 activity by glucose. We first examined whether DBC1 regulates SIRT1 activity in HEPG2 cells by transfecting cells with a control and a DBC1-specific siRNA. We found that SIRT1 activity increased in cells treated with DBC1 siRNA (Figure 3A). This increase in activity was prevented by cotransfection of DBC1 and SIRT1 siRNA (Figure 3A), demonstrating that DBC1 regulates SIRT1 activity in these cells. The same result was observed when we measured endogenous p53 acetylation as a surrogate of cellular SIRT1 activity. Cells transfected with DBC1 siRNA showed a decrease in the acetylation of endogenous p53 protein (Figure 3B), which indicates that endogenous SIRT1 activity increased. The decrease in p53 acetylation was blocked by treating the DBC1 siRNA–transfected cells with nicotinamide. We then tested the role of DBC1 in the regulation of SIRT1 activity by glucose. Cells that were incubated with low glucose (5 mM) for 24 hours showed higher (3-fold) SIRT1 activity than did cells incubated for 24 hours with high glucose (30 mM; Figure 3C). Treatments with the different glucose concentrations did not affect SIRT1 or DBC1 protein levels (Figure 3C). In cells treated with high glucose, SIRT1 activity was higher in DBC1 siRNA–treated cells than in control siRNA–treated cells (Figure 3D), which suggests that the low SIRT1 activity observed under high-glucose conditions could be explained by a DBC1-mediated mechanism. In contrast, DBC1 siRNA had no effect on SIRT1 activity in cells incubated in low glucose (Figure 3D). To further demonstrate that changes in SIRT1 activity resulted from DBC1 siRNA treatment, we normalized the data to their respective high- or low-glucose controls. The actual values for SIRT1 activity in the control condition (nonspecific siRNA) corresponded to 73 arbitrary fluorescence units (AFUs) and 154 AFUs for cells in high and low glucose, respectively. This result suggests that under low-glucose conditions, DBC1 was not bound to SIRT1, which increased SIRT1 activity. We propose that DBC1 is a regulator of SIRT1 activity during different metabolic conditions.
Figure 3. DBC1 controls glucose-dependent SIRT1 activity in human hepatocytes.
(A) HEPG2 cells were transfected with DBC1 siRNA, DBC1 siRNA plus control siRNA, and DBC1 siRNA plus SIRT1 siRNA. NAD-dependent SIRT1 activity was measured, and expressed as a percentage of control (nonspecific siRNA). The efficiency of transfection was confirmed by Western blot. (B) HEPG2 cells were transfected with DBC1 siRNA and DBC1; 5 mM nicotinamide was added 16 hours before fixation. Endogenous p53 acetylation levels were studied by immunofluorescence. Original magnification, ×600. (C) HEPG2 cells were cultured in serum-free media for 24 hours and then incubated for 24 hours in low (5 mM) or high (30 mM) glucose. SIRT1 activity was expressed in AFUs. n = 3. *P < 0.05, t test. (D) SIRT1 activity in HEPG2 cells transfected with DBC1 siRNA and then treated with high and low glucose as described in C. SIRT1 activity measured with DBC1 siRNA was normalized to the respective controls (nonspecific siRNA transfection) with high and low glucose. Data are shown as percent increase in SIRT1 activity over the respective controls (with high or low glucose, set as 100%). The fluorescence values in the control siRNA condition corresponded to 73 and 154 AFUs for cells in high and low glucose, respectively. Shown is 1 representative experiment of 3. The siRNA transfection efficiency was confirmed by Western blot. Except where indicated, data are mean ± SD.
Dbc1 KO mice show increased endogenous SIRT1 activity in multiple tissues.
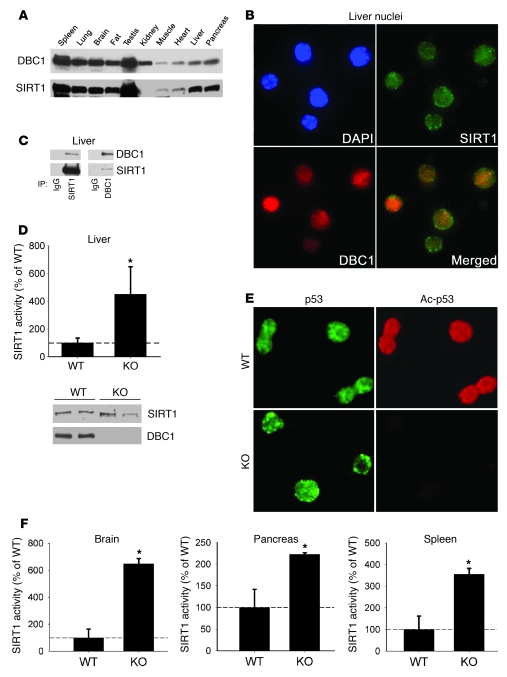
The evidence that SIRT1 activity in cells is tightly regulated by DBC1 prompted us to explore whether this regulation also occurs in mouse tissues. Tissue distribution analysis showed a correlation between expression levels of both proteins in mouse tissues (Figure 4A). Both proteins were highly expressed in spleen, lung, brain, fat, testis, liver, and pancreas. We also found that, in the liver, these proteins colocalized within the nucleus (Figure 4B) and coimmunoprecipitated in nuclear extracts (Figure 4C), which confirmed that DBC1 and SIRT1 interact in mouse liver.
Figure 4. SIRT1 and DBC1 interact in mouse tissue, and Dbc1 KO mice show increased endogenous SIRT1 activity in vivo.
(A) Western blots showing the expression of SIRT1 and DBC1 in different mouse tissues. (B) Isolated mouse liver nuclei immunofluorescence for SIRT1 and DBC1, showing that both proteins colocalized inside the nucleus. Original magnification, ×400. (C) Coimmunoprecipitation of SIRT1 and DBC1 from isolated nuclear extracts from liver. Shown is DBC1-SIRT1 coimmunoprecipitation using SIRT1 (left) and DBC1 (right) antibodies. Normal rabbit IgG was used as control for the immunoprecipitation. (D) Endogenous NAD-dependent SIRT1 activity was measured in isolated nuclear extracts from the livers of WT and Dbc1 KO mice. SIRT1 activity is expressed as a percentage of WT activity. Western blots of SIRT1 and DBC1 from these nuclear fractions served as control for protein levels. (E) Endogenous p53 acetylation was analyzed in isolated liver nuclei. Total p53 antibody was used as control. Original magnification, ×400. (F) Endogenous NAD-dependent SIRT1 activity was measured in nuclear extracts from brain, pancreas, and spleen of WT and Dbc1 KO mice. SIRT1 activity is expressed as a percentage of WT activity. Data are mean ± SD. *P < 0.05, t test.
We used Dbc1 KO mice to further investigate the role of DBC1 in the regulation of SIRT1 in vivo. We confirmed that DBC1 protein was completely absent in livers from Dbc1 KO mice (Figure 4D). Compared with WT mice, Dbc1 KO mice showed a 4-fold increase in endogenous SIRT1 activity in the liver (Figure 4D). In addition, the Dbc1 KO mice showed decreased levels of p53 acetylation (Figure 4E), although the levels of total p53 were similar. Taken together, these results show that SIRT1 activity is regulated by endogenous DBC1 in the liver. To further explore the role of DBC1 as a regulator of SIRT1, we measured SIRT1 activity from other tissues from WT and Dbc1 KO mice, including pancreas, brain, and spleen. In all of these tissues, SIRT1 activity was significantly higher in samples from Dbc1 KO mice than in samples from WT mice, although the levels of activation varied among the different tissues (Figure 4F). We propose that DBC1 is a general regulator of SIRT1 activity in mouse tissues.
Regulation of DBC1-SIRT1 interaction and SIRT1 activity in liver under different metabolic conditions.
