Abstract
MCF-7/AdrVp is a multidrug-resistant human breast cancer subline that displays an ATP-dependent reduction in the intracellular accumulation of anthracycline anticancer drugs in the absence of overexpression of known multidrug resistance transporters such as P glycoprotein or the multidrug resistance protein. RNA fingerprinting led to the identification of a 2.4-kb mRNA that is overexpressed in MCF-7/AdrVp cells relative to parental MCF-7 cells. The mRNA encodes a 663-aa member of the ATP-binding cassette superfamily of transporters that we term breast cancer resistance protein (BCRP). Enforced expression of the full-length BCRP cDNA in MCF-7 breast cancer cells confers resistance to mitoxantrone, doxorubicin, and daunorubicin, reduces daunorubicin accumulation and retention, and causes an ATP-dependent enhancement of the efflux of rhodamine 123 in the cloned transfected cells. BCRP is a xenobiotic transporter that appears to play a major role in the multidrug resistance phenotype of MCF-7/AdrVp human breast cancer cells.
Keywords: mitoxantrone, anthracyclines, transporter proteins
The development of resistance to multiple chemotherapeutic drugs occurs frequently during the treatment of advanced carcinoma of the breast. Two transmembrane xenobiotic transporter proteins, P glycoprotein (Pgp) and the multidrug resistance protein (MRP), cause multidrug resistance when transfected into drug-sensitive cells in culture (1–5). Despite these findings, the role these transporters play in clinical drug resistance exhibited by human breast cancer is unclear; hence, alternate or additional drug resistance mechanisms operative in this disease have been sought. To address this problem, Chen et al. (6) selected human breast carcinoma MCF-7 cells for resistance to the anthracycline doxorubicin in the presence of verapamil, an inhibitor of Pgp. The resultant multidrug-resistant subline, MCF-7/AdrVp, exhibits marked crossresistance to certain other anthracyclines [daunorubicin and 3′-deamino-3′-(3-cyano-4-morpholinyl) doxorubicin] and to the anthracenedione mitoxantrone but remains sensitive to vinca alkaloids, paclitaxel (6, 7), and cis-platin. MCF-7/AdrVp cells do not overexpress Pgp or MRP, despite a marked reduction in the intracellular accumulation of the anthracycline daunorubicin and the fluorescent dye rhodamine 123 compared with MCF-7 cells (6, 7). MCF-7/AdrVp cells do not display the altered subcellular distribution of drug (7) seen in certain cells that overexpress MRP. Although the decreased accumulation of daunorubicin is not reversed by the classical Pgp antagonist cyclosporin A, depletion of ATP results in complete abrogation of the enhanced efflux of both daunorubicin and rhodamine (7). These findings led to the hypothesis that an ATP-dependent xenobiotic transporter may contribute significantly to the multidrug-resistance phenotype of MCF-7/AdrVp cells. To test this hypothesis, we sought mRNA species that were differentially overexpressed in MCF-7/AdrVp cells compared with MCF-7 cells by using the technique of RNA fingerprinting.
MATERIALS AND METHODS
Cell Lines.
MCF-7 human breast carcinoma cells, their drug-resistant subline MCF-7/AdrVp, and a partial-revertant subline (MCF-7/AdrVpPR), cultured in the absence of adriamycin, were obtained from Antonio Fojo (Medicine Branch, National Cancer Institute). The cells were maintained in culture as described (8). The MCF-7/AdrVp subline was continuously maintained in the presence of 1 μg/ml doxorubicin (Pharmacia & Upjohn) and 2.5 μg/ml verapamil (Sigma).
RNA Fingerprinting.
RNA Fingerprinting was performed by using the protocol in the Delta RNA Fingerprinting kit (CLONTECH), a modification of the differential-display technique (9, 10). The sequence of the P6 arbitrary primer was 5′-ATTAACCCTCACTAAATGCTGGGTG-3′; the sequence of the T9 oligo(dT) primer was 5′-CATTATGCTGAGTGATATCTTTTTTTTTGG-3′. PCR products that represented differentially expressed cDNAs were excised from the differential-display gels, eluted by boiling in 40 μl of distilled, deionized H2O for 5 min, amplified by PCR for 20 cycles using the original primers, and separated on 2% agarose/ethidium bromide gels to confirm the size of the reamplified product. The reamplified PCR products were ligated into the multiple cloning site of TA cloning vector pCR2.1 (Invitrogen) according to the manufacturer’s protocol; after ligation, the vector was transfected into the TOP10F strain of Escherichia coli. Individual bacterial colonies were picked, and plasmid DNA was isolated (Wizard miniprep; Promega). To confirm that the cloned PCR products isolated by TA cloning were overexpressed in the RNA fingerprinting reaction mixture from the drug resistant cells, a “reverse” Northern analysis was performed. Plasmid DNA isolated from 12 different colonies of transfected E. coli was fixed in duplicate to Zeta Probe-GT (Bio-Rad, Richmond, CA) membranes in a slot-blot apparatus. One of the duplicate membranes was probed with the 33P-labeled PCR mixture that amplified MCF-7 cDNA by using the original “P” and “T” primers in the RNA Fingerprinting kit. The other membrane was probed with the original 33P-labeled parallel PCR reaction mixture that amplified the cDNA produced from MCF-7/AdrVp cells using standard Northern blot conditions of hybridization, after which the binding of probe was assessed by using autoradiography.
