Abstract
DNA methyltransferase 1 (DNMT1) catalyzes DNA methylation and is overexpressed in many human diseases, including cancer. The tobacco-specific carcinogen NNK also induces DNA methylation. However, the role of DNMT1-mediated methylation in tobacco carcinogenesis remains unclear. Here we used human and mouse lung cancer samples and cell lines to determine a mechanism whereby NNK induced DNMT1 expression and activity. We determined that in a human lung cell line, glycogen synthase kinase 3β (GSK3β) phosphorylated DNMT1 to recruit β-transducin repeat–containing protein (βTrCP), resulting in DNMT1 degradation, and that NNK activated AKT, inhibiting GSK3β function and thereby attenuating DNMT1 degradation. NNK also induced βTrCP translocation to the cytoplasm via the heterogeneous nuclear ribonucleoprotein U (hnRNP-U) shuttling protein, resulting in DNMT1 nuclear accumulation and hypermethylation of the promoters of tumor suppressor genes. Fluorescence immunohistochemistry (IHC) of lung adenomas from NNK-treated mice and tumors from lung cancer patients that were smokers were characterized by disruption of the DNMT1/βTrCP interaction and DNMT1 nuclear accumulation. Importantly, DNMT1 overexpression in lung cancer patients who smoked continuously correlated with poor prognosis. We believe that the NNK-induced DNMT1 accumulation and subsequent hypermethylation of the promoter of tumor suppressor genes may lead to tumorigenesis and poor prognosis and provide an important link between tobacco smoking and lung cancer. Furthermore, this mechanism may also be involved in other smoking-related human diseases.
Introduction
Epigenetic disorders give rise to several human diseases, including various cancers, neuron disorders, psychosis, and cardiovascular diseases, many of which are mediated by altered expression and activity of DNA methyltransferase 1 (DNMT1; refs. 1–8). DNMT1 is involved in DNA methylation (4). Cancer cells undergo changes in 5′-methylcytosine distribution, including global DNA hypomethylation and region-specific hypermethylation of promoter CpG islands associated with tumor suppressor genes (TSGs; ref. 9). Aberrant promoter hypermethylation of CpG islands associated with TSGs can lead to transcriptional silencing and result in tumorigenesis (4). DNMT1 is reported to be especially overexpressed in lung and liver cancer patients that are smokers (10, 11).
The key ingredient of tobacco smoke carcinogen, nitrosamine 4-(methylnitro-samino)-1-(3-pyridyl)-1-butanone (also known as nicotine-derived nitrosamine ketone; NNK) systemically induces tumors of the lung in rats, mice, and hamsters and also plays a major role in lung carcinogenesis (12, 13). As previously reported in mouse and rat studies, NNK exposure not only leads to gene mutation, but also induces hypermethylation of multiple TSG promoters in liver or lung tumors, such as cyclin-dependent kinase inhibitor 2A (p16, inhibits CDK4) (p16INK4A), death-associated protein kinase 1 (Dapk1), retinoic acid receptor β (Rarb), and runt-related transcription factor 3 (Runx) (14–17). Clinical studies indicated that smoking is associated with promoter hypermethylation at more than 20 TSGs in lung tumors (18, 19). However, the mechanism underlying the promoter hypermethylation of TSGs relevant to tobacco carcinogenesis has not been elucidated.
NNK has been shown to induce alterations in many signaling pathways, such as the EGFR, AKT, MAPK, ERK1/2, and NFκB pathways (13, 20–22). Therefore, we proposed that NKK induces overexpression of DNMTs through some signaling pathways, thereby leading to hypermethylation at multiple TSG promoters. Lung cancer is one of the most common cancers worldwide and is the leading cause of cancer mortality in industrialized countries (23). About 85%–90% of lung cancer cases are caused by cigarette smoking (12). Using lung cancer as a model, we performed cell, animal, and clinical studies to analyze the molecular mechanisms of DNMT1 overexpression in relation to NNK.
Results
DNMT1 overexpression strongly correlates with smoking status and poor prognosis of lung cancer patients.
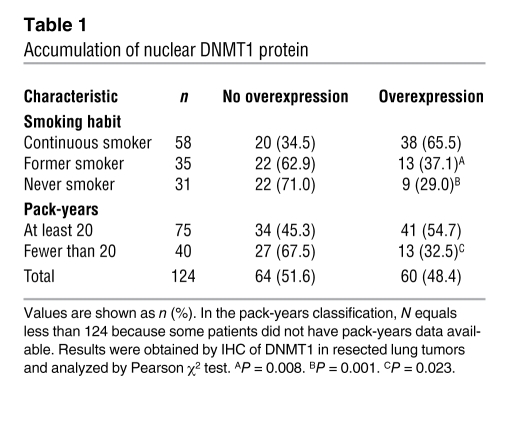
We examined whether DNMT1 expression correlated with the smoking status of lung cancer patients (see Methods for details of smoking habit and pack-year categorizations). IHC for DNMT1 protein expression was performed on 124 lung tumor samples. DNMT1 protein was highly expressed and localized in nuclei in tumor tissue compared with the surrounding nonneoplastic stroma tissue of patients that continuously smoked, but its nuclear expression level was relatively low in tumor tissue of patients that never smoked or formerly smoked (P = 0.001 and P = 0.008, respectively; Figure 1A and Table 1). DNMT1 protein was also highly expressed in patients with 20 or more pack-years of cigarette smoking compared with those with fewer than 20 pack-years (P = 0.023; Table 1).
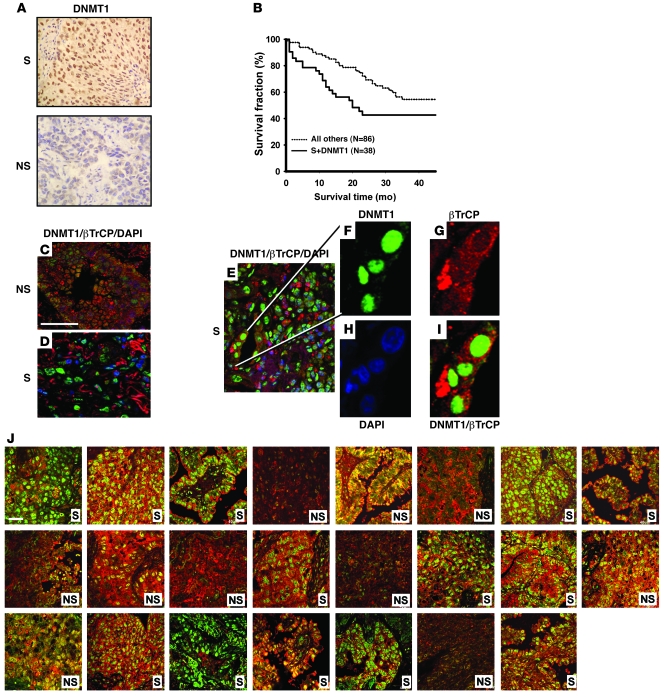
Figure 1. DNMT1 nuclear overexpression correlates with poor prognosis and shows mutually exclusive localization with βTrCP expression in lung cancer patients who continuously smoked.