We tested whether DBC1 regulates SIRT1 activity in the liver under different metabolic loads. Under normal feeding conditions, we observed that at least 50% of total liver SIRT1 seems to be bound to DBC1, as assessed by coimmunoprecipitation studies (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI39319DS1). We observed no change in SIRT1 or DBC1 protein expression after 24 hours of starvation (Figure 5A). However, a significant increase in SIRT1 activity, correlating with a decrease in the interaction between SIRT1 and DBC1 after starvation, was observed (Figure 5, C and E). In fact, the interaction between these proteins was nearly absent after starvation. Furthermore, when Dbc1 KO mice were starved for 24 hours, there was a small, statistically nonsignificant (P = 0.21) increase in SIRT1 activity, whereas WT mice showed a 5-fold increase in activity (Figure 5G).
Figure 5. SIRT1 activity and DBC1-SIRT1 interaction correlate under different metabolic conditions in the liver.
(A–F) Mice were starved for 24 hours (A, C, and E) or placed on a HFD for 4 weeks (B, D, and F). Control mice were fed a normal chow diet. (A and B) Western blot analysis showed that SIRT1 and DBC1 protein levels did not change in the liver after starvation or HFD. Each lane represents a different mouse liver. (C and D) SIRT1 activity was measured from liver nuclear extracts. Data were normalized to the activity measured in the normal chow diet (n = 6). (E and F) Coimmunoprecipitation from liver nuclear extracts using DBC1 antibody. Normal rabbit IgG was used as control for the immunoprecipitation (n = 3). DBC1 and SIRT1 band intensities were calculated by densitometry as described in Methods. The ratio between SIRT1 and DBC1 values was used to calculate changes in interaction. (G) WT and Dbc1 KO mice were put on starvation for 24 hours, and liver SIRT1 activity was measured from nuclear extracts. Shown is 1 representative experiment of 3. SIRT1 activity in WT and Dbc1 KO mice after starvation was also normalized to that of the respective fed states, shown at right as percent increase over control (set as 100%). Data are mean ± SD. *P < 0.05, t test.
In contrast, we observed a decrease in liver SIRT1 activity after 4 weeks of HFD compared with mice fed a normal chow diet (Figure 5D). Both SIRT1 and DBC1 levels remained unchanged in the liver after this treatment (Figure 5B). Consistent with the results described above, this decrease in SIRT1 activity observed after 4 weeks of HFD correlated with an increase in DBC1-SIRT1 association (Figure 5F). These data suggest that DBC1 modulates SIRT1 activity during different metabolic conditions in mouse liver.
Role of DBC1 in the development of experimental liver steatosis.
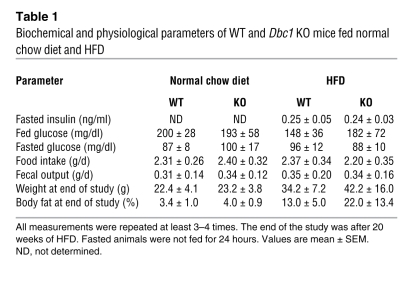
Adult WT and Dbc1 KO mice maintained on normal chow diet (4% total calories derived from fat, 3.04 kcal/g) did not differ in body weight, fat content, food intake, fecal output, or glucose tolerance (Table 1 and Supplemental Figure 2). Furthermore, there were no statistically significant differences in energy expenditure (EE) between WT and Dbc1 KO mice, although EE and activity tended to be higher in the Dbc1 KO mice (Supplemental Figure 3). All other biochemical parameters, including insulin, leptin, adiponectin, and plasma glucose, did not differ between the WT and Dbc1 KO mice fed normal chow (Table 1 and data not shown).
Table 1 .
Biochemical and physiological parameters of WT and Dbc1 KO mice fed normal chow diet and HFD
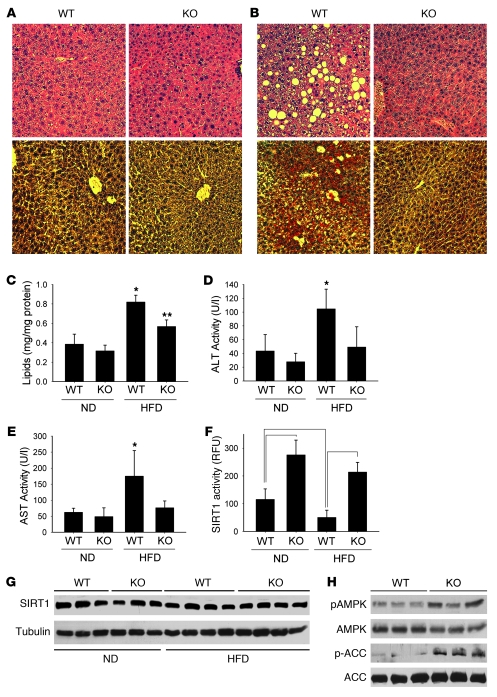
To further evaluate the role of DBC1 in metabolism and to determine whether DBC1 has a role in the development of experimental liver steatosis, we challenged mice with a high-calorie HFD (60% calories from fat, 5.05 kcal/g). When Dbc1 KO mice were fed a HFD, weight accumulation was nearly the same as — or higher than — that in WT mice (Table 1). In contrast, there was a substantial beneficial effect in Dbc1 KO mice for the development of liver steatosis after 20 weeks of HFD. After that period, WT mice presented clear signs of liver steatosis, with high levels of lipid deposition (white and red spots in H&E and Oil red O staining, respectively; Figure 6, A and B). In contrast, Dbc1 KO mice showed no signs of liver steatosis after 20 weeks of HFD (Figure 6, A and B). Quantification of lipid content in liver revealed no significant differences between WT and Dbc1 KO mice fed a normal diet; however, after 20 weeks on a HFD, we found significantly lower lipid content in the liver of Dbc1 KO mice compared with WT mice (Figure 6C). In addition, WT mice fed a HFD for 20 weeks had significant liver injury, as measured by increasing levels of plasma alanine aminotransferase (ALT) and aspartate aminotransferase (AST; Figure 6, D and E). In contrast, Dbc1 KO mice showed normal levels of AST and ALT after 20 weeks of HFD feeding.
Figure 6. Dbc1 KO mice are protected against HFD-induced liver steatosis.
(A and B) WT and Dbc1 KO mice were fed normal chow (A) or put on HFD for 20 weeks (B). Liver sections were stained with H&E (top) or Oil Red O–hematoxylin (bottom). Original magnification, ×200. (C) Lipid content from liver homogenates of mice fed normal chow diet (ND) or HFD. *P < 0.05, **P < 0.05, ANOVA. (D and E) ALT (D) and AST (E) activities were measured in plasma from mice fed normal chow or HFD. *P < 0.05, ANOVA. Dbc1 KO mice showed no significant increase in the activity of either protein compared with the groups fed normal chow. (F) SIRT1 activity in liver nuclear extracts from mice fed normal chow or HFD. Statistically significant differences (P < 0.05, ANOVA) are shown by brackets. RFU, relative fluorescence units. (G) Western blots for SIRT1 from WT and Dbc1 KO mouse livers in mice fed normal chow or HFD for 20 weeks. Each lane represents a different mouse liver. (H) Western blots for phosphorylated ACC (Ser80), ACC, phosphorylated AMPK (Thr172), and AMPK from WT and Dbc1 KO mice livers after 20 weeks of HFD. Each lane represents a different mouse liver. Data are mean ± SD.