Construction of cDNA Library.
A cDNA library was constructed from MCF-7/AdrVp RNA by using the CapFinder PCR cDNA library construction kit (CLONTECH) according to the manufacturer’s protocol. The CapFinder technique is designed specifically to produce full-length double-stranded cDNA. The library was screened with the RNA Fingerprinting PCR product of interest by using the manufacturer’s recommended protocol. Positive clones were isolated and subjected to secondary and tertiary screening, with additional testing by Northern blot hybridization using RNA obtained from MCF-7, MCF-7/AdrVp, and MCF-7/AdrVpPR cells. Multiple clones had 2.4-kb inserts, the approximate size of the BCRP mRNA suggested by Northern blotting. Four 2.4-kb inserts were ligated into the pCR2.1 plasmid (see above); sequencing of the 2.4-kb cDNA insert was performed by using an automated DNA sequencer (Perkin–Elmer). All DNA sequences were confirmed by sequencing in the reverse direction.
Data Analysis.
Analyses of cDNA and deduced protein sequences were accomplished using protein and nucleotide-sequence databases that were accessed by using the Wisconsin sequence analysis package, Version 8 (Genetics Computer Group, Madison, WI) which are available through the Frederick Cancer Research Center’s Supercomputing Facility (Frederick, MD). Statistical analyses were accomplished with the minitab statistical software (minitab release 8 extended; Minitab, State College, PA).
Reverse Transcription–PCR (RT-PCR).
The program oligo (Version 5.0; National Biosciences, Plymouth, MN) was used to help determine suitable primers for detection of the human homologue of the Drosophila white gene (w) by RT-PCR. The upper primer began at 5′ position 2,136 of human w mRNA and had the sequence 5′-CGACCGACGACACAGA-3′; the lower primer began at 3′ position 2,590 and had the sequence 5′-CTTAAAATGAATGCGATTGAT-3′. The expected PCR product was 475 bp in length. Random hexamers were used to prime the reverse transcription reaction, which was followed by 25 cycles of PCR. An RT-PCR assay for β-actin was also performed; reaction conditions for this assay have been reported (11).
Transfection and Enforced Expression of BCRP in MCF-7 cells.
The full-length breast cancer resistance protein (BCRP) cDNA was inserted into the multiple cloning site of expression vector pcDNA3 (Invitrogen). After the pcDNA3–BCRP construct was sublconed, DNA sequence analysis was performed to confirm the insert of the selected clone was in a sense orientation to the cytomegalovirus (CMV) promoter of the pcDNA3 vector. MCF-7 cells were transfected with pcDNA3–BCRP by using the calcium phosphate precipitation method (12), selected by culture with geneticin (G418, 1 mg/ml), and subcloned by limiting dilution in 96-well flat-bottomed culture plates (Sarstedt, Newton, NC). Subclones were tested for expression of BCRP mRNA by using Northern blot analysis. As a control, MCF-7 cells were also transfected with the empty pcDNA3 vector and selected by growth in medium containing 1 mg/ml G418.
Pharmacokinetics of Intracellular Drugs and Effect of ATP Depletion.
The intracellular accumulation and retention of daunorubicin in MCF-7 cells was determined by using flow cytometry as described (8). Cells cultured in 25-cm2 flasks (Corning Costar) were exposed to 1 μg/ml daunorubicin for up to 180 min (accumulation phase) or exposed to daunorubicin for 180 min, washed free of drug with ice-cold saline solution, and resuspended in prewarmed culture medium in the absence of drug (retention phase). At the time intervals indicated in the figure, aliquots of cells were trypsinized off of the plates, and intracellular daunorubicin content was measured (8). Controls for binding of anthracycline to plasma membrane were accomplished by incubating cells with daunorubicin on ice for an appropriate period of time, whereupon cellular daunorubicin content was determined by using flow cytometry. The value for membrane binding was subtracted from the value obtained for intracellular drug content of cells incubated with drug at 37°C. Intracellular drug content is expressed in fluorescence units (FU) per cell. Fluorescence units are arbitrary numbers between 1 and 10,000. Cell volumes were measured by using a Coulter Channelyzer as described (8).
MCF-7 cells were depleted of ATP by incubation in glucose-free DMEM containing 50 mM 2-deoxy-d-glucose and 15 mM sodium azide for 20 min (37°C). Rhodamine 123 was added (0.5 μg/ml final concentration) for an additional 30 min. The cells were placed on ice, washed free of rhodamine, and incubated under ATP-depleting conditions for an additional 30 min, and rhodamine retention was determined by flow cytometry (excitation 488 nm, emission 520 nm).