(A) Representative IHC analysis of DNMT1 in paraffin sections from lung cancer patients who were smokers (S) and those who did not smoke (NS). Increased DNMT1 nuclear positive immunoreactivity was found in the patient who smoked. Original magnification, ×200. (B) Kaplan-Meier survival curves for overall postoperative survival showed that lung cancer patients who continuously smoked and had overexpression of DNMT1 protein had a worse prognosis compared with all other patient groups (P = 0.007, log-rank test). (C–I) Representative fluorescence IHC and confocal microscopy of DNMT1 (green), βTrCP (red), and DAPI (blue) in paraffin sections of tumors from lung cancer patients who did not (C) and did continuously smoke (D–I). βTrCP protein was mainly localized in the cytoplasm, which was distinct from the nuclear localization of DNMT1 protein in patients who smoked. Original magnification, ×630 (C–E); ×1,500 (F–I). Scale bar: 40 μm. (J) Representative figures of double fluorescence IHC for DNMT1 and βTrCP in a tissue array including 23 lung cancer patients. Original magnification, ×630. Scale bar: 40 μm.
Table 1 .
Accumulation of nuclear DNMT1 protein
In addition, Kaplan-Meier survival curves for postoperative overall survival showed that lung cancer patients who both continuously smoked and had DNMT1 protein overexpression had a worse prognosis than did all other lung cancer patients (P = 0.007; Figure 1B). A multivariate Cox regression model was used to analyze the association between poor survival with clinical parameter or molecular risk factor including age, smoking, tumor type, tumor stage, and DNMT1 overexpression. The combination of lung cancer, continuous smoking, and DNMT1 overexpression was identified as an independent prognostic factor for poor survival (P = 0.011; Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI40706DS1).
NNK increases DNMT1 protein expression and activity.
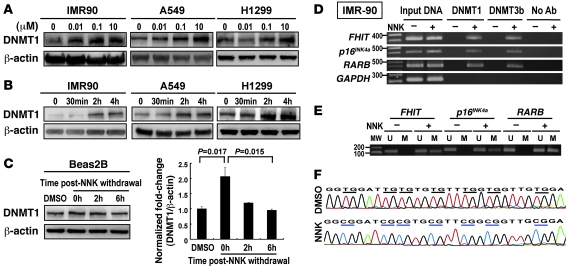
We examined the effect of NNK on DNMT1 expression in IMR90 normal lung cells and in the A549 and H1299 lung carcinoma cell lines. NNK effectively induced DNMT1 protein expression in both dose- and time-dependent manners (Figure 2, A and B). Because patients who formerly smoked showed less nuclear accumulation of DNMT1 protein than did patients who continuously smoked, the data suggested NNK-induced DNMT1 may be reversible. Therefore, we examined the effect of smoking cessation on DNMT1 protein levels in Beas-2B bronchial epithelial cells and found that DNMT1 protein returned to the basal (DMSO control) level 2–6 hours after discontinuation of NNK treatment (Figure 2C). We then investigated whether NNK-induced DNMT could concomitantly activate DNMT binding to TSG promoters and result in 5′CpG hypermethylation of the bound TSG promoters. Using chromatin immunoprecipitation–PCR (ChIP-PCR) analysis, we found that binding of DNMT1 to the tested TSG promoters was substantially increased by NNK (Figure 2D). In addition, induction of DNMT protein expression by NNK enhanced the methylation levels of these promoters in IMR90 cells, as measured by methylation-specific PCR (MSP) and bisulfite sequencing assays (Figure 2, E and F).
Figure 2. NNK-induced DNMT1 protein binds to promoters and results in hypermethylation of TSGs.
(A and B) Expression levels of DNMT1 in human lung cell lines were analyzed by Western blotting. NNK increased protein levels of DNMT1 in (A) dose-dependent and (B) time-dependent manners in the 3 cell lines tested. β-Actin antibody was used as an internal control. (C) DNM1 protein returned to basal levels 2–6 hours after discontinuation of NNK treatment in Beas-2B bronchial epithelial cells. (D) ChIP-PCR using DNMT1 and DNMT3b antibodies for amplification of FHIT, p16INK4a, and RARB promoters before and after NNK treatment for 24 hours in IMR90 cells. Binding of DNMT1 and DNMT3b proteins to their target TSG promoters was induced by NNK. (E and F) MSP for promoter hypermethylation status in the FHIT, p16INK4a, and RARB promoters (E) and bisulfite sequencing of the p16INK4a promoter (F) before and after NNK treatment for 48 hours in IMR90 cells. Hypermethylated genes were defined as those that produced amplified methylation products in MSP or CG-dinucleotide sequences in bisulfite sequencing assays. M, methylated; U, unmethylated.
NNK prolongs DNMT1 protein stability through AKT signaling, which is associated with the ubiquitin-proteosome system.
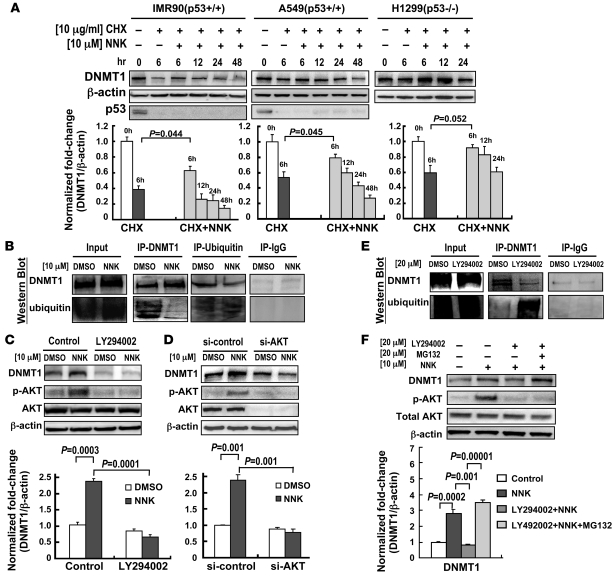
Interestingly, NNK did not affect the mRNA expression level of DNMT1 in cell and clinical models (Supplemental Figure 1 and Supplemental Table 2), which suggests that NNK-induced DNMT1 protein levels may result from protein stabilization. Therefore, we measured the changes of DNMT1 protein half-life upon NNK treatment in cells treated with the protein synthesis inhibitor cycloheximide (CHX), and found that NNK prolonged DNMT1 protein half-life compared with that of untreated cells (Figure 3A). Next, we examined whether the stabilization of NNK-induced DNMT1 was mediated through the ubiquitin-proteasome pathway. IP–Western blot showed reduced ubiquitination of DNMT1 protein after NNK treatment (Figure 3B).
Figure 3. NNK enhances DNMT1 protein stability through the AKT signaling pathway, which is associated with the ubiquitin-proteosome system.
(A) NNK prolonged DNMT1 protein half-life. DNMT1 protein levels were determined by Western blotting and densitometry. β-Actin was used as a loading control, and p53 served as a positive control for CHX treatment because its half-life is known to be short. White bars, cells treated with DMSO; dark gray bars, cells treated with CHX for 6 hours; light gray bars, cells treated with CHX and NNK for the indicated times. (B) Ubiquitination of DNMT1 decreased upon NNK treatment. A549 cells were treated with NNK or DMSO control for 2 hours, after which cell lysates were immunoprecipitated with anti-DNMT1 or anti-ubiquitin antibody and then Western blotted. Normal IgG was used as a negative control. (C and D) A549 cells were pretreated with or without the PI3K/AKT pathway inhibitor LY294002 (C) or AKT siRNA (D), then treated with NNK. Western blotting and densitometry showed that each reduced levels of NNK-induced DNMT1. (E) LY294002 treatment increased ubiquitination of DNMT1, leading to low levels of DNMT1 protein in A549 cells. (F) Western blotting and densitometry of A549 cells treated with NNK with or without pretreatment with LY294002 and the proteasome inhibitor MG132 showed altered DNMT1 protein levels. Data are mean ± SEM (n = 3).