To explore the mechanisms involved in this differential lipid accumulation between WT and Dbc1 KO mice, we analyzed levels and activity of several enzymes involved in lipid accumulation in the liver. In WT mice, we observed a significant decrease in SIRT1 activity after 20 weeks on a HFD compared with mice on a normal diet (Figure 6F). In contrast, Dbc1 KO mice on a HFD showed no significant decrease in SIRT1 activity in comparison to mice on a normal diet (Figure 6F). We found no changes in expression of SIRT1 in the livers of mice fed a normal diet or HFD in either WT or Dbc1 KO mice (Figure 6G). We also analyzed 2 key enzymes previously implicated in the development of liver steatosis that are known to be regulated by SIRT1. First, we found an increase in AMPK phosphorylation in livers from Dbc1 KO mice compared with WT mice fed HFD (Figure 6H). We also observed that the enzyme acetyl-CoA carboxylase (ACC), a key regulator and rate-limiting step for the de novo synthesis of lipids from glucose, was phosphorylated in the livers of Dbc1 KO mice under the same metabolic conditions. Phosphorylation of ACC leads to its inactivation, which decreases synthesis of lipids, increases β-oxidation, and promotes protection against liver steatosis. In fact, SIRT1 has been previously shown to induce activation of AMPK, which then phosphorylates ACC, thereby inhibiting its activity and preventing the development of fat accumulation in cultured hepatocytes and in mouse liver in vivo (24). The phosphorylation of these 2 proteins was unchanged between WT and Dbc1 KO when the mice were fed a normal diet (Supplemental Figure 4). We also tested the expression of other key regulators of lipid metabolism in the liver. Fatty acid synthase (FAS) levels were unchanged at the mRNA and protein levels in WT and Dbc1 KO mice, both with normal chow and with HFD. The transcription factor SREBP1c showed no difference at the mRNA level between WT and Dbc1 KO mice fed a HFD (Supplemental Figure 4). We propose that activation of SIRT1 and AMPK, and subsequent inactivation of ACC, in the livers of Dbc1 KO mice fed a HFD could be involved in the protection against liver steatosis.
DBC1 regulates inflammation.
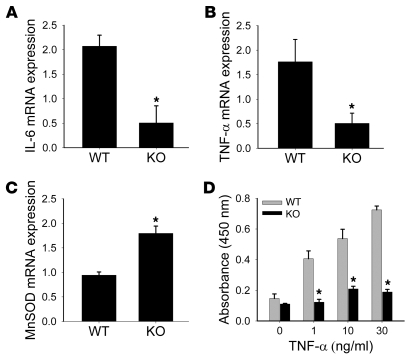
Pfluger et al. previously showed that SIRT1 overexpression leads to decreased liver inflammation during HFD (4). We investigated whether Dbc1 KO mice showed a similar phenotype. WT mice on HFD presented high hepatic expression of the inflammatory cytokines TNF-α and IL-6 (Figure 7, A and B). In contrast, Dbc1 KO mice fed a HFD demonstrated reduced expression of both inflammatory cytokines (Figure 7, A and B). Furthermore, as previously described for a transgenic mouse overexpressing liver-specific SIRT1 (4), Dbc1 KO mice had increased expression of the antioxidant enzyme manganese superoxide dismutase (MnSOD; Figure 7C). To further evaluate the role of DBC1 in the inflammatory response in the liver, we isolated Kupffer cells from WT and Dbc1 KO mice. Kupffer cells were treated with LPS, and the TNF-α released into the media was measured. Kupffer cells from Dbc1 KO mice showed decreased TNF-α release in response to LPS treatment compared with cells from WT mice, demonstrating that DBC1 regulates the function of the professional resident inflammatory cells in the liver (Supplemental Figure 5). Yeung et al. have shown that SIRT1 decreases inflammatory response through interaction and deacetylation of p65 NF-κB (11). To test the potential mechanism by which DBC1 regulates inflammation, we treated WT and Dbc1 KO MEFs with different doses of TNF-α. The Dbc1 KO MEFs showed decreased p65 NF-κB activation in response to TNF-α compared with WT MEFs (Figure 7D). We propose that the mechanism of protection against inflammation in Dbc1 KO is mediated by SIRT1-dependent inactivation of NF-κB (11).
Figure 7. Dbc1 KO mice and cells are protected against inflammation.
(A–C) Semiquantitative RT-PCR from mouse livers after 20 weeks of HFD for IL-6 (A), TNF-α (B), and MnSOD (C) mRNA. Total levels were normalized to 18s rRNA expression levels. (D) WT and Dbc1 KO–derived MEFs were treated with different doses of TNF-α for 6 hours. NF-κB activation (p65) was measured from nuclear cell extracts. *P < 0.05, t test. Data are mean ± SD.
DBC1 controls oleate/palmitate-induced cellular lipid accumulation through a SIRT1-dependent mechanism.
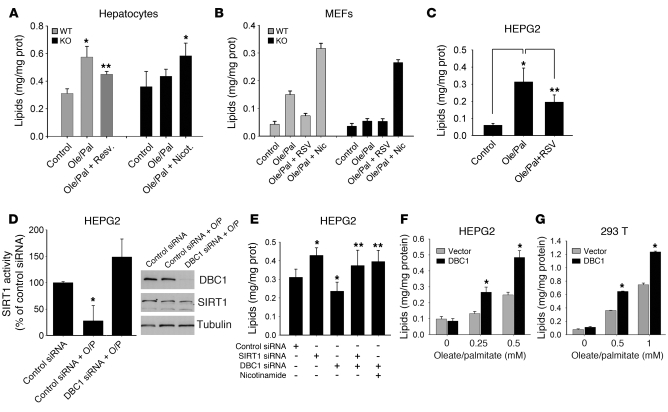
We further investigated whether the prevention of cellular lipid accumulation in the Dbc1 KO mice was a SIRT1-dependent process, using established cellular models to induce cellular steatosis while minimizing cellular apoptosis (45). We isolated and cultured primary hepatocytes from WT and Dbc1 KO mice and then treated these cells with lipids to induce steatosis. Incubation of WT hepatocytes with 0.5 mM oleate/palmitate (2:1 ratio) increased intracellular lipid accumulation (Figure 8A). This lipid accumulation was partially reversed when cells were also incubated in the presence of 100 μM resveratrol, a putative in vivo SIRT1 activator. However, when hepatocytes from the Dbc1 KO mice were incubated with 0.5 mM oleate/palmitate, no significant lipid accumulation was observed (Figure 8A). This protection against lipid accumulation was reversed when the cells were incubated in the presence of the SIRT1 inhibitor nicotinamide (5 mM). Similar results were obtained when we repeated these experiments in MEFs (Figure 8B and Supplemental Figure 6). WT MEFs accumulated lipids when incubated in the presence of 1 mM oleate/palmitate (2:1 ratio). The lipid accumulation was partially reversed by 100 μM resveratrol and increased by the presence of 5 mM nicotinamide in the media. On the other hand, Dbc1 KO MEFs did not accumulate lipids when incubated with 1 mM oleate/palmitate (Figure 8B and Supplemental Figure 6). However, in the presence of 5 mM nicotinamide, Dbc1 KO MEFs accumulated lipids similarly to the WT cells (Figure 8B).
Figure 8. DBC1 controls oleate/palmitate-induced cellular steatosis through a SIRT1-dependent mechanism.
(A) Lipid accumulation levels in primary hepatocytes isolated from WT and Dbc1 KO mice. An average of 3 different experiments is shown. *P < 0.05, **P < 0.1 versus control, t test. (B) Lipid accumulation in MEFs derived from WT and Dbc1 KO mice. Shown is 1 representative experiment of 4. (C) Lipid accumulation in HEPG2 cells. *P < 0.05, **P < 0.1, t test. n = 3. (D) Lipid accumulation in HEPG2 cells. Cells were transfected with control or DBC1 siRNA and then incubated with oleate/palmitate. SIRT1 activity was measured; values were normalized to those from control siRNA transfection. Protein levels were evaluated by Western blot. *P < 0.05, t test. n = 3. (E) Lipid accumulation in HEPG2 cells. Cells were transfected with control, SIRT1, and DBC1 siRNAs and then incubated with oleate/palmitate. *P < 0.05, **P < 0.1 versus control siRNA, t test. n = 4. (F and G) Effect of DBC1 overexpression on lipid accumulation in HEPG2 (F) and 293T (G) cells. Cells were transfected with empty vector or myc-DBC1 for 48 hours. During the last 16 hours of transfection, cells were incubated with different concentrations of oleate/palmitate (2:1 ratio). *P < 0.05, t test. n = 3. Data are mean ± SD.