RESULTS
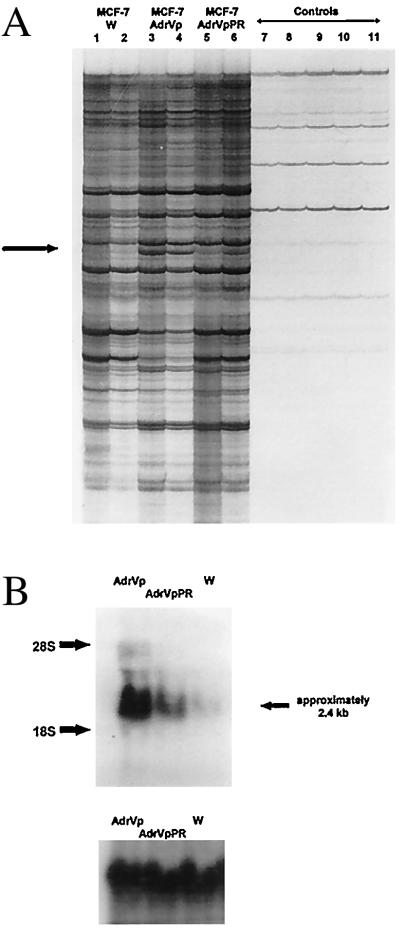
To test the hypothesis outlined in the Introduction, we employed the technique of RNA fingerprinting, a modification of the method of differential display that uses PCR and degenerate-primer pairs to amplify cellular mRNA. For the PCR reactions, cDNA was prepared from MCF-7, MCF-7/AdrVp, and MCF-7/AdrVpPR cells, the latter of which have partially reverted to drug sensitivity by culture in the absence of selecting agents. When RNA fingerprinting was performed with the primers P6 and T9, a PCR product was found reproducibly in excess in reactions that used cDNA from MCF-7/AdrVp cells as template, compared with those reactions with cDNA from MCF-7 or MCF-7/AdrVpPR cells (Fig. 1A). The PCR product overexpressed in MCF-7/AdrVp cells was excised from the dried gel, purified, and ligated into a TA cloning vector. Screening of the TA vector clones by the reverse Northern blot method led to the isolation of a single clone whose PCR product insert identified, by standard Northern analysis, a 2.4-kb mRNA species that was markedly overexpressed in MCF-7/AdrVp cells, compared with MCF-7 (Fig. 1B, Top). The partially revertant MCF-7/AdrVpPR subline had intermediate expression of the 2.4-kb mRNA species (Fig. 1B).
Figure 1.
(A) RNA fingerprinting of MCF-7 cells. Total cellular RNA was treated with DNase, reverse-transcribed into cDNA, and amplified by PCR using upstream and downstream primers and radiolabeled [33P]dATP. The figure depicts a portion of the autoradiograph of a 5% polyacrylamide gel electrophoresis of the PCR mixture by using the primer pair P6 and T9. Lanes 1, 3, and 5 are reaction mixtures in which cDNA diluted 1:10 was added; lanes 2, 4, and 6 represent reaction mixtures in which cDNA diluted 1:40 was added. Lanes 7 and 8 are H2O controls, in which sterile water was added to the PCR mixture in place of cDNA. Lanes 9, 10, and 11 are RNA controls, in which 0.02 μg of cellular RNA from MCF-7/W, MCF-7/AdrVp, or MCF-7/AdrVpPR cells was added instead of cDNA. These RNA controls served to indicate contamination of the RNA with genomic DNA. The arrow indicates a PCR product representing an mRNA species overexpressed in MCF-7/AdrVp cells, compared with MCF-7/W or MCF-7/AdrVpPR cells. This product was excised from the gel for further amplification by PCR. (B) Northern blot hybridization (Upper) of mRNA from MCF-7/W, MCF-7/AdrVp, or MCF-7/AdrVpPR cells by using the 795-bp PCR product obtained from RNA fingerprinting studies and isolated by TA cloning as a probe after labeling with [32P]dCTP (Prime-a-Gene labeling kit, Promega). To control for equivalence in sample loading, the blot was stripped and rehybridized with a radiolabeled probe for 18S RNA (Lower).
The differentially expressed PCR product in the TA clone was sequenced and found to be a 795-bp cDNA. Protein database searches of the deduced amino acid sequence revealed a high degree of homology to members of the ATP-binding cassette (ABC) family of transport proteins. The 795-bp cDNA fragment was radiolabeled and used as a probe to screen a cDNA library prepared from MCF-7/AdrVp cells. A clone of λ bacteriophage containing a 2.4-kb cDNA fragment was identified and isolated. The cDNA insert was completely sequenced and found to be 2,418 bp in length. Analysis of the cDNA for ORFs using the program frames contained in the GCG software package indicated the presence of a long ORF that began at position 239 and ended with the stop codon TAA at position 2,204–2,206. The deduced amino acid sequence of this ORF is shown in Fig. 2A. The protein encoded by this sequence has been designated breast cancer resistance protein (BCRP). A comparison of the BCRP amino acid sequence using the GCG software fasta revealed a high degree of homology to at least 50 ABC transport proteins. The highest match was PIR2:G02068, the human homologue of the Drosophila white (w) gene, which has 638 amino acids and is 29.3% identical to BCRP. The w gene in Drosophila functions in the cellular transport of guanine and tryptophan, which are retinal-pigment precursors (13–16). The human homologue of w (17) is not overexpressed in MCF-7/AdrVp cells compared with MCF-7 cells as detected by an RT-PCR assay (data not shown).