Because the AKT pathway was notably stimulated by NNK in all cell lines tested (Supplemental Figure 2), and a previous study reported that the AKT pathway stabilizes DNMT1 protein via decreased ubiquitination (24), it is possible that DNMT1 protein stability is induced by NNK through the AKT pathway. To test this hypothesis, we first measured DNMT1 protein levels during NNK treatment in the presence of the PI3K/AKT pathway inhibitor LY294002, which we found to potently block NNK-induced DNMT1 protein overexpression in A549 cells (Figure 3C). We also found that AKT siRNA specifically reduced NNK-induced DNMT1 protein expression (Figure 3D). To verify that reduced ubiquitination of DNMT1 protein was mediated by AKT, we performed IP–Western blot in LY294002-treated A549 cells and found that LY294002 increased ubiquitination of DNMT1, leading to low DNMT1 protein levels (Figure 3E). In addition, the reduction of NNK-mediated DNMT1 stability by LY294002 was abolished by the proteasome inhibitor MG132 (Figure 3F). Similar results were observed in other cell lines (data not shown). Collectively, these results clearly indicate that AKT is a mediator induced by NNK to block the ubiquitin-proteasome degradation of DNMT1.
NNK activates AKT, then inhibits GSK3β/β-transducin repeat–containing protein–mediated protein degradation, leading to DNMT1 protein accumulation.
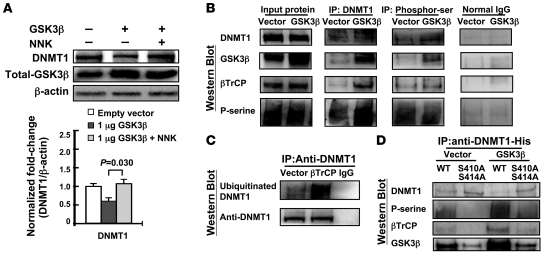
It has previously been shown that AKT inactivates GSK3β Ser/Thr kinase. GSK3β phosphorylates its substrate protein and recruits an E3-ubiqutin ligase, β-transducin repeat–containing protein (βTrCP), leading to substrate degradation (25, 26). Therefore, we investigated whether GSK3β is involved in NNK-induced DNMT1 protein stability. Knockdown of GSK3β in cells or treatment with a GSK3β inhibitor enhanced expression of DNMT1 protein (Supplemental Figure 3). Exogenous GSK3β promoted DNMT1 protein degradation in a dose-dependent manner in A549 cells (Supplemental Figure 4A), and this effect was attenuated after treatment with NNK (Figure 4A). Moreover, NNK treatment inactivated GSK3β, manifested by increased phosphorylation of GSK3β at Ser9 (Supplemental Figure 3A). Confocal microscopic analysis of immunofluorescent staining confirmed that expression levels of DNMT1 and the inactive form of GSK3β, p-GSK3βSer9, increased upon NNK treatment (Supplemental Figure 5). These data suggested that NNK-induced DNMT1 accumulation was mediated by inactivation of GSK3β. Using IP–Western blot, we demonstrated that exogenous GSK3β apparently interacted with DNMT1 and increased its phosphorylation (Figure 4B). In addition, exogenous GSK3β promoted binding of DNMT1 by βTrCP (Figure 4B). Because AKT phosphorylates GSK3β at Ser9 and inhibits the function of GSK3β, these results suggested the involvement of AKT/GSK3β/βTrCP pathway in DNMT1 protein degradation.
Figure 4. GSK3β and βTrCP interact with DNMT1 protein and enhance DNMT1 degradation.
(A) A549 cells were transfected with pCMV-SPORT6-GSK3β, then treated with or without NNK. The GSK3β pathway promoted DNMT1 protein degradation, which was attenuated by NNK treatment. Data are mean ± SEM (n = 3). (B) Cell lysates were immunoprecipitated with anti-DNMT1 or anti–phospho-Ser antibody and then Western blotted. DNMT1 protein formed complexes with GSK3β and βTrCP proteins. Increase DNMT1 phosphorylation was mediated by exogenous GSK3β, which increased the interaction between DNMT1 and βTrCP. Normal IgG was used as a negative control. (C) βTrCP increased the ubiquitination level of DNMT1. A549 cells were transfected with pCMV-SPORT6-βTrCP for 24 hours, after which cell lysates were immunoprecipitated with anti-DNMT1 and then western blotted. (D) Site-directed mutagenesis of both Ser410 (S410A) and Ser414 (S414A) on DNMT1 protein decreased the phosphorylation level of DNMT1 protein by GSK3β and disrupted the interaction between βTrCP and DNMT1. Cells were transfected with WT or mutant His-tag DNMT1 expression vector and exogenous GSK3β or vector control. Cell lysates were immunoprecipitated with anti–His-tag antibody and then Western blotted.
Interestingly, using the ScanProsite tool (http://www.expasy.ch/tools/scanprosite/), we found that DNMT1 protein peptides contained an ESGXXS sequence, similar to the E3-ubiquitin ligase βTrCP conserved binding motif DSGXXS (27). To determine whether βTrCP was involved in DNMT1 ubiquitination, A549 cells were transfected with exogenous βTrCP, and IP–Western blot showed that βTrCP strongly increased the ubiquitination level of DNMT1 (Figure 4C). In addition, exogenous βTrCP promoted DNMT1 protein degradation in a dose-dependent manner (Supplemental Figure 4B). Next, we examined whether the ESGXXS sequence of DNMT1 was necessary for βTrCP binding using site-directed mutagenesis of Ala replacement for both Ser410 and Ser414 on DNMT1 protein. These amino acid changes appeared to disrupt the interaction between βTrCP and DNMT1 (Figure 4D). To delineate the GSK3β-interacting domain of DNMT1 and the phosphorylation sites on DNMT1, we examined the phosphorylation efficiency of exogenous GSK3β in WT and mutant DNMT1 at Ser410 and Ser414. Mutant DNMT1 showed lower phosphorylation by GSK3β than did WT DNMT1, suggestive of Ser410 and Ser414 phosphorylation of DNMT1 by GSK3β (Figure 4D). Together, these results strongly suggest that GSK3β may phosphorylate DNMT1 at Ser410 and Ser414 and recruit the E3-ubiqutin ligase βTrCP, leading to proteasomal degradation of DNMT1.
NNK treatment enhances AKT downstream proteins, promotes hnRNP-U/βTrCP translocation to the cytoplasm, and induces DNMT1 accumulation in the nucleus.
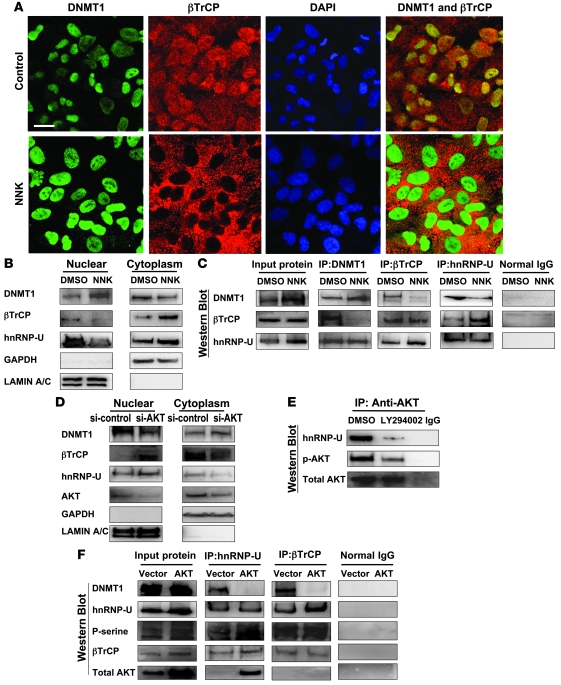
To further confirm the interaction and expression of DNMT1 and βTrCP, we performed immunofluorescence confocal microscopy. In untreated cells, DNMT1 was located predominantly in the nucleus, and βTrCP was seen in both nucleus and cytoplasm. Importantly, mutually exclusive localization of DNMT1 and βTrCP, along with increased nuclear DNMT1 levels, were observed in cells treated with NNK (Figure 5A), which suggests that NNK induced E3-ubiqutin ligase βTrCP translocation to cytoplasm, thus causing DNMT1 accumulation in the nucleus. In addition, cell fractionation assay showed that NNK-induced βTrCP protein was decreased in the nucleus and increased in the cytoplasm, the inverse of the DNMT1 staining (Figure 5B).