To further verify these results, we investigated the effect of DBC1 and SIRT1 manipulation on lipid accumulation in HEPG2 cells. Lipid accumulation in HEPG2 cells was induced by incubating cells with 0.5 mM oleate/palmitate (2:1 ratio) for 24 hours (Figure 8C). Treated cells presented cytosolic lipid deposition, as revealed by Oil red O staining (Supplemental Figure 6). This intracellular lipid accumulation was partially reversed by 100 μM resveratrol (Figure 8C). In HEPG2 cells, SIRT1 activity decreased in the presence of 0.5 mM oleate/palmitate (Figure 8D). This decrease in SIRT1 activity by oleate and palmitate was prevented by transfecting cells with DBC1 siRNA (Figure 8D). These data suggest that, as observed for high-glucose treatment (Figure 3), lipid treatment decreases SIRT1 activity by a DBC1-dependent mechanism. Next, we measured the effect of manipulating the expression of both SIRT1 and DBC1 on lipid accumulation in HEPG2 cells. Cells transfected with SIRT1 siRNA showed an increase in lipid accumulation compared with control siRNA (Figure 8E). In contrast, transfection of HEPG2 cells with DBC1 siRNA produced the opposite effect, decreasing lipid accumulation. This protective effect of DBC1 siRNA on oleate/palmitate-induced cellular lipid accumulation was eliminated when cells were cotransfected with SIRT1 siRNA or treated with 5 mM nicotinamide (Figure 8E), which indicates that the effect of DBC1 knockdown on lipid accumulation is dependent on the expression and activity of SIRT1.
Finally, we tested whether overexpression of DBC1 increases oleate/palmitate-induced cellular lipid accumulation. We overexpressed DBC1 for 48 hours in HEPG2 and 293T cells. At 20 hours before harvesting, cells were treated with different concentrations of oleate/palmitate, and lipid accumulation was compared between cells transfected with DBC1 and cells transfected with empty vector. For both cell lines, HEPG2 and 293T, we observed an increase in lipid accumulation when DBC1 was overexpressed (Figure 8, F and G, and Supplemental Figure 6). Because SIRT1 and HDAC3 deacetylases share some protein targets, we decided to rule out the possibility that HDAC3 affected lipid accumulation. By overexpressing HDAC3 along with SIRT1 and DBC1 in 293T cells, we found that although SIRT1 overexpression protected against lipid accumulation, HDAC3 overexpression had no effect on lipid accumulation (Supplemental Figure 6). All these results clearly showed that DBC1 plays a key role in hepatic cellular steatosis by its regulation of SIRT1 activity.
Discussion
In this study, we assessed the role of DBC1 in regulating SIRT1 under different metabolic conditions and established a critical role for DBC1 in the pathogenesis of liver steatosis and liver inflammation that we believe to be novel. We showed that DBC1 tightly regulated SIRT1 activity in different metabolically relevant cell types and modulated glucose and lipid-induced SIRT1 activity. We also demonstrated that DBC1 and SIRT1 interacted in multiple tissues and that DBC1 regulated SIRT1 activity in vivo in these tissues. In addition, we found that DBC1-SIRT1 interaction was modulated by different metabolic conditions in the liver. An increase in the association between these proteins occurred in mice fed a HFD, and a decrease occurred after 24 hours of starvation. These changes in DBC1-SIRT1 association paralleled changes in SIRT1 activity: lower in the livers of mice fed a HFD and higher after 24 hours of starvation. Finally, we demonstrated that Dbc1 KO mice were protected against the development of HFD-induced steatosis, showing decreased inflammation, fat infiltration, and liver injury. We provided strong evidence that this protective effect of DBC1 was dependent on SIRT1 activation; however, other mechanisms may also be involved.
Researchers have previously proposed that changes in intracellular NAD+ levels or the NAD+/NADH ratio are the primary mechanisms regulating SIRT1 activity (8, 20, 46). Some reports have shown an increase in NAD+ levels in the liver upon starvation without changes in NADH levels (8), whereas others have shown an increase in NAD+ and NADH with a net decrease in the NAD+/NADH ratio in mice under caloric restriction (34). The differences in SIRT1 activity observed here could not be explained by changes in NAD+ levels inside the cell for 2 reasons. First, cell and tissue homogenates were prepared and diluted so that NAD+ contamination from the cell extracts was minimal and insufficient to support SIRT1 activity. As shown in Figure 2, when exogenous NAD+ was absent, the signal detected was not significantly higher than the blank. These results do not imply that NAD+ changes inside the cell are not involved in the regulation of SIRT1 activity, but they do suggest that other factors, such as posttranslational modifications (32, 33) or protein-protein interaction (38–40), are also playing an important role in the regulation of SIRT1 activity. A previous study reported changes in endogenous SIRT1 activity in the liver independent of the NAD+ concentration in the reaction media (47). Second, we could not detect any significant difference in NAD+, NADH, or the NAD+/NADH ratio in the liver after starvation or HFD. Of note about this result is that we measured total intracellular NAD+ levels. We cannot rule out the possibility that local NAD+ changes could be occurring in different subcellular compartments, specifically in the nucleus, that could not be detected by the whole cellular NAD+ preparation. Strong evidence suggests that regulation of NAD+ levels might be compartmentalized inside the cells (37, 48, 49). However, to our knowledge, no reports have been published that measured changes in NAD+ levels inside the nucleus, where SIRT1 is primarily localized. This observation also raises the question of whether the changes in intracellular NAD+ levels that have been shown in different experimental situations (8, 20, 50) are indeed being sensed by nuclear SIRT1 or are occurring in another organelle. This important question needs to be addressed to understand the role of NAD+ as a limiting factor in the regulation of SIRT1 activity in vivo. Zhang et al. recently showed that once SIRT1 binds to chromatin, it recruits and binds to the enzyme NMNAT-1, which is responsible for the final step in NAD+ biosynthesis (48). This observation suggests that NAD+ levels might be tightly regulated in a site-specific manner in the nucleus. However, more direct measurements should be made to address this issue. Nonetheless, we believe that regulation of SIRT1 activity is more complex than was originally thought. Our data clearly showed that SIRT1 activity was regulated by DBC1 during different metabolic states, independent of changes in NAD+ levels. Whether NAD+ levels change inside the nucleus and whether this change can be sensed by SIRT1 in vivo needs further investigation.
Investigators have also proposed that SIRT1 levels change in mouse liver upon starvation. However, these data are still contradictory. Although some studies found an increase in SIRT1 levels (8), others found a decrease (34) or no change (27). Our data demonstrated that although SIRT1 and DBC1 levels did not change in the liver after different metabolic states, there were changes in the interaction between DBC1 and SIRT1 (Figure 5). This, in turn, led to the modulation of SIRT1 activity. However, how the DBC1-SIRT1 interaction is regulated and which molecular pathways are involved remains unknown. One possibility is that SIRT1 is modified posttranslationally during different metabolic conditions. Yang et al. recently described that SUMOylation of human SIRT1 increases its activity (32). It is possible that SUMOylation of SIRT1 could be implicated in some of the effects observed in human-derived cells. However, this is unlikely in the case of mouse liver, because mouse SIRT1 lacks the lysine residue, K734, that was shown to be SUMOylated in humans. Studies in human cells also identified 2 potential phosphorylation sites for human SIRT1 (51, 52) and suggested that human SIRT1 could be phosphorylated by PI3K. Indeed, a recent report shows that SIRT1 is phosphorylated in at least 2 different sites by cell cycle–dependent kinases and that this phosphorylation increases SIRT1 activity (33). On the other hand, DBC1 may be modified during different metabolic conditions, altering its interaction with SIRT1. Further studies are necessary to determine how DBC1-SIRT1 interaction is regulated in different metabolic conditions. At the animal level, we found that Dbc1 KO mice have higher SIRT1 activity compared with WT mice in all tissues tested. This observation confirms that DBC1 is indeed a SIRT1 inhibitor in vivo. Interestingly, we found that the increase in activity in the Dbc1 KO mice varied among different tissues. We believe that this finding could be explained by the possibility that the amount of DBC1 that binds to SIRT1 varies in different tissues. Furthermore, SIRT1 activity may be regulated by other factors, including phosphorylation, SUMOylation, and interaction with other proteins. These aspects make cellular and tissue-specific regulation of SIRT1 a complex issue; a generalization about the amount of DBC1 bound to SIRT1 seems difficult to achieve with our present knowledge. Furthermore, no reliable method is yet available for determining the exact amount or percentage of interaction between endogenous proteins in vivo. Nonetheless, using coimmunoprecipitation, we observed that at least 50% (and, in one mouse, up to 80%) of endogenous SIRT1 that was immunoprecipitated may be bound to DBC1, which indicates that DBC1 is one of the primary regulators of SIRT1 function.