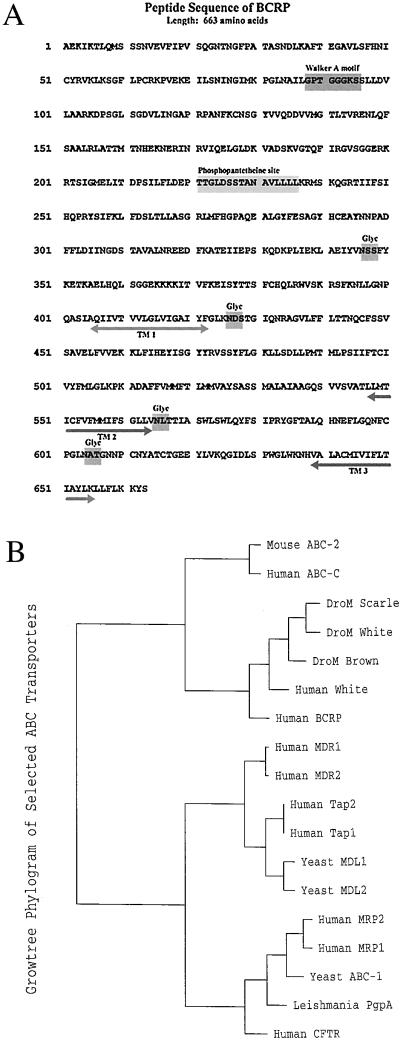
Figure 2.
(A) Deduced amino acid sequence of BCRP with display of motifs. The deduced sequence was obtained from the nucleotide sequence of full-length BCRP cDNA by applying the program translate, which is part of the GCG software package. TM 1, TM 2, and TM 3 refer to potential transmembrane regions; Glyc refers to potential sites of N-glycosylation. (B) Phylogram of the evolution of the amino acid sequence of BCRP in relation to certain other members of the ABC family of transport proteins. The phylogram was created by using the multiple-sequence alignment program pileup, which is part of the GCG software package, using a gap weight of 3.0 and a gap length weight of 0.1. The GCG program distances was then applied to create pairwise evolutionary distances between the aligned sequences, by using the Kimura protein-distance correction method. A graphical phylogenetic tree was then created from the distance matrix with the GCG program growtree.
Analysis of the BCRP peptide sequence with the GCG program motifs demonstrated a single Walker “A” ATP/GTP binding region (18) at amino acids 88–95 and a phosphopantetheine attachment site at amino acids 221–236 (Fig. 2A). As the prosthetic group of acyl carrier proteins in some multienzyme complexes, phosphopantetheine serves in the attachment of activated fatty acid and amino acid groups (19).
Examination of BCRP structure with GCG programs pepplot and plotstructure revealed a relatively hydrophilic amino-terminal domain (amino acids 1–400) that contains the ATP-binding sequence and a relatively hydrophobic carboxyl-terminal domain (amino acids 401–663) containing at least three putative transmembrane domains and four potential N-glycosylation sites (Fig. 2A). The transmembrane domains were estimated with a program to predict helices in integral membrane proteins (20). Analysis of the BCRP sequence by the GCG program dotplot demonstrated that the peptide is homologous with one-half of the duplicated Pgp or MRP molecule, except that Pgp or MRP have the configuration NH2-[transmembrane domains]-[ATP binding 1]-[transmembrane domains]-[ATP binding 2]-COOH, whereas BCRP is NH2-[ATP binding]-[transmembrane domains]-COOH. The phylogenetic relationship of BCRP to other members of the ABC transporter superfamily was determined by using the GCG programs pileup, distances, and growtree. This analysis revealed BCRP is only distantly related to Pgp or MRP (Fig. 2B).
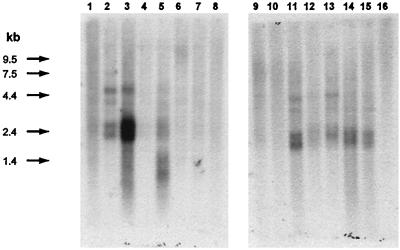
The BCRP cDNA was used as a probe in Northern blots to examine the expression of BCRP mRNA in selected normal human tissues (Fig. 3). The greatest expression was seen in placental tissue with considerably lower levels of expression in brain, prostate, small intestine, testis, ovary, colon, and liver. BCRP transcripts were below the level of detection in heart, lung, skeletal muscle, kidney, pancreas, spleen, thymus, and peripheral-blood leukocytes.
Figure 3.
Tissue distribution of BCRP mRNA. This analysis was performed by using commercially prepared human multiple-tissue Northern blots (MTN Blot and MTN Blot II, CLONTECH) and the manufacturer’s recommended procedure. The 795-bp PCR product obtained from RNA fingerprinting was used as probe after labeling with [32P]dCTP. Tissues tested were: heart (lane 1), brain (lane 2), placenta (lane 3), lung (lane 4), liver (lane 5), skeletal muscle (lane 6), kidney (lane 7), pancreas (lane 8), spleen (lane 9), thymus (lane 10), prostate (lane 11), testis (lane 12), ovary (lane 13), small intestine (lane 14), colon (lane 15), and peripheral-blood leukocytes (lane 16).