Figure 5. NNK enhances hnRNP-U/βTrCP translocation to the cytoplasm and induces DNMT1 accumulation in the nucleus, but this effect is attenuated by AKT inhibition.
(A) Immunofluorescence confocal microscopy to localize DNMT1 (green), βTrCP (red), and nucleus (blue; DAPI). The merged figures showed colocalization of DNMT1 with βTrCP protein in A549 cells treated with DMSO control. Mutually exclusive localization of DNMT1 and βTrCP was observed in cells treated with NNK. Original magnification, ×630. Scale bar: 20 μm. (B) Cell fractionation assay to analyze distribution of nuclear and cytoplasmic proteins, using anti-DNMT1, anti-βTrCP, anti–hnRNP-U, and anti-GSK3βSer9 antibodies. Anti-GAPDH and anti–lamin A/C antibodies were used as cytoplasmic and nuclear protein markers, respectively. The distribution of βTrCP and hnRNP-U proteins was contrary to that of DNMT1 protein. (C) DNMT1 and βTrCP interaction was interrupted by NNK treatment. In contrast, βTrCP and hnRNP interaction was increased by NNK treatment. (D) Cell fractionation assay was performed in A549 cells with or without AKT siRNA. Inhibition of AKT induced both hnRNP-U and βTrCP proteins predominantly located in the nucleus, where DNMT1 protein level decreased. (E) IP assay using anti-AKT antibody showed that hnRNP formed a complex with AKT, and the interaction was attenuated by LY294002. (F) An increase of hnRNP-U phosphorylation was mediated by exogenous AKT, which increased the interaction between hnRNP-U and βTrCP. In addition, AKT decreased the interaction of DNMT1 with hnRNP-U and βTrCP.
To determine how βTrCP was transported from nucleus to cytoplasm, we studied the shuttling protein hnRNP-U, which was previously reported to interact with βTrCP and account for its nuclear localization (27). IP–Western blot assays showed that DNMT1 indeed interacted with hnRNP-U and βTrCP in the untreated cells. However, βTrCP/DNMT1 interaction was weakened after treatment with NNK. In addition, binding between hnRNP-U and βTrCP was enhanced in cells treated with NNK (Figure 5C). Importantly, the localization and expression level of hnRNP-U decreased in the nucleus and increased in the cytoplasm after NNK treatment, as assessed by cell fractionation assay (Figure 5B). Together, these data strongly suggest that hnRNP-U shuttling protein mediates βTrCP translocation from the nucleus to the cytoplasm by NNK treatment, leading to DNMT1 accumulation in the nucleus.
To determine whether AKT plays a role in hnRNP-U/βTrCP nucleocytoplasmic shuttling, AKT was knocked down by AKT siRNA in cells and used for cell fractionation assay. In AKT knockdown cells, both hnRNP-U and βTrCP proteins were predominantly located in the nucleus, where DNMT1 protein level was decreased (Figure 5D). In addition, IP–Western blot assay demonstrated that hnRNP-U formed a complex with AKT, and the interaction was attenuated by LY294002 (Figure 5E). Importantly, NNK-induced AKT increased phosphorylation of hnRNP-U and interaction between hnRNP-U and βTrCP, whereas it decreased the interaction of DNMT1 with βTrCP and hnRNP-U (Figure 5F). These results firmly support involvement of the PI3K/AKT pathway in hnRNP-U/βTrCP nucleocytoplasmic shuttling and DNMT1 accumulation in the nucleus.
NNK induces DNMT1, p-AKTser473, p-GSK3βser9, cytoplasmic hnRNP-U, and cytoplasmic βTrCP protein expression level in mouse lung adenoma tissues.
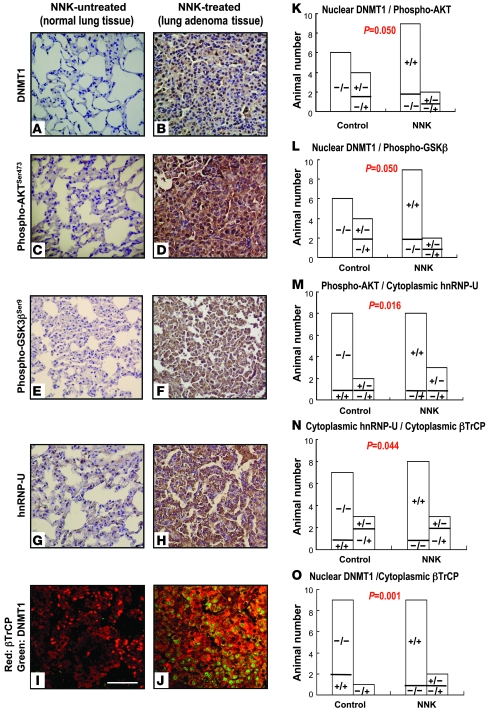
To further confirm the finding of NNK-induced DNMT1 by AKT/GSK3β/βTrCP/hnRNP-U in an animal model, we treated 11 A/J mice with NNK. Normal lung tissues of untreated mice and adenoma lung tissues of NNK-treated mice were examined for protein expression and localization of DNMT1, p-AKTSer473, p-GSK3βSer9, hnRNP-U, and βTrCP using IHC, double fluorescence IHC, and confocal microscopy (Figure 6, A–J). Protein expression levels of DNMT1, p-AKTSer473, and p-GSK3βSer9 were concordantly increased in lung adenoma tissue of NNK-treated mice (Figure 6, B, D, and F). High cytoplasmic expression of hnRNP-U and βTrCP proteins was also observed (Figure 6, H and J). Concordance analysis showed coincident cytoplasmic expression of hnRNP-U and βTrCP proteins in lung adenoma tissue of NNK-treated mice (P = 0.044; Figure 6N). We also found high staining of nuclear DNMT1 (P = 0.050; Figure 6K) and cytoplasmic hnRNP-U (P = 0.016; Figure 6M) when p-AKTSer473 staining was high. In addition, expression of p-GSK3βSer9 (P = 0.050; Figure 6L) and cytoplasmic βTrCP (P = 0.001; Figure 6O) was associated with nuclear DNMT1 expression in all mice analyzed. The mutually exclusive localization of DNMT1 and βTrCP observed in lung adenoma of NKK-treated mice compared with normal lung tissue of untreated mice (Figure 6, J and I) confirming in an animal model that NNK induced E3-ubiqutin ligase βTrCP translocation to the cytoplasm, leading to DNMT1 accumulation in the nucleus.
Figure 6. Representative IHC and fluorescence IHC of mouse tissue slides.