In the present study, we demonstrated that Dbc1 KO mice were protected against the development of HFD-induced liver steatosis. Pfluger et al. showed that increased expression of SIRT1 in the liver protects against the development of fatty liver disease (4). Resveratrol, a compound that activates SIRT1 in vitro and has also been postulated to do so in vivo, protects against the development of alcoholic and HFD-induced liver steatosis (6, 24, 25). These findings, in association with our cellular studies, demonstrate that DBC1 is a key regulator of SIRT1 in the liver. The data also suggest that increased binding between SIRT1 and DBC1, and the resultant decrease in SIRT1 activity during the consumption of a HFD, could contribute to liver steatosis. Mechanistically, SIRT1 is known to stimulate AMPK phosphorylation, which in turn phosphorylates and inactivates ACC, the rate-limiting enzyme in lipid synthesis (24). The increased phosphorylation of both AMPK and ACC observed in the Dbc1 KO mice fed a HFD may explain why the Dbc1 KO mice were protected against the development of fatty liver steatosis. In addition, Dbc1 KO mice had reduced expression of the inflammatory markers IL-6 and TNF-α and increased expression of the antioxidant enzyme MnSOD. We also found that the absence of DBC1 decreased activation of NF-κB by TNF-α. From these results, we propose that during HFD feeding, the inhibition of SIRT1 activity by DBC1 plays a role in the inflammatory response triggered during the development of fatty liver disease (4). Whether the antiinflammatory effect is mediated at the level of hepatocytes or at the level of the professional systemic inflammatory cells is unknown and is the focus of ongoing research in our laboratory. However, we found that Dbc1 KO Kupffer cells had impaired TNF-α secretion (Supplemental Figure 5). From these data, we propose that DBC1 may have a key role in the development of HFD-induced liver steatosis through a mechanism that involves SIRT1 inhibition. This mechanism, in turn, triggers an increase in lipid synthesis by the liver and the onset of the inflammatory response observed during fatty liver disease. DBC1 has also been recently shown to associate and regulate other targets such as the estrogen receptor, androgen receptor, and SUV39H1 methyltransferase (53–55). Whether the effects of DBC1 on these proteins have any physiological relevance or account for any of the phenotypes seen in our studies is not known and deserves further evaluation.
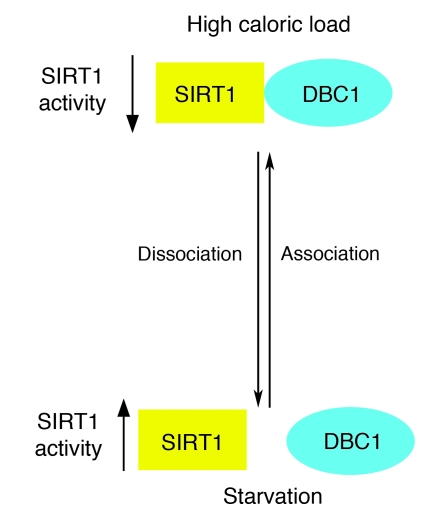
In summary, we showed for the first time to our knowledge that DBC1 interacted and regulated SIRT1 activity in tissues. We also showed that DBC1 regulated endogenous SIRT1 activity in metabolically active cells and played a role in energy-dependent modulation of SIRT1 activity. These findings lead us to propose a role for DBC1 as a molecular switch that regulates SIRT1 activity under different metabolic conditions (Figure 9). Under starvation or low-caloric diets, DBC1 remains dissociated from SIRT1, promoting an increase in SIRT1 activity. In the presence of high-caloric diets, DBC1 binds to SIRT1 and inhibits its deacetylase activity. Our finding that DBC1 inhibition had significant effects on SIRT1 activity makes it an attractive candidate for pharmacological targeting. DBC1 pharmacological inhibition could be a novel approach to improve treatment of metabolic disorders, such as metabolic syndrome, type II diabetes, liver steatosis, and various diseases of aging.
Figure 9. Working model for the regulation of DBC1-SIRT1 interactions under different metabolic conditions.
Methods
Reagents and antibodies.
Unless otherwise specified, all reagents and chemicals were purchased from Sigma-Aldrich. SIRT1 antibodies for Western blot and immunoprecipitation were from Upstate Biotechnology and Abcam, and those for immunofluorescence were from Santa Cruz Biotechnology Inc. CD38 antibody was from Santa Cruz Biotechnology Inc. PEPCK antibody was from Cayman Chemical Co. DBC1 and NAMPT antibodies were from Bethyl Laboratories. Antibodies for p53, acetylated p53, phosphorylated ACC, ACC, and tubulin were from Abcam. Phosphorylated AMPK (Ser172) and AMPK antibodies were from Cell Signaling Technology.
Animal handling and experiments.
All mice used in this study were maintained in the Mayo Clinic Animal Breeding facility. All experimental protocols were approved by the Institutional Animal Care and Use Committee at Mayo Clinic (protocol no. A33209), and all studies were performed according to the methods approved in the protocol. For the metabolic studies, mice were placed on either a normal control diet (diet no. 3807; KLIBA-NAFAG) or a HFD (AIN-93G, modified to provide 60% of calories from fat; Dyets Inc.) ad libitum and monitored for 6–20 weeks. Animals were kept in individual cages for the entire study. Body weight was recorded weekly, and food intake was measured for 7 consecutive days. For the starvation experiments, mice were housed for 24 hours without food, but with water ad libitum. Mice with ad libitum access to both normal food and water were used as controls. In all experiments, groups consisted of at least 3–4 mice.
Generation and characterization of Dbc1 KO mice.
The gene trap embryonic stem cell line XN876 was obtained from Bay Genomics. The Dbc1 gene in this cell line was disrupted by a gene-trap vector inserted between exon 6 and exon 7 of the Dbc1 gene (geneID 219158). The exact insertion site was mapped by genomic PCR and DNA sequencing. Primers for all subsequent genotyping were designed as follows: DBC1 WT, 5′-ACAAGGTCTTAAGTAGCTTGGGCTGGC-3′; DBC1 neo, 5′-GACAGTATCGGCCTCAGGAAGATCG-3′; DBC1 forward, 5′-GTACCTGGAGGGTTTATGTTACCT-3′. The DBC1 ES cells (129/Ola) were injected into C57BL/6 blastocysts to generate chimeric mice. Chimeras were then backcrossed with C57BL/6 mice to generate Dbc1+/– mice, which were used for subsequent breeding. All MEFs were generated from E13.5 or E14.5 embryos using standard procedures. For all the experiments involving Dbc1 KO mice, WT littermates were used as controls.
Cell culture.