To evaluate BCRP function in vitro, the full-length cDNA was inserted into the multiple cloning site of the expression vector pcDNA3. MCF-7 cells were transfected with the vector containing the full-length BCRP cDNA (pcDNA3–BCRP) or with the empty vector as a control. After selection with geneticin (G418), the pcDNA3–BCRP-transfected cells were cloned by using limiting dilution; multiple clones were tested for BCRP expression by using Northern blot hybridization (Fig. 4A). Two BCRP-overexpressing clones (clones 6 and 8) and one clone that did not overexpress BCRP (clone 19) were selected for studies of BCRP function in comparison to cells transfected only with empty vector and to parental MCF-7 cells. The expression of BCRP mRNA in clone 6 was less than in clone 8 (Fig. 4A).
Figure 4.
(A) Northern analysis of the expression of BCRP mRNA in subclones of MCF-7/W cells stably transfected with expression vector pcDNA3-BCRP or in MCF-7/W cells transfected with empty vector pcDNA3 (uncloned-vector control) after selection in medium containing G418. The subclones were isolated by plating the transfected cells by limiting dilution in 96-well flat-bottomed culture flasks. The 795-bp PCR product originally obtained from the differential-display studies was radiolabeled and used as probe. Ethidium bromide stains of 1% agarose gel electrophoresis of the total cellular RNA used for the Northern blot demonstrated approximate equivalency of sample loading (results not shown). (B) Daunorubicin (DNR) accumulation and retention of MCF-7 cells transfected with empty pcDNA3 vector (vector control) or with clone 6 or clone 8 isolated from MCF-7/W cells after transfection with pcDNA3-BCRP. The data points are the mean of duplicate determinations; the vertical bars represent the upper or lower range for that data point. The cell volumes, measured by Coulter Channelyzer are 2,515 ± 56, 3,074 ± 112, and 2,459 ± 56 μm3 for MCF-7/BCRP-clone 6, MCF-7/BCRP-clone 8, and MCF-7/pcDNA3 vector control cells, respectively. These values are comparable to our previous measurements of MCF-7 cell volumes (8). (C) Effects of ATP depletion on the retention of rhodamine 123 by transfectant MCF-7/pcDNA3 (empty vector control) or MCF-7/BCRP clone 8 cells. The cells were incubated in complete medium or under ATP-depleting conditions (see Materials and Methods) for 20 min, and rhodamine 123 (0.5 μg/ml final concentration) was added for an additional 30 min. The cells were washed free of rhodamine and returned to culture either in complete medium or under ATP-depleting conditions for an additional 30 min. Rhodamine retention, expressed as arbitrary fluorescence units (FU) per cell, was determined by using flow cytometry (excitation 488 nm, emission 520 nm). The vertical lines over each bar represent the SD of four replicate determinations. Viability (trypan blue dye exclusion) of the cells in each treatment group was >90% at the completion of the study. (D) Representative sulforhodamine-B cytotoxicity (21) studies for mitoxantrone, daunorubicin, doxorubicin, cis-platin, paclitaxel, or vincristine against MCF-7/W (∗) or MCF-7/pcDNA3-BCRP clone 8 (■) cells. These data are typical of those used to obtain LC50 values that comprise the data displayed in Table 1. The vertical bars for each data point represent ± SD of six replicate determinations.
Daunorubicin accumulation and retention were examined in transfected cells by using flow cytometry. The BCRP-overexpressing clones 6 and 8 reproducibly displayed diminished accumulation and retention of daunorubicin when compared with the vector-transfected controls (Fig. 4B). The intracellular steady-state concentrations of daunorubicin in clones 8 and 6, respectively, were approximately 30% or 50% of that attained in the vector control cells. This was not the result of differences in cell volume, because the volumes of the BCRP-overexpressing sublines tested were not lower than the empty vector-transfected control cells. In previous studies (8), the intracellular steady-state accumulation of daunorubicin in MCF-7/AdrVp cells was approximately 25% of that in MCF-7/W cells, which is comparable to the accumulation of daunorubicin that we observed in transfectant clone 8 in the current studies.
The transport function of BCRP appears to depend on ATP (Fig. 4C). The intracellular retention of rhodamine 123 in BCRP-overexpressing clone-8 cells was diminished to 45% of that of vector control cells. Depletion of ATP increased rhodamine retention in clone 8 cells to levels comparable to those of vector control cells cultured in complete medium but had no effect on the rhodamine retention in the vector control cells (Fig. 4C).