IHC and fluorescence IHC were performed on paraffin sections of normal lung tissue from untreated mice and lung adenoma of NNK-treated mice. Protein expression levels of nuclear DNMT1 (A and B), p-AKTSer473 (C and D), the inactive form of p-GSK3βSer9 (E and F), cytoplasmic hnRNP (G and H), and cytoplasmic βTrCP (red, I and J; merged image with DNMT1, green) increased in lung adenoma tissue of NNK-treated mice compared with the normal lung tissue of untreated mice. Original magnification, ×200 (A–H); ×630 (I and J). Scale bar: 40 μm. (K–O) Concordance analysis by Pearson c2 test showed a strong correlation between nuclear DNMT1 and p-AKTSer473 (K), p-GSK3βSer9 (L), and cytoplasmic βTrCP (O); between p-AKTSer473 and cytoplasmic hnRNP (M); and between cytoplasmic hnRNP and cytoplasmic βTrCP (N) in control and NNK-treated mice. Symbols within bars denote expression of the respective proteins; for example, “+/–” in K denotes mice with positive nuclear DNMT1 expression and negative p-AKTSer473 expression.
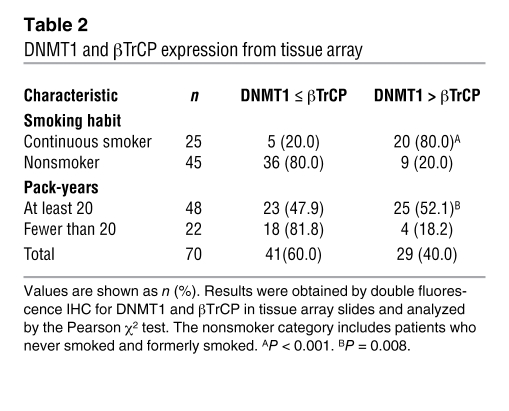
DNMT1 and βTrCP interaction is disrupted in lung tumor tissue of patients who smoked.
To determine whether the localization of DNMT1 and βTrCP proteins differs between lung cancer patients who did and did not smoke, we analyzed the protein expression and localization of DNMT1 and βTrCP in lung tumor tissue from smoker and nonsmoker patients by double fluorescence IHC and confocal microscopy in tissue array from 70 lung cancer patients. In tumors from patients who did not smoke, DNMT1 colocalized with βTrCP in the nucleus and cytoplasm (yellow-brown; Figure 1C). However, βTrCP protein (red) was mainly localized in the cytoplasm, which was distinct from the nuclear localization of DNMT1 protein (green), in patients who smoked (Figure 1, D–I), confirming that mutually exclusive localization of DNMT1 and βTrCP led to DNMT accumulation in tumors of patients who smoked. Statistical analysis indicated that the expression of nuclear DNMT1 was higher than nuclear βTrCP in patients who smoked compared with those who did not (Figure 1J and Table 2; P < 0.001), as well as in patients with at least 20 pack-years of cigarette smoking compared with those with fewer than 20 pack-years (Table 2; P = 0.008). These data are in agreement with the findings in NNK-treated cell and animal models.
Table 2 .
DNMT1 and βTrCP expression from tissue array
Discussion
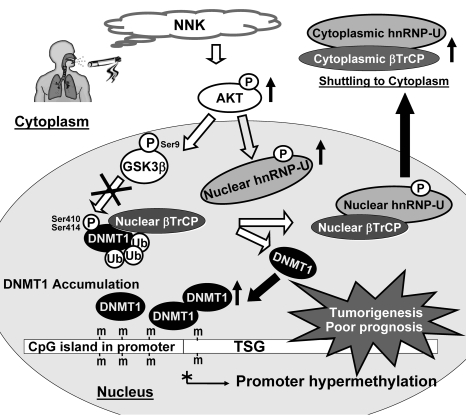
Epigenetic disorders give rise to several significant human diseases, many of which are mediated by altered DNMT1 expression and activity (1–6). In mouse and rat studies, exposure to the tobacco-specific carcinogen NNK not only leads to liver or lung tumor formation, but also induces hypermethylation of multiple TSG promoters (14–17). Unraveling how NNK induces DNMT1-mediated promoter hypermethylation will help us understand the process of carcinogenesis and may also reveal the etiology of other human diseases. Using cell, animal, and clinical models, we identified that DNMT1 was phosphorylated by GSK3β and that DNMT1 formed complexes with GSK3β, βTrCP, and hnRNP-U proteins. In addition, our study revealed what we believe to be a new mechanism of NNK-induced DNMT1 protein stability by AKT/hnRNP-U/βTrCP nucleocytoplasmic shuttling. Furthermore, the NNK-induced DNMT1 proteins bound to promoters of various TSGs and caused promoter hypermethylation of the bound TSGs, which ultimately led to tumorigenesis and poor prognosis in clinical manifestation (Figure 7).
Figure 7. Proposed model to illustrate the accumulation of nuclear DNMT1 by NNK-induced AKT/GSK3β/βTrCP/hnRNP-U signaling leading to promoter hypermethylation and tumorigenesis.
NNK induces DNMT1 protein accumulation in the nucleus through AKT/GSK3β/βTrCP signaling and AKT/hnRNP-U/βTrCP nucleocytoplasmic shuttling. βTrCP is an E3 ubiquitin ligase that specifically interacts with and degrades DNMT1. NNK induces activation of AKT, then promotes GSK3β phosphorylation at Ser9 to form inactive GSK3β, which subsequently attenuates the ability of βTrCP to degrade DNMT1 protein. In addition, NNK activates AKT to induce interaction between phosphorylated hnRNP-U and βTrCP, which disrupts βTrCP/DNMT1 interaction. The hnRNP-U/βTrCP complex translocates to the cytoplasm, leading to DNMT1 accumulation in the nucleus. Furthermore, these NNK-induced DNMT1 proteins bind to promoters of various TSGs and result in promoter hypermethylation, which ultimately leads to tumorigenesis and poor prognosis.
It was previously shown that AKT inactivates GSK3β, which is also known to phosphorylate several substrates to create a recognition motif for βTrCP and lead to degradation of substrate protein via the ubiquitin-proteasome pathway (25). Our data demonstrated that DNMT1 protein indeed formed a complex with GSK3β and βTrCP (Figure 4, B and E). In addition, inhibition of GSK3β during NNK treatment increased DNMT1 protein levels in both cell (Supplemental Figure 3) and animal models (Figure 6). Exogenous GSK3β expression coincided with DNMT1 phosphorylation increase and binding with βTrCP (Figure 4B). However, mutation at Ser410 and Ser414 abolished the phosphorylation of DNMT1 by GSK3β and DNMT1/βTrCP interaction, leading to increased DNMT (Figure 4E). Interestingly, Ser410 and Ser414 of DNMT1 are located at a peptide containing ESGXXS sequence, which is similar to the βTrCP conserved binding motif DSGXXS. In addition, SXXXS is the recognition and phosphorylation site by GSK3β (28). Mutation at Ser410 and Ser414 attenuated GSK3β/DNMT1 interaction (Figure 4E). These findings strongly suggest that GSK3β phosphorylates DNMT1, at least at Ser410 and Ser414, to facilitate recruitment of βTrCP and cause proteasomal degradation of DNMT1.
To search for the βTrCP-interacting domain on DNMT1, we examined the induction level by NNK and βTrCP interaction ability of 3 cDNA deletion constructs of DNMT1, containing amino acids 1–585, 585–1,128, or 1,116–1,632 of DNMT1. The N-terminal domain of 1–585 amino acids was induced by NNK treatment (Supplemental Figure 6A). This region was also important for βTrCP interaction, as shown by IP-Western assay (Supplemental Figure 6B). These findings are in agreement with the notion that DNMT1 protein accumulation was dependent on the N-terminal 120 amino acids, which seemed to function as a destruction domain in cells causing ubiquitination and proteasome-mediated degradation (29). We will further determine whether this region can be ubiquitinated by βTrCP.