293T cells were grown in RPMI, 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Immortalized MEFs derived from WT and Dbc1 KO mice, were grown in DMEM (5 g/l glucose), 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. INS cells were cultured in RPMI-1640 medium containing 11.1 mM glucose supplemented with 10% FBS, 1 mM pyruvate, 10 mM HEPES, 50 μM 2-mercaptoethanol, 100 U/ml penicillin, and 100 μg/ml streptomycin. HEPG2 cells were cultured in DMEM (1 g/l glucose), 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin. Primary hepatocytes were isolated as previously described (56). Cells were incubated for 6 hours in DMEM (1 g/l glucose), 10% FBS, 100 U/ml penicillin, and 100 μg/ml streptomycin before treatments.
Glucose treatment in cells.
Before glucose treatments, HEPG2 cells were incubated in serum-free medium for 24 hours. Cells were transferred to serum-free DMEM medium containing either 5 mM glucose (low glucose) or 30 mM glucose (high glucose) and incubated for 24 hours before harvesting.
Lipid treatment in cells.
Cells were incubated with a mixture of oleic acid and palmitic acid in a 2:1 ratio in culture media supplemented with 1% fatty acid–free BSA (Sigma-Aldrich). Lipids were used at concentrations shown to induce steatosis, but not apoptosis (45). Incubations with lipids were performed for 16–24 hours. Primary hepatocytes were incubated overnight in the presence of 0.5 mM oleate/palmitate (2:1 ratio). MEFs were incubated overnight with oleate/palmitate (2:1 ratio) at 1 mM final concentration. HepG2 cells were incubated for 16 hours in the presence of 0.5 mM oleate/palmitate (2:1 ratio). Resveratrol and nicotinamide were used at 100 μM and 5 mM, respectively, in all cases.
siRNA.
siRNA against DBC1 was synthesized by Dharmacon. The siRNA duplexes were 21 bp as follows: DBC1 siRNA sense strand, 5′-AAACGGAGCCUACUGAACAUU-3′. SIRT1 siRNA was from Dharmacon SmartPool (L003540-00-0005). Nontargeting siRNA (D001210-03-20; Dharmacon) was used as control. Transfection was performed twice, 24 hours apart, with 200 nM siRNA using DharmaFECT1 reagent according to the manufacturer’s instructions. Cells were harvested or fixed 72 hours after the first transfection. Treatment with nicotinamide (5 mM) was performed overnight before harvesting the cells.
Nuclear isolation.
Liver nuclei were isolated as previously described (42), with minor modifications. All steps were performed on ice. Briefly, livers were cut into small pieces and homogenized 10 times with a Dounce homogenizer in buffer containing 0.25 M sucrose; 50 mM Tris-HCl, pH 7.4; 25 mM KCl; 5 mM MgCl2 (TKM solution); and 1 mM PMSF (homogenizing medium). Homogenate was filtered through cheesecloth and centrifuged twice at 800 g for 10 minutes. The resuspended pellet was added to the top of tubes that contained an equal volume of cushion composed (from top to bottom) of 0.5 ml 1.0 M sucrose solution, 0.5 ml 1.5 M sucrose solution, and 1.5 ml 2.1 M sucrose solution, all prepared in TKM buffer. Tubes were centrifuged at 100,000 g for 60 minutes, and the resulting pellet was resuspended in homogenizing medium and sedimented at 800 g for 10 minutes. The final pellet containing the purified nuclei was resuspended in homogenizing medium (5 strokes) at a protein concentration of about 2.5 mg/ml and stored at 4°C until used.
Immunoprecipitation and Western blot.
Mouse tissues and cultured cells were lysed in NETN buffer (20 mM Tris-HCl, pH 8.0; 100 mM NaCl; 1 mM EDTA; and 0.5% NP-40) supplemented with 5 mM NaF, 50 mM 2-glycerophosphate, and a protease inhibitor cocktail (Roche). Homogenates were incubated at 4°C for 20 minutes under constant agitation, then centrifuged at 10,000 g for 10 minutes at 4°C. For each immunoprecipitation, 1 mg protein was used. Samples were incubated with 20 μl Protein A-G (Santa Cruz Biotechnology Inc.) and 1–2 μg antibody for 1–2 hours at 4°C under constant rotation. Nonspecific IgG (Santa Cruz Biotechnology Inc.) was used as control. Finally, immunoprecipitates were washed 2–3 times with cold NETN before addition of 2× Laemmli buffer. Immunoprecipitation from isolated liver nuclei was performed as described above, except the lysis buffer contained 20 mM HEPES, pH 7.9; 125 mM NaCl; 0.1% NP-40; 0.1% Triton X-100; 1 mM EDTA; 5 mM nicotinamide; 5 μM trichostatin A (TSA); 5 mM NaF; 50 mM 2-glycerophosphate; and a protease inhibitor cocktail (Roche). For each immunoprecipitation, 50–100 μg protein was used. Cell and tissue lysates and immunoprecipitates were analyzed by Western blot with indicated antibodies. Western blots were developed using SuperSignal West Pico chemiluminescent substrate (Pierce). Films were scanned and bands quantified by densitometry using Image J (http://rsbweb.nih.gov/ij/).
Immunofluorescence imaging.
Cells were cultured on coverslips. In the case of isolated liver nuclei, they were allowed to settle onto glass coverslips treated with poly-L-lysine (0.01 mg/ml). Samples were fixed for 30 minutes with 4% paraformaldehyde prepared in PBS, permeabilized with 0.1% Triton X-100 in PBS for 15 minutes, washed 3 times with PBS, and incubated for 1 hour at room temperature in blocking buffer (PBS plus 2% BSA). Coverslips were incubated with the primary antibody overnight at 4°C, washed 3 times with PBS at room temperature, and then incubated with the secondary antibody (anti-rabbit Cy5, anti-goat Cy3; Jackson ImmunoResearch Laboratories) for 1 hour at room temperature. Samples were mounted in antifade medium (ProLong Gold; Invitrogen). Images were obtained with a Nikon microscope and a Plan Apo ×60 oil, 1.42 NA lens. Images were processed with Image J.
SIRT 1 activity in cells and mouse tissues.
SIRT1 activity was determined with a SIRT1 Fluorometric Kit (Biomol International) according to the manufacturer’s instructions. This assay uses a small lysine-acetylated peptide, corresponding to K382 of human p53, as a substrate. The lysine residue is deacetylated by SIRT1, and this process is dependent on the addition of exogenous NAD+. The fluorescence values obtained in the absence of NAD+ did not differ from the blank. Addition of exogenous NAD+ was necessary, most likely because endogenous NAD+ was lost during sample preparation. SIRT1 inhibitors nicotinamide (2 mM), suramin (100 μM), and sirtinol (100 μM) were used to confirm the specificity of the reaction. Samples (cultured cells or isolated nuclei) were homogenized in NETN buffer as described above. Homogenates were then incubated for 10 minutes at 37°C to allow degradation of any contaminant NAD+. Next, 10 mM DTT was added to the medium, and homogenates were incubated again for 10 minutes at 37°C. The homogenates (20–30 μg protein/well) were then incubated in SIRT1 assay buffer in the presence of either 100 μM Fluor de Lys–SIRT1 substrate (Enzo Life Sciences) and 5 μM TSA to determine the SIRT1-independent activity, or with 100 μM Fluor de Lys–SIRT1 substrate, 5 μM TSA, and 200 μM NAD+ to determine the SIRT1-dependent activity. After 60 minutes of incubation at 37°C, the reaction was terminated by adding a solution containing Fluor de Lys Developer (Enzo Life Sciences) and 2 mM nicotinamide. Plates were incubated at 37°C for 1 hour. Values were determined by reading fluorescence on a fluorometric plate reader (Spectramax Gemini XPS; Molecular Devices) with an excitation wavelength of 360 nm and an emission wavelength of 460 nm. Calculation of net fluorescence included the subtraction of a blank consisting of buffer containing no NAD+ and expressed as a percentage of control. The SIRT1-dependent activity was calculated after subtracting fluorescence values obtained in the absence of NAD+. In all cases, we confirmed the linearity of the reaction over time.