The sensitivities of the various transfected sublines to chemotherapeutic agents were tested by the sulforhodamine-B cytotoxicity assay (21). The BCRP-overexpressing clones 6 and 8 displayed resistance to mitoxantrone, daunorubicin, and doxorubicin compared with non-BCRP-overexpressing clone 19 cells, MCF-7 cells, or the empty vector-transfected controls (Table 1; Fig. 4D). Like MCF-7/AdrVp cells, the MCF-7/BCRP transfectant clones 6 and 8 displayed the greatest degree of resistance to mitoxantrone. The pattern of crossresistance displayed by the BCRP-overexpressing transfected cells is very similar to the pattern displayed by MCF-7/AdrVp cells; however, MCF-7/AdrVp cells have a greater relative resistance to all cytotoxic drugs within the phenotype. The BCRP-overexpressing clones 6 and 8 are sensitive to cis-platin, paclitaxel, and vincristine, as are MCF-7/AdrVp cells (Table 1). BCRP transfectant clone 19, which did not overexpress BCRP, met the minimal statistical criterion (P = 0.0497) for significance of a low degree of resistance to cis-platin (resistance factor = 2.7) in comparison to MCF-7/W but not in comparison to cells transfected with the empty vector (MCF-7/pcDNA3; see Table 1). With this exception, the sensitivity of clone 19 was not statistically different from MCF-7/W or MCF-7/pcDNA3 for the drugs tested.
Table 1.
Effects of chemotherapeutic drugs on BCRP-transfected MCF-7 cells
| Drug | Cell type
|
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MCF-7/W
|
MCF-7/pcDNA3
|
MCF-7/BCRPclone19
|
MCF-7/BCRP clone6
|
MCF-7/BCRP clone8
|
MCF-7/AdrVp
|
|||||||||||||
| Median LC50, nM [Range, nM] | N | RF | Median LC50, nM [Range, nM] | N | RF | Median LC50, nM [Range, nM] | N | RF | Median LC50, nM [Range, nM] | N | RF | Median LC50, nM [Range, nM] | N | RF | Median LC50, nM [Range, nM] | N | RF | |
| Mitoxantrone | 41 | 7 | 1.0 | 40 | 6 | 1.0 | 54 | 1 | 1.3 | 370*† | 4 | 9.0 | 1,300*† | 5 | 32 | 160,000 | 2 | 3,902 |
| [20–105] | [15–115] | [—] | [130–700] | [280–3,600] | [120,000–200,000] | |||||||||||||
| Daunorubicin | 42 | 7 | 1.0 | 68 | 5 | 1.6 | 54 | 1 | 1.6 | 180*† | 4 | 4.3 | 350*† | 5 | 8.3 | 1,700 | 3 | 41 |
| [8–105] | [21–110] | [—] | [150–380] | [110–600] | [1,500–1,800] | |||||||||||||
| Doxorubicin | 56 | 5 | 1.0 | 49 | 4 | 0.9 | 70 | 4 | 1.3 | 168* | 4 | 3.0 | 1,050*† | 4 | 19 | 8,650 | 2 | 155 |
| [14–110] | [16–150] | [25–102] | [81–600] | [205–5,300] | [6,800–10,500] | |||||||||||||
| Cisplatin | 2,600 | 4 | 1.0 | 3,900 | 3 | 1.5 | 6,900 | 3 | 2.7 | 7,000 | 3 | 27 | 3,700 | 3 | 1.5 | 2,875 | 2 | 1.1 |
| [1,600–4,000] | [3,000–4,050] | [4,700–6,900] | [3,080–13,000] | [3,500–13,000] | [1,050–4,700] | |||||||||||||
| Paclitaxel | 1.9 | 4 | 1.0 | 5.1 | 2 | 2.7 | 0.9 | 3 | 0.5 | 1.9 | 3 | 1.0 | 3.0 | 3 | 1.6 | 1.9 | 4 | 1.0 |
| [0.9–33] | [3–7.1] | [0.8–28] | [1.4–21] | [1.8–36] | [1.3–3.6] | |||||||||||||
| Vincristine | 0.26 | 3 | 1.0 | 0.39 | 2 | 1.5 | 0.63 | 3 | 2.4 | 0.37 | 3 | 1.4 | 0.28 | 3 | 1.1 | 0.9 | 3 | 3.5 |
| [0.08–0.65] | [0.28–0.51] | [0.09–0.76] | [0.35–0.91] | [0.25–0.59] | [0.19–1.3] | |||||||||||||
Sensitivity of selected MCF-7 sublines to antineoplastic agents determined by sulforhodamine-B cytotoxicity assay (21). Experiments such as those displayed in Figure 4D were used to obtain the LC50. For each drug and cell type, the table displays the median LC50 and range of LC50 measurements (nM), the number of experimental determinations of LC50 that were performed (N), and the resistance factor (RF). The RF was calculated by dividing the median LC50 for a given drug against a transfected cell line by the median LC50 of that drug against nontransfected MCF-7/W cells. For each drug tested, the LC50 for the BCRP-transfected cells was examined for statistically significant difference from the LC50 of MCF-7/W or MCF-7/pcDNA3 by the Mann–Whitney test, using minitab statistical software minitab release 8 extended, Minitab, State College, PA) and a 95% confidence interval. The values of P for the statistically significant differences are as follows Mitoxantrone: MCF-7/W vs. MCF-7/BCRPclone6, P = 0.0107, MCF-7/W vs. MCF-7/BCRPclone8, P = 0.0058, MCF-7/pcDNA3 vs. MCF-71BCRPclone6, P = 0.0142, MCF-7/pcDNA3 vs. MCF-7/BCRPclone8, P = 0.0081; daunorubicin: MCF-7/W vs. MCF-7/BCRPclone6, P = 0.0107, MCF-7/W vs. MCF-7/BCRPclone8, P = 0.0058, MCF-7/pcDNA3 vs. MCF-7/BCRPclone6, P = 0.0195, MCF-7/pcDNA3 vs. MCF-7/BCRPclone8, P = 0.0163; doxorubicin: MCF-7/W vs. MCF-7/BCRPclone6, P = 0.0373, MCF-7/W vs. MCF-7/BCRPclone8, P = 0.02, MCF-7/pcDNA3 vs. MCF-7/BCRPclone8, P = 0.0304; cis-platin: MCF-7/W vs. MCF-7/BCRPclone19, P = 0.0497.