A previous study showed that hnRNP-U interacted with βTrCP and was responsible for its nuclear localization (27). Our data also indicated that hnRNP-U interacted with βTrCP and DNMT1. In addition, both hnRNP-U and βTrCP were translocated to the cytoplasm, whereas DNMT1 was predominately located in the nucleus, in cells treated with NNK (Figure 5) and in lung adenoma tissue of NNK-treated mice (Figure 6). Our data suggest a mechanism whereby phosphorylated hnRNP-U mediated by AKT activation (Figure 5, E and F) disrupts the DNMT1/βTrCP interaction and leads to DNMT1 nuclear accumulation after NNK treatment. Whether nuclear exportation of βTrCP after NNK treatment is exclusively mediated by hnRNP-U or occurs in cooperation with other shuttling proteins will be investigated in the future. In addition, the phosphorylated domain of hnRNP-U that modulates the βTrCP interaction needs to be identified.
Our study provides cell, animal, and clinical evidence that NNK induces DNMT1 through the AKT/GSK3β/βTrCP/hnRNP-U pathway and elucidates a mechanism involved in smoking-related lung cancer. In support of our present study is the prior finding that active Akt is detected in airway epithelial cells and lung tumors from NNK-treated A/J mice as well as in human lung cancers derived from patients who smoked (30). However, other pathways that regulate NNK-induced DNMT protein levels, such as NFκB signaling (Supplemental Figure 2), may be important. In addition, NNK is a β-adrenergic receptor (β-AR) agonist that phosphorylates EGFR in lung cells (22). EGFR activation triggers the PI3K/AKT pathway in lung, head and neck, and pancreatic cancers (31). It will be interesting to examine whether β-AR– and EGFR-mediated AKT activation is involved in NNK-induced DNMT protein. It will also be worthwhile to extend our finding to determine whether NNK induces other DNMTs, such as DNMT3b, through a similar mechanism (Figure 2).
That DNMT1 overexpression may serve as a prognostic factor in smoking-related lung cancer is an important finding. Patients who formerly or never smoked showed a better prognosis than did patients who continuously smoked (Figure 1). Interestingly, we found that tumors from patients who quit smoking before receiving surgery showed significantly lower nuclear accumulation of DNMT1 protein compared with tumors from patients who continuously smoked (Table 1), although there was no correlation between time since smoking cessation and DNMT1 protein expression level. In addition, withdrawal of NNK treatment reversed the accumulated DNMT1 to the basal level within 2–6 hours in the cell model (Figure 2C). The findings of our study support patient care options, such as quitting smoking, that we expect to restore DNMT1 function and improve survival. In addition, drugs aimed at DNMT protein depletion, such as AKT inhibitor or a demethylation reagent, may prove a good therapeutic strategy for cancer treatment. Whether the amount of nuclear accumulation of DNMT1 in adenoma of animal models and/or in patient tumors of a clinical study correlate dose-dependently with NNK treatment and cigarette exposure is worthy of investigation. Collectively, the results of our present study shed light on the relevance of DNMT overexpression in tobacco carcinogenesis. NNK-induced DNMT1 accumulation by AKT/GSK3β/βTrCP/hnRNP-U signaling pathway was not due to the effect of NNK treatment on the cell cycle (Supplemental Figure 7). Such a mechanism may also be important for other types of cancer and human disease.
Methods
Clinical characterization of patients.
Tumor lung tissues were obtained from 124 lung cancer patients recruited at the Taipei and Taichung Veterans General Hospitals between 1993 and 2004, after appropriate institutional review board permission and informed patient consent were secured. Clinical parameters are provided in Supplemental Table 3. Among these patients, paraffin blocks of tumor tissue from 70 patients were made into tissue array. Information on the smoking history of the patients was obtained from hospital records. Patients were categorized as those who did not smoke (never smoked and formerly smoked) and those who continuously smoked (both regular and occasional continuous smoking). Patients were also categorized by smoking history based on pack-years, calculated as the number of packs smoked per day multiplied by the number of years smoked. The 2 groups in this categorization smoked 20 or more pack-years or fewer than 20. Followup of 124 patients was performed at 2-month intervals in the first year after surgery and at 3-month intervals thereafter at outpatient clinics or by routine phone calls. The end of the followup period was December 2008 for all patients. Overall survival was calculated from the date of surgery to the date of death or of the final followup. The mean followup period was 35 months (range, 1–61 months).
Animal and NNK treatment.
Female A/J mice were acquired from the animal center of National Cheng Kung University Medical College after appropriate institutional review board permission was obtained. Mice (8 weeks old) were randomly divided into 2 groups, one treated i.p. with 2 mg NNK (Toronto Research Chemicals) in 0.1 ml saline per mouse, and the other with saline only as control. The experiment was terminated 24 weeks after treatment; all surviving mice were sacrificed under ether anesthesia. Normal lung and adenoma lung tissue samples were resected, fixed with formalin, embedded in paraffin for histological examination, and stained with H&E for microscopic evaluation by a pathologist.
Cell culture and NNK in combination with different inhibitor treatments.
IMR90 and Beas-2B normal lung cell lines and A549 and H1299 lung cancer cell lines were used in the current study. These lung cell lines were cultured at 37°C, 5% CO2, in an incubator with DMEM (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin-streptomycin. For the dose-dependent study, cells were treated with NNK at indicated doses for 2 hours at 37°C. For the time-course study, cells were treated with 10 μM NNK for the indicated times at 37°C. To examine the effect of smoking cessation on DNMT1 protein levels in Beas-2B bronchial epithelial cells, cells were treated with 10 μM NNK for 6 hours, which was then replaced with fresh medium without NNK for the indicated times.
Other inhibitor compounds, including the translational inhibitor CHX, the proteosome inhibitor MG132, and the specific GSK3β inhibitor SB415286, were purchased from Sigma-Aldrich. The PIK3/AKT pathway inhibitor LY294002 was obtained from Calbiochem. To examine the effect of NNK on DNMT1 protein half-life in lung cells, cells were treated with 10 μg/ml CHX for 6 hours, then with 10 μM NNK for 6–48 hours. For AKT and degradation pathway analysis, cells were treated with 10 μM NNK, 20 μM LY294002, and 20 μM MG132, individually or together. For GSK3β inhibitor treatment, cells were treated with 25 μM SB415286 with or without 10 μM NNK, then subjected to Western blot analysis.
Antibodies.
The antibodies used included DNMT1, AKT, phospho–AKTSer473, NFκB, phospho-NFκB, GSK3β, phospho-GSK3βSer9, βTrCP, Erk1/2, phospho-Erk1/2, ubiquitin, phospho-p38, p53, hnRNP-U, His, lamin A/C, GAPDH, and β-actin. A complete list of antibodies used and the reaction conditions in the present study are shown in Supplemental Table 4.
IHC assay for tissues from animals and patients.
Normal lung and adenoma lung tissues from mice exposed to NNK were analyzed using IHC assay to detect the expression levels of DNMT1, p-AKTSer473, p-GSK3βSer9, βTrCP, and hnRNP-U protein. Routinely formalin-fixed tissues were incubated with primary antibody, biotinylated secondary antibody, and finally with streptavidin–horseradish peroxidase for 20 minutes (DAKO LSAB kit K0675). The chromagen used was 3,3′-diaminobenzidine tetrahydrochloride.
Lung tumor sections from the 124 lung cancer patients were also analyzed with IHC using the polyclonal anti-DNMT1 antibody to detect DNMT1 protein expression. The evaluation of IHC staining was conducted blindly, without prior knowledge of the clinical and pathologic characteristics of the cases. The surrounding nonneoplastic stroma served as an internal negative control for each slide. Staining for nuclear DNMT1 and for cytoplasmic p-AKTSer473, p-GSK3ser9, βTrCP, and hnRNP-U was scored as follows: 0, negative; 1, low; 2, medium; 3, strong. A score of 2 or 3 in greater than 50% of the stained area indicated overexpression.