CD38 NADase activity.
NADase activity was determined by using etheno-NAD as described previously (35). Briefly, livers were homogenized in a medium containing 0.25 M sucrose; 40 mM Tris–HCl, pH 7.4; and a protease inhibitor cocktail. The homogenate was then centrifuged at 1,000 g for 10 minutes at 4°C, and the supernatant was used for activity measurement. Homogenates were adjusted to a protein concentration of 1 mg/ml and put in a fluorometer cuvette at 37°C under constant agitation. The reaction was triggered by adding 0.2 mM etheno-NAD to the medium. The change in fluorescence (Δf) over time was measured at 300 nm excitation and 410 nm emission. CD38 NADase activity was expressed as Δf/Δt per mg protein.
Detection of intracellular NAD+ levels in cells and tissues by cycling assay.
NAD+ was isolated as previously described (35). Briefly, mouse tissues were frozen in liquid N2, pulverized into a powder, and extracted with 10% trichloroacetic acid (TCA) at 4°C. TCA was removed by adding 2 volumes of 1,1,2-trichloro-1,2,2-trifluoroethane-trioctylamine (3:1 ratio). Samples were agitated, and the mixture was allowed to separate into phases at room temperature. The aqueous superior layer containing the NAD was then removed and adjusted to pH 8 with 1 M Tris. NAD+ was determined by the cycling assay, as described previously (35). Briefly, 0.1 ml sample was incubated with 0.1 ml solution containing 0.1 M NaH2PO4/NaHPO4, pH 8; 0.76% ethanol; 4 μM flavine mononucleotide; 40 μg/ml alcohol dehydrogenase; 0.04 U/ml diaphorase; and 8 μM resazurin. For the standard curve, β-NAD+ from yeast was used. Fluorescence was monitored periodically in a fluorescence plate reader (Spectramax Gemini XPS; Molecular Devices). NAD+ values were expressed as nanomoles NAD+ per milligram tissue.
Lipid accumulation and measurement in cells and tissue.
Cells were incubated in the presence of a mixture of oleic acid and palmitic acid in a 2:1 ratio in culture media supplemented with 1% fatty acid–free BSA. All lipid incubations were performed for 24 hours. MEFs were incubated for 24 hours in DMEM (1 g/l glucose) plus 10% FBS before lipid incubations. Nicotinamide (5 mM) and resveratrol (100 μM) were included during incubation with lipids. In cells transfected with plasmids or siRNA, lipids were added in the last 24 hours of transfection. Lipid content in the cells and tissue was measured according to Zang et al. (57), with minor modifications. Samples were homogenized in lysis buffer as described above. Lipid content was measured by using the Infinity reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions.
Biochemical markers and hormones.
Leptin and adiponectin were measured with an ELISA kit (R&D Systems). ALT and AST liver enzyme activity from mouse plasma was measured by spectrophotometer (Randox Laboratories Ltd.) according to the manufacturer’s instructions.
Oxygen consumption, carbon dioxide production, and activity.
These measurements were performed as previously described (58). Each mouse was placed in a 30 cm × 10 cm cylindrical chamber for 24 hours prior to the measurement of 24-hour activity and EE. Oxygen consumption and carbon dioxide production were measured by a customized, high-precision, single-chamber indirect calorimeter (Columbus Instruments), as we have reported previously (58). Thermogenesis was calculated from oxygen consumption and carbon dioxide production. Calibration of the calorimeter was performed at the beginning of each measurement day. We set the calorimeter to deliver room air to the chamber at 0.60–0.65 l/min, and we collected samples every minute (with a 5-minute reference period every 30 samples), with a sample flow of 0.4 l/min. We calculated resting EE by averaging the EE values associated with 0 activity counts for the minute of the EE measurement and the prior 4 minutes. Activity EE was calculated by subtracting resting EE from total daily EE. Simultaneous physical activity measurements were performed using customized high-precision racks of collimated infrared activity sensors (Opto-M Varimex Minor, Columbus Instruments) placed around the acrylic calorimetry chamber. There were 45 collimated beams of infrared light crossing the 30-cm-diameter cage, allowing the detection of 1 inch of movement in 3 orthogonal axes. Photosensors registered an activity unit each time a beam was interrupted (ambulatory counts defined as nonrepetitive horizontal beam breaks; total counts calculated as combined horizontal, ambulatory, and vertical counts; stationary counts calculated as ambulatory counts minus horizontal counts).
Fecal analysis.
Daily fecal outputs were determined through the use of metabolic cages and were measured by collecting all feces from each individual mouse (3 or 4 mice per day) for 7 days. Feces were allowed to air dry for at least 72 hours before weighing.
Liver staining.
Livers were excised and immediately placed in 4% formalin for posterior processing by standard staining techniques. For the Oil red O staining, excised livers were embedded in embedding media and snap frozen in liquid nitrogen. Liver sections were fixed in 4% formalin and then stained with Oil red O for 30 minutes. After differentiation with 85% propylene glycol, sections were counterstained with Harris hematoxylin.
RNA isolation and RT-PCR analysis.
Total RNA from liver was isolated using the RNeasy Protect Mini Kit (Qiagen) according to the manufacturer’s instructions. RT-PCR was performed with the RT-PCR Kit (Qiagen). The following primers were used: IL-6 forward, 5′-AGTTGCCTTCTTGGGACTGA-3′; IL-6 reverse, 5′-TCCACGATTTCCCAGAGAAC-3′; TNF-α forward, 5′-CGTCAGCCGATTTGCTATCT-3′; TNF-α reverse, 5′-CGGACTCCGCAAAGTCTAAG-3′; MnSOD forward, 5′-CCGAGGAGAAGTACCACGAG-3′; MnSOD reverse, 5′-GCTTGATAGCCTCCAGCAAC-3′; 18s rRNA forward, 5′-CCGCAGCTAGGAATAATGGA-3′; 18s rRNA reverse, 5′-GAGTCAAATTAAGCCGCAGG-3′. PCR products were visualized by 2% agarose gel and stained with ethidium bromide. Bands were scanned and densitometry was performed with Image J. Results were normalized to 18s rRNA levels.
TNF-α treatments and NF-κB activation.
WT and Dbc1 KO mouse-derived MEFs were treated with hTNF-α (EMD Chemicals) at different concentrations for 6 hours in complete media. NF-κB activation (p65 subunit) was measured with the TransAM NF-κB p65 activation assay (Active Motif) according to the manufacturer’s instructions. Nuclear extracts were made according to manufacturer recommendations.
Statistics.
Values are presented as mean ± SEM of 3–5 experiments, unless otherwise indicated. The significance of differences between means was assessed by ANOVA or 2-tailed Student’s t test, as indicated. A P value less than 0.05 was considered significant.
Supplementary Material
Acknowledgments
This work was supported in part by grants from the American Federation of Ageing Research and from the Mayo Foundation, by the Strickland Career Development Award, and by National Institute of Diabetes and Digestive and Kidney Diseases, NIH, grant DK-084055. The contribution by C. Escande constitutes part of his PhD work.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(2):545–558. doi:10.1172/JCI39319.
Colleen M. Novak’s present address is: Department of Biological Sciences and School of Biomedical Sciences, Kent State University, Kent, Ohio, USA.