Differs significantly from MCF-7/W, P < 0.05 (Mann–Whitney test).
Differs significantly from MCF-7/pcDNA3 (empty vector control, P < 0.05, Mann–Whitney U test).
DISCUSSION
The data presented here strongly support the conclusion that the novel ABC-transport protein family member BCRP is an ATP-dependent xenobiotic transporter that plays a major role in the drug-resistance phenotype of MCF-7/AdrVp cells. The overexpression of BCRP mRNA in MCF-7/AdrVp cells, which is diminished in MCF-7/AdrVpPR, suggests an important role for BCRP in resistance to cytotoxic agents. Furthermore, the enforced overexpression of BCRP in MCF-7 cells diminished daunorubicin cellular accumulation and imparted a pattern of drug crossresistance to the transfected cells that was virtually identical to that of MCF-7/AdrVp cells. The degree of BCRP overexpression in transfectant clones 6 and 8 correlates with the alterations in the intracellular steady-state level of daunorubicin and their degree of resistance to mitoxantrone, daunorubicin, and doxorubicin.
A major difference between the BCRP-overexpressing transfectant clones and the original MCF-7/AdrVp subline is that the degree of drug resistance in the latter is greater than in the transfected cells; however, the steady-state BCRP mRNA levels in the transfectants are comparable to those of MCF-7/AdrVp cells (Fig. 4A). A number of possibilities may contribute to this difference. Alterations in protein stability, localization, or both may contribute to the full drug-resistant phenotype, or the expression of other proteins may be required. Recently, we reported members of the carcinoembryonic antigen (CEA) family, primarily the nonspecific cross-reacting antigen and CEA itself, are markedly overexpressed on the cell surface of MCF-7/AdrVp and MCF-7/AdrVpPR cells compared with drug-sensitive MCF-7 cells (22). A high density of these acidic glycoproteins on the cell surface may protonate drugs such as mitoxantrone, daunorubicin, or doxorubicin, which will prevent entry into the cell. Indeed, Kawaharata et al. (23) reported that the enforced expression of CEA in transfected NIH 3T3 cells resulted in both diminished accumulation of and resistance to doxorubicin in the transfected cells. Hence, the relative overexpression of CEA family members on the MCF-7/AdrVp cell surface could act in concert with BCRP to cause greater resistance to mitoxantrone, doxorubicin, and daunorubicin than could BCRP alone. This hypothesis could be tested by cotransfecting the MCF-7/BCRP-clone 8 subline with an expression vector containing the nonspecific cross-reacting antigen or CEA.
Another possible explanation for the greater degree of resistance of MCF-7/AdrVp cells compared with the transfectants is that BCRP may participate in a multiprotein transporter complex. The translocation pathway of typical ABC transporters consists of two ATP-binding domains and two highly hydrophobic domains that contain membrane-spanning regions. This configuration can be accomplished in a single large molecule (e.g., MRP or Pgp). Alternatively, the active complex of certain ABC transporters can be formed by the heterodimerization of two nonidentical proteins, each containing a single ATP-binding and hydrophobic region. The w and brown (b) proteins of Drosophila and the Tap-1 and Tap-2 proteins that transport major histocompatibility class I peptides are examples of ABC-transport protein family members that exhibit such a cooperative interaction. The presence of the phosphopantetheine attachment site on BCRP suggests that BCRP may be part of a multiprotein complex. Possibly BCRP has one or more protein cofactors that function as a much more efficient transporter in a heteromeric state. The activation or overexpression of this cofactor in MCF-7/AdrVp relative to MCF-7 cells could explain the greater drug resistance in the MCF-7/AdrVp subline relative to the BCRP transfectants.
Acknowledgments
We thank Drs. Susan Bates and Antonio Fojo of the Medicine Branch, National Cancer Institute, National Institutes of Health, for the gift of MCF-7/AdrVp sublines. We also thank Dr. Gary Smythers of the Supercomputing Facility of the Frederick Cancer Research Center, National Cancer Institute, National Institutes of Health for help with the GCG software and identifying potential transmembrane domains of BCRP. We are grateful to Dr. De-Qi Xu for assistance in preparing the cDNA library from MCF-7/AdrVp cells. We thank Drs. Raji Sridhara and Julie Eiseman for assistance with the statistical analysis of data. This work was supported in part by Public Health Service Grant CA52178 to D.D.R. from the National Cancer Institute, National Institutes of Health, Department of Health and Human Services, and by a Veteran’s Affairs merit review grant to D.D.R.