Double fluorescence IHC and confocal microscopic analysis in cultured cells and animal and patient tissue.
Immunofluorescence and confocal microscopic analysis of DNMT1, βTrCP, and DAPI were performed in A549 cells with or without NNK treatment, in paraffin sections of lung adenoma from NNK-treated mice and normal lung tissue from untreated mice, and in tissue array of 70 tumors from lung cancer patients who did and did not continuously smoke. Sections were incubated with primary antibody followed by incubation with the fluorescent secondary antibodies (Invitrogen) and DAPI for 1 hour, after which images were obtained with a confocal microscope (FV1000; Olympus).
Plasmid, siRNA, and transfection.
pOTB7-GSK3β and pCMV-SPORT6-βTrCP were purchased from Open Biosystems. pOTB7-GSK3β was subcloned into pCMV-SPORT6 vector. At 24 hours after transfection, proteins were extracted and then immunoblotted. AKT in cells was depleted by siRNA treatment. The AKT and GSK3β siRNA duplexes (see Supplemental Table 5) were obtained from Invitrogen, and transfection was performed with Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocols. At 48 hours after transfection, cells were analyzed by Western blotting.
Protein extraction and Western blot analysis.
Cells were lysed and centrifuged. Samples containing equal amounts of protein (50 μg) were separated by 6% SDS-PAGE and electroblotted onto Immobilon-P membranes (Millipore Co.). Immunoblotting was performed using antibodies against β-actin as a loading control. Each Western blot analysis was repeated at least 3 times. Quantification was performed with MultiGauge software to obtain the mean and SD values.
Cell fractionation assay.
Cells were lysed in membrane lysis buffer (10 mM HEPES, pH 8.0; 1.5 mM MgCl2; 10 mM KCl; 1.0 mM DTT; and 1% Igepal CA-630), then vortexed and centrifuged. The supernatant was the cytoplasmic fraction. Pellets were resuspended in nuclear lysis buffer (20 mM HEPES, pH 8.0; 1.5 mM MgCl2; 0.2 mM EDTA; 420 mM NaCl; 1.0 mM DTT; and 25% glycerol), then sonicated twice on ice and centrifuged. The supernatant was the nuclear fraction. For immunoblot analysis, 50 μg protein from either the nuclear fraction or the cytosolic fraction was used.
Immunoprecipitation assay.
Catch and Release Reversible Immunoprecipitation System kit (Millipore) was used for protein-protein interaction analysis. Cell protein lysates (1 mg) were incubated with the appropriate antibodies or normal mouse IgG. Affinity ligand was added (10 μl), and wash buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 20 mM α-glycerol-phosphate; 1% NP-40; and 5 mM EDTA) was added to a final volume of 500 μl. After incubation at 4°C overnight, immune complexes were washed thrice with wash buffer. Proteins were eluted by boiling in sample buffer, separated by 6% SDS-PAGE, then blotted with appropriate antibodies.
ChIP-PCR assay.
Cells were crosslinked with 1% formaldehyde for 15 minutes at 37°C and stopped by the addition of glycine to a final concentration of 0.125 M. Lysates were sonicated on ice to shear DNA to lengths between 200 and 800 bp. Subsequent steps were performed with the ChIP assay kit (Millipore) according to the manufacturer’s instructions. Chromatin was immunoprecipitated for 16 hours at 4°C using the DNMT1 antibody (Supplemental Table 4). FHIT, p16INK4a, and RARB primers used for PCR are listed in Supplemental Table 5.
MSP and bisulfite sequencing assays.
The promoter methylation status of the FHIT, p16INK4a, and RARB genes were determined by chemical treatment with sodium bisulfite, MSP analysis, and bisulfite sequencing assay. Primer sequences are shown in Supplemental Table 5. All PCRs were performed with positive controls for both unmethylated and methylated alleles and no DNA control during MSP assay. Hypermethylated genes were defined as those that produced amplified methylation products in MSP or CG-dinucleotide sequences in bisulfite sequencing assays.
Semiquantitative RT-PCR assay.
Expression levels of DNMT1 were detected by RT-PCR assay using β-actin as an internal control. The primers used and their PCR conditions are shown in Supplemental Table 5. The number of cycles, primer amount, and cDNA used were determined to provide quantitative amplification during RT-PCR. The average level of mRNA for DNMT genes in tumor lung tissues was 1.3- to 1.4-fold higher than that in normal lung tissues. To improve the specificity of analysis, we considered tumors with 1.5-fold higher expression than that of normal cells to have an abnormal pattern.
Flow cytometry and cell cycle synchronization assays.
Cell cycle distribution was determined by flow cytometry. A549 cells (2 × 106) were incubated in medium containing 10% FBS and treated with DMSO or 10 μM NNK for 24 hours. Cells were trypsinized and fixed with 70% ethanol overnight at –20°C. Fixed cells were stained with a solution containing 20 μg/ml propidium iodide, 200 μg/ml RNase A, and 0.1% Triton X-100 for 20 minutes in the dark. Cell cycle distribution was performed by FACScan flow cytometry (BD Biosciences) and calculated using ModFIT LT version 2.0 software (BD Biosciences). To induce specific cell cycle in A549 cells, M phase was enriched by 24 hours of treatment with 10 μM nocodazole (Sigma-Aldrich), G1 phase was induced 5 hours after nocodazole release, and S phase was induced by treatment with 10 μg/ml aphidicolin (Sigma-Aldrich) for 24 hours.
Statistics.
SPSS (SPSS Inc.) was used for all statistical analysis. Pearson χ2 test was used to compare the frequency of DNMT1 and βTrCP alterations between lung cancer patients with different smoking statuses and pack-years and to study relationships between variables in tumor samples. Pearson χ2 test was also used to compare the relationship among DNMT1, p-AKTSer473, p-GSK3βSer9, cytoplasmic hnRNP-U, and cytoplasmic βTrCP protein expression levels in mouse lung tissues with or without NNK treatment. To analyze the difference of DNMT1 protein levels among various treatments in cell model studies, 2-tailed Student’s t test was used. Type III censoring was performed on subjects who were still alive at the end of the study. Survival curves were calculated by the Kaplan-Meier method, and comparison was performed by the log-rank test. A P value less than 0.05 was considered statistically significant. A Cox proportional hazards regression model was used for the multivariable analysis of independent prognostic factors for survival.
Supplementary Material
Acknowledgments
This work was supported in part by grant NSC95-2320-B-006-087-MY3 from the National Science Council, grant DOH98-TD-G-111-024 from the Department of Health, grant 98-EC-17-A-19-S2-0069 from Ministry of Economic Affairs, and grant EO-097-PP-03 from National Health Research Institute (The Executive Yuan, Republic of China). The authors thank Ying-Jan Wang (Department of Environmental and Occupational Health, Medical College of National Cheng Kung University) for assistance with NNK treatment of A/J mice and Li-June Juan (Genomic Research Center, Academia Sinica, Taipei, Republic of China) for help with DNMT cDNA expression vector constructions.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2010;120(2):521–532. doi:10.1172/JCI40706.