References
- 1.Guarente L. Sirtuins as potential targets for metabolic syndrome. Nature. 2006;444(7121):868–874. doi: 10.1038/nature05486. [DOI] [PubMed] [Google Scholar]
- 2.Guarente L. Sirtuins in aging and disease. Cold Spring Harb Symp Quant Biol. 2007;72:483–488. doi: 10.1101/sqb.2007.72.024. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh HS. The anti-aging, metabolism potential of SIRT1. Curr Opin Investig Drugs. 2008;9(10):1095–1102. [PubMed] [Google Scholar]
- 4.Pfluger PT, Herranz D, Velasco-Miguel S, Serrano M, Tschop MH. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105(28):9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deng XQ, Chen LL, Li NX. The expression of SIRT1 in nonalcoholic fatty liver disease induced by high-fat diet in rats. Liver Int. 2007;27(5):708–715. doi: 10.1111/j.1478-3231.2007.01497.x. [DOI] [PubMed] [Google Scholar]
- 6.Ajmo JM, Liang X, Rogers CQ, Pennock B, You M. Resveratrol alleviates alcoholic fatty liver in mice. Am J Physiol Gastrointest Liver Physiol. 2008;295(4):G833–G842. doi: 10.1152/ajpgi.90358.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.You M, Cao Q, Liang X, Ajmo JM, Ness GC. Mammalian sirtuin 1 is involved in the protective action of dietary saturated fat against alcoholic fatty liver in mice. J Nutr. 2008;138(3):497–501. doi: 10.1093/jn/138.3.497. [DOI] [PubMed] [Google Scholar]
- 8.Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434(7029):113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 9.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107(2):149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 10.Brunet A, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303(5666):2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 11.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23(12):2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeong J, et al. SIRT1 promotes DNA repair activity and deacetylation of Ku70. Exp Mol Med. 2007;39(1):8–13. doi: 10.1038/emm.2007.2. [DOI] [PubMed] [Google Scholar]
- 13.Fulco M, et al. Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell. 2003;12(1):51–62. doi: 10.1016/S1097-2765(03)00226-0. [DOI] [PubMed] [Google Scholar]
- 14.Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403(6771):795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 15.Landry J, et al. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci U S A. 2000;97(11):5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banks AS, et al. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab. 2008;8(4):333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belden WJ, Dunlap JC. SIRT1 is a circadian deacetylase for core clock components. Cell. 2008;134(2):212–214. doi: 10.1016/j.cell.2008.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277(47):45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 19.Boily G, et al. SirT1 regulates energy metabolism and response to caloric restriction in mice. PLoS ONE. 2008;3(3):e1759. doi: 10.1371/journal.pone.0001759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bordone L, et al. Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic beta cells. PLoS Biol. 2006;4(2):e31. doi: 10.1371/journal.pbio.0040031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cohen HY, et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 22.Feige JN, et al. Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab. 2008;8(5):347–358. doi: 10.1016/j.cmet.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 23.Gerhart-Hines Z, et al. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26(7):1913–1923. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hou X, et al. SIRT1 regulates hepatocyte lipid metabolism through activating AMP-activated protein kinase. J Biol Chem. 2008;283(29):20015–20026. doi: 10.1074/jbc.M802187200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lagouge M, et al. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127(6):1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 26.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107(2):137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 27.Kanfi Y, Peshti V, Gozlan YM, Rathaus M, Gil R, Cohen HY. Regulation of SIRT1 protein levels by nutrient availability. FEBS Lett. 2008;582(16):2417–2423. doi: 10.1016/j.febslet.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 28.Nemoto S, Fergusson MM, Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha}. J Biol Chem. 2005;280(16):16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 29.Chen WY, Wang DH, Yen RC, Luo J, Gu W, Baylin SB. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123(3):437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang C, et al. Interactions between E2F1 and SirT1 regulate apoptotic response to DNA damage. Nat Cell Biol. 2006;8(9):1025–1031. doi: 10.1038/ncb1468. [DOI] [PubMed] [Google Scholar]
- 31.Abdelmohsen K, et al. Phosphorylation of HuR by Chk2 regulates SIRT1 expression. Mol Cell. 2007;25(4):543–557. doi: 10.1016/j.molcel.2007.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang Y, et al. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat Cell Biol. 2007;9(11):1253–1262. doi: 10.1038/ncb1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sasaki T, et al. Phosphorylation regulates SIRT1 function. PLoS ONE. 2008;3(12):e4020. doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen D, et al. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22(13):1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aksoy P, White TA, Thompson M, Chini EN. Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun. 2006;345(4):1386–1392. doi: 10.1016/j.bbrc.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 36.Yang H, Lavu S, Sinclair DA. Nampt/PBEF/Visfatin: a regulator of mammalian health and longevity? Exp Gerontol. 2006;41(8):718–726. doi: 10.1016/j.exger.2006.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang H, et al. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130(6):1095–1107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim EJ, Kho JH, Kang MR, Um SJ. Active regulator of SIRT1 cooperates with SIRT1 and facilitates suppression of p53 activity. Mol Cell. 2007;28(2):277–290. doi: 10.1016/j.molcel.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 39.Kim JE, Chen J, Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451(7178):583–586. doi: 10.1038/nature06500. [DOI] [PubMed] [Google Scholar]
- 40.Zhao W, Kruse JP, Tang Y, Jung SY, Qin J, Gu W. Negative regulation of the deacetylase SIRT1 by DBC1. Nature. 2008;451(7178):587–590. doi: 10.1038/nature06515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rodgers JT, Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci U S A. 2007;104(31):12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Aksoy P, et al. Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun. 2006;349(1):353–359. doi: 10.1016/j.bbrc.2006.08.066. [DOI] [PubMed] [Google Scholar]
- 43.Revollo JR, Grimm AA, Imai S. The regulation of nicotinamide adenine dinucleotide biosynthesis by Nampt/PBEF/visfatin in mammals. Curr Opin Gastroenterol. 2007;23(2):164–170. doi: 10.1097/MOG.0b013e32801b3c8f. [DOI] [PubMed] [Google Scholar]
- 44.Fulco M, et al. Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell. 2008;14(5):661–673. doi: 10.1016/j.devcel.2008.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gomez-Lechon MJ, Donato MT, Martinez-Romero A, Jimenez N, Castell JV, O’Connor JE. A human hepatocellular in vitro model to investigate steatosis. Chem Biol Interact. 2007;165(2):106–116. doi: 10.1016/j.cbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 46.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120(4):473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 47.Nakahata Y, et al. The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell. 2008;134(2):329–340. doi: 10.1016/j.cell.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang T, et al. Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem. 2009;284(30):20408–20417. doi: 10.1074/jbc.M109.016469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziegler M. New functions of a long-known molecule. Emerging roles of NAD in cellular signaling. Eur J Biochem. 2000;267(6):1550–1564. doi: 10.1046/j.1432-1327.2000.01187.x. [DOI] [PubMed] [Google Scholar]
- 50.Ramsey KM, et al. Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science. 2009;324(5927):651–654. doi: 10.1126/science.1171641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beausoleil SA, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101(33):12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24(10):1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 53.Fu J, et al. Deleted in breast cancer 1, a novel androgen receptor (AR) coactivator that promotes AR DNA-binding activity. J Biol Chem. 2009;284(11):6832–6840. doi: 10.1074/jbc.M808988200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li Z, Chen L, Kabra N, Wang C, Fang J, Chen J. Inhibition of SUV39H1 methyltransferase activity by DBC1. J Biol Chem. 2009;284(16):10361–10366. doi: 10.1074/jbc.M900956200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Trauernicht AM, Kim SJ, Kim NH, Boyer TG. Modulation of estrogen receptor alpha protein level and survival function by DBC-1. Mol Endocrinol. 2007;21(7):1526–1536. doi: 10.1210/me.2007-0064. [DOI] [PubMed] [Google Scholar]
- 56.Malhi H, Bronk SF, Werneburg NW, Gores GJ. Free fatty acids induce JNK-dependent hepatocyte lipoapoptosis. J Biol Chem. 2006;281(17):12093–12101. doi: 10.1074/jbc.M510660200. [DOI] [PubMed] [Google Scholar]
- 57.Zang M, et al. AMP-activated protein kinase is required for the lipid-lowering effect of metformin in insulin-resistant human HepG2 cells. J Biol Chem. 2004;279(46):47898–47905. doi: 10.1074/jbc.M408149200. [DOI] [PubMed] [Google Scholar]
- 58.Barbosa MT, et al. The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J. 200721133629–3639. . [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.