ABBREVIATIONS
- RT-PCR
reverse transcription–PCR
- Pgp
P-glycoprotein
- MRP
multidrug resistance protein
- ABC
ATP-binding cassette
- BCRP
breast cancer resistance protein
- w
Drosophila white gene
Note added in Proof
A blast-n search of the nonredundant database of GenBank human expressed sequence tag (EST) entries for homology to the BCRP cDNA sequence revealed 46 “blast hits.” The highest score was clone EST57481, which was predicted to represent 1 of 21 new genes of the human ABC family, as reported previously (24). EST157481 is almost identical to bases 1,187–2,016 of the BCRP cDNA sequence as recorded in the GenBank database.
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF09895).
References
- 1.Kessel D, Botterill V, Wodinsky I. Cancer Res. 1968;28:938–941. [PubMed] [Google Scholar]
- 2.Biedler J L, Riehm H. Cancer Res. 1970;30:1174–1184. [PubMed] [Google Scholar]
- 3.Ling V, Thompson L H. J Cell Physiol. 1974;83:103–116. doi: 10.1002/jcp.1040830114. [DOI] [PubMed] [Google Scholar]
- 4.Gros P, Ben Neryah Y B, Croop J M, Housman D E. Nature (London) 1986;323:728–731. doi: 10.1038/323728a0. [DOI] [PubMed] [Google Scholar]
- 5.Cole S P C, Bhardwaj G, Gerlach J H, Mackie J E, Grant C E, Almquist K C, Stewart A J, Kurz E U, Duncan A M V, et al. Science. 1992;258:1650–1654. doi: 10.1126/science.1360704. [DOI] [PubMed] [Google Scholar]
- 6.Chen Y-N, Mickley L A, Schwartz A M, Acton E M, Hwang J, Fojo A T. J Biol Chem. 1990;265:10073–10080. [PubMed] [Google Scholar]
- 7.Lee J S, Scala S, Matsumoto Y, Dickstein B, Robey R, Zhan Z, Altenberg G, Bates S E. J Cell Biochem. 1997;65:513–526. [PubMed] [Google Scholar]
- 8.Doyle L A, Ross D D, Sridhara R, Fojo A T, Kaufmann S H, Lee E J, Schiffer C A. Br J Cancer. 1995;71:52–58. doi: 10.1038/bjc.1995.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang P, Pardee A. Science. 1992;257:967–971. doi: 10.1126/science.1354393. [DOI] [PubMed] [Google Scholar]
- 10.Liang P, Averboukh L, Keyomarsi K, Sager R, Pardee A. Cancer Res. 1992;52:6966–6968. [PubMed] [Google Scholar]
- 11.Ross D D, Doyle L A, Schiffer C A, Lee E J, Grant C E, Cole S P C, Deeley R G, Yang W, Tong Y. Leukemia. 1996;10:48–55. [PubMed] [Google Scholar]
- 12.Ausubel F, Kingston R, Moore D, Seidman J, Smith J, Struhl K, editors. Current Protocols in Molecular Biology. Vol. 1. New York: Wiley; 1988. , Chap. 9. [Google Scholar]
- 13.Morgan T H. Science. 1910;32:120–122. doi: 10.1126/science.32.812.120. [DOI] [PubMed] [Google Scholar]
- 14.Bingham P M, Levis R, Rubin G M. Cell. 1981;25:693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- 15.O’Hare K, Murphy C, Levis R, Rubin G M. J Mol Biol. 1984;180:437–455. doi: 10.1016/0022-2836(84)90021-4. [DOI] [PubMed] [Google Scholar]
- 16.Pepling M, Mount S M. Nucleic Acids Res. 1990;18:1633. doi: 10.1093/nar/18.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen H, Rossier C, Lalioti M D, Lynn A, Chakravarti A, Perrin G, Antonarkis S E. Am J Hum Genet. 1996;59:66–75. [PMC free article] [PubMed] [Google Scholar]
- 18.Walker J E, Saraste M, Runswick M J, Gay N J. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pugh E L, Wakil S J. J Biol Chem. 1965;240:4727–4733. [PubMed] [Google Scholar]
- 20.Rao J K M, Garos P. Biochim Biophys Acta. 1986;899:179–214. [Google Scholar]
- 21.Skehan P, Storeng R, Scudiero D, Monks A, McMahon J, Vistica D, Warren J T, Bokesch H, Kenny S, Boyd M R. J Natl Cancer Inst. 1990;82:1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 22.Ross D D, Gao Y, Yang Y, Leszyk J, Shively J, Doyle L A. Cancer Res. 1997;57:5460–5464. [PubMed] [Google Scholar]
- 23.Kawaharata H, Hinoda Y, Itoh F, Endo T, Oikawa S, Nakazato H, Imai K. Int J Cancer. 1997;72:377–382. doi: 10.1002/(sici)1097-0215(19970717)72:2<377::aid-ijc29>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 24.Allikmets R, Gerrard B, Hutchinson A, Dean M. Hum Mol Genet. 1996;5:1649–1655. doi: 10.1093/hmg/5.10.1649. [DOI] [PubMed] [Google Scholar]