References
- 1.Jamaluddin MS, Yang X, Wang H. Hyperhomocysteinemia, DNA methylation and vascular disease. Clin Chem Lab Med. 2007;45(12):1660–1666. doi: 10.1515/CCLM.2007.350. [DOI] [PubMed] [Google Scholar]
- 2.Costa E, Dong E, Grayson DR, Guidotti A, Ruzicka W, Veldic M. Reviewing the role of DNA (cytosine-5) methyltransferase overexpression in the cortical GABAergic dysfunction associated with psychosis vulnerability. Epigenetics. 2007;2(1):29–36. doi: 10.4161/epi.2.1.4063. [DOI] [PubMed] [Google Scholar]
- 3.Kanai Y. Alterations of DNA methylation and clinicopathological diversity of human cancers. Pathol Int. 2008;58(9):544–558. doi: 10.1111/j.1440-1827.2008.02270.x. [DOI] [PubMed] [Google Scholar]
- 4.Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature. 2004;429(6990):457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 5.Abdolmaleky HM, et al. Methylomics in psychiatry: Modulation of gene-environment interactions may be through DNA methylation. Am J Med Genet B Neuropsychiatr Genet. 2004;127B(1):51–59. doi: 10.1002/ajmg.b.20142. [DOI] [PubMed] [Google Scholar]
- 6.Mastroeni D, Grover A, Delvaux E, Whiteside C, Coleman PD, Rogers J. Epigenetic changes in Alzheimer’s disease: Decrements in DNA methylation. Neurobiol Aging. doi: 10.1016/j.neurobiolaging.2008.12.005. [published online ahead of print December 29, 2008]. doi:10.1016/j.neurobiolaging.2008.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rhee I, et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416(6880):552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 8.Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J Biol Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- 9.Laird PW. Cancer epigenetics. Hum Mol Genet. 2005;14(Spec No 1):R65–R76. doi: 10.1093/hmg/ddi113. [DOI] [PubMed] [Google Scholar]
- 10.Lin RK, Hsu HS, Chang JW, Chen CY, Chen JT, Wang YC. Alteration of DNA methyltransferases contributes to 5’CpG methylation and poor prognosis in lung cancer. Lung Cancer. 2007;55(2):205–213. doi: 10.1016/j.lungcan.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 11.Hammons GJ, et al. Increased expression of hepatic DNA methyltransferase in smokers. Cell Biol Toxicol. 1999;15(6):389–394. doi: 10.1023/A:1007658000971. [DOI] [PubMed] [Google Scholar]
- 12.Hecht SS. Cigarette smoking and lung cancer: chemical mechanisms and approaches to prevention. Lancet Oncol. 2002;3(8):461–469. doi: 10.1016/S1470-2045(02)00815-X. [DOI] [PubMed] [Google Scholar]
- 13.Akopyan G, Bonavida B. Understanding tobacco smoke carcinogen NNK and lung tumorigenesis. Int J Oncol. 2006;29(4):745–752. [PubMed] [Google Scholar]
- 14.Pulling LC, Klinge DM, Belinsky SA. p16INK4a and beta-catenin alterations in rat liver tumors induced by NNK. Carcinogenesis. 2001;22(3):461–466. doi: 10.1093/carcin/22.3.461. [DOI] [PubMed] [Google Scholar]
- 15.Vuillemenot BR, Hutt JA, Belinsky SA. Gene promoter hypermethylation in mouse lung tumors. Mol Cancer Res. 2006;4(4):267–273. doi: 10.1158/1541-7786.MCR-05-0218. [DOI] [PubMed] [Google Scholar]
- 16.Hutt JA, et al. Life-span inhalation exposure to mainstream cigarette smoke induces lung cancer in B6C3F1 mice through genetic and epigenetic pathways. Carcinogenesis. 2005;26(11):1999–2009. doi: 10.1093/carcin/bgi150. [DOI] [PubMed] [Google Scholar]
- 17.Pulling LC, Vuillemenot BR, Hutt JA, Devereux TR, Belinsky SA. Aberrant promoter hypermethylation of the death-associated protein kinase gene is early and frequent in murine lung tumors induced by cigarette smoke and tobacco carcinogens. Cancer Res. 2004;64(11):3844–3848. doi: 10.1158/0008-5472.CAN-03-2119. [DOI] [PubMed] [Google Scholar]
- 18.Kim JS, Kim H, Shim YM, Han J, Park J, Kim DH. Aberrant methylation of the FHIT gene in chronic smokers with early stage squamous cell carcinoma of the lung. Carcinogenesis. 2004;25(11):2165–2171. doi: 10.1093/carcin/bgh217. [DOI] [PubMed] [Google Scholar]
- 19.Piperi C, Vlastos F, Farmaki E, Martinet N, Papavassiliou AG. Epigenetic effects of lung cancer predisposing factors impact on clinical diagnosis and prognosis. J Cell Mol Med. 2008;12(5A):1495–1501. doi: 10.1111/j.1582-4934.2008.00309.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tsurutani J, et al. Tobacco components stimulate Akt-dependent proliferation and NFkappaB-dependent survival in lung cancer cells. Carcinogenesis. 2005;26(7):1182–1195. doi: 10.1093/carcin/bgi072. [DOI] [PubMed] [Google Scholar]
- 21.Carlisle DL, Liu X, Hopkins TM, Swick MC, Dhir R, Siegfried JM. Nicotine activates cell-signaling pathways through muscle-type and neuronal nicotinic acetylcholine receptors in non-small cell lung cancer cells. Pulm Pharmacol Ther. 2007;20(6):629–641. doi: 10.1016/j.pupt.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 22.Laag E, Majidi M, Cekanova M, Masi T, Takahashi T, Schuller HM. NNK activates ERK1/2 and CREB/ATF-1 via beta-1-AR and EGFR signaling in human lung adenocarcinoma and small airway epithelial cells. Int J Cancer. 2006;119(7):1547–1552. doi: 10.1002/ijc.21987. [DOI] [PubMed] [Google Scholar]
- 23.Minna JD, Roth JA, Gazdar AF. Focus on lung cancer. Cancer Cell. 2002;1(1):49–52. doi: 10.1016/S1535-6108(02)00027-2. [DOI] [PubMed] [Google Scholar]
- 24.Sun L, et al. Phosphatidylinositol 3-kinase/protein kinase B pathway stabilizes DNA methyltransferase I protein and maintains DNA methylation. Cell Signal. 2007;19(11):2255–2263. doi: 10.1016/j.cellsig.2007.06.014. [DOI] [PubMed] [Google Scholar]
- 25.Taketo MM. Shutting down Wnt signal-activated cancer. Nat Genet. 2004;36(4):320–322. doi: 10.1038/ng0404-320. [DOI] [PubMed] [Google Scholar]
- 26.Sharma M, Chuang WW, Sun Z. Phosphatidylinositol 3-kinase/Akt stimulates androgen pathway through GSK3beta inhibition and nuclear beta-catenin accumulation. J Biol Chem. 2002;277(34):30935–30941. doi: 10.1074/jbc.M201919200. [DOI] [PubMed] [Google Scholar]
- 27.Davis M, et al. Pseudosubstrate regulation of the SCF(beta-TrCP) ubiquitin ligase by hnRNP-U. Genes Dev. 2002;16(4):439–451. doi: 10.1101/gad.218702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frame S, Cohen P. GSK3 takes centre stage more than 20 years after its discovery. Biochem J. 2001;359(Pt 1):1–16. doi: 10.1042/0264-6021:3590001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Agoston AT, et al. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J Biol Chem. 2005;280(18):18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 30.West KA, et al. Rapid Akt activation by nicotine and a tobacco carcinogen modulates the phenotype of normal human airway epithelial cells. J Clin Invest. 2003;111(1):81–90. doi: 10.1172/JCI16147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laurent-Puig P, Lievre A, Blons H. Mutations and response to epidermal growth factor receptor inhibitors. Clin Cancer Res. 2009;15(4):1133–1139. doi: 10.1158/1078-0432.CCR-08-0905. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.