SUMMARY
Trimethylation on histone H3 lysine 27 (H3K27me3) by Polycomb repressive complex 2 (PRC2) regulates the balance between self-renewal and differentiation of embryonic stem cells (ESCs). The mechanisms by which the activity and recruitment of PRC2 are controlled are largely unknown. Here we demonstrate that the founding member of the Jumonji-family, JMJ (JUMONJI or JARID2), is tightly associated with PRC2, colocalizes with PRC2 and H3K27me3 on chromatin, and modulates PRC2 function. In vitro JMJ inhibits PRC2 methyltransferase activity, a finding consistent with increased H3K27me3 marks at PRC2 targets in Jmj−/− ESCs. Paradoxically, JMJ is required for efficient binding of PRC2, indicating that the interplay of PRC2 and JMJ fine-tunes deposition of the H3K27me3 mark. During differentiation, activation of genes marked by H3K27me3 and lineage commitments are delayed in Jmj−/− ESCs. Our results demonstrate that dynamic regulation of Polycomb complex activity orchestrated by JMJ balances self-renewal and differentiation, further highlighting the involvement of chromatin dynamics in cell-fate transitions.
INTRODUCTION
Eukaryotic gene transcription is influenced by chromatin structure, in large part established through modification of histone tails (Kouzarides, 2007). Methylation of lysine residues in histones is dynamically regulated by the opposing activities of histone lysine methyltransferases (KMTs) and demethylases (KDMs) (Cloos et al., 2008). Recently several KDMs with exquisite substrate specificity have been implicated in diverse processes, including embryonic patterning, stem cell self-renewal, differentiation, neuronal development and spermatogenesis (Nottke et al., 2009). Mutations or deregulation of KDMs are often linked to human cancers and diseases (van Haaften et al., 2009; Yamane et al., 2007).
The majority of KDMs are characterized by the presence of a JmjC domain, a conserved region first recognized in the founding member of the Jumonji family, JUMONJI (JMJ or JARID2). A catalytic triad (H, D/E, H), which is predicted to mediate cofactor Fe(II) binding, is highly conserved in JmjC-containing proteins and required for KDM activity (Klose and Zhang, 2007). JMJ, however, has amino acid substitutions in this subregion, and is predicted to be enzymatically inactive (Cloos et al., 2008). Nonetheless, JMJ is critical in embryonic development. Jmj−/− mouse embryos develop various defects in neuronal, cardiac, liver and hematopoietic tissues with penetrance dependent on the genetic background (Jung et al., 2005; Takeuchi et al., 2006). Developmental defects have been associated with increased numbers of progenitor cells and depletion of differentiating or mature cells, suggesting a role for JMJ in regulating proliferation and differentiation. Despite the activities of JMJ-related proteins in chromatin regulation and the multiple roles of JMJ in development, the function of JMJ is poorly understood.
Polycomb group (PcG) proteins are required for establishing and maintaining cellular memory. Four PcG proteins, including two EZH proteins (EZH1 / EZH2), EED and SUZ12, comprise the core of the Polycomb repressive complex 2 (PRC2), a methyltransferase complex that acts specifically on histone H3 lysine 27 (H3K27) (Margueron et al., 2008; Shen et al., 2008). In embryonic stem cells (ESCs), PRC2 is critically required for both maintenance and execution of pluripotency (Boyer et al., 2006; Pasini et al., 2007; Shen et al., 2008). PRC2 and tri-methylation on H3K27 (H3K27me3) occupy a set of genes controlling differentiation and prevent full expression of these genes until lineage commitment. Despite its central role in development, little is known regarding how the activity of PRC2 is regulated, PRC2 is targeted to distinct regions during cell fate transitions, or the pluripotent state is maintained to preserve plasticity, such that the influence of the H3K27me3 mark may be attenuated when gene expression is required.
To address these issues, we characterized the PRC2 complex in mouse ESCs and identified closely associated proteins in a PRC2 interactome. Here we demonstrate that JMJ is a novel subunit of the PRC2 complex, colocalizes with PRC2 on chromatin, and regulates H3K27me3 modification by modulating PRC2 activity. Fine-tuned, dynamic regulation of H3K27me3 by JMJ and PRC2 balances the stem cell state and differentiation.
RESULTS
JMJ interacts with PRC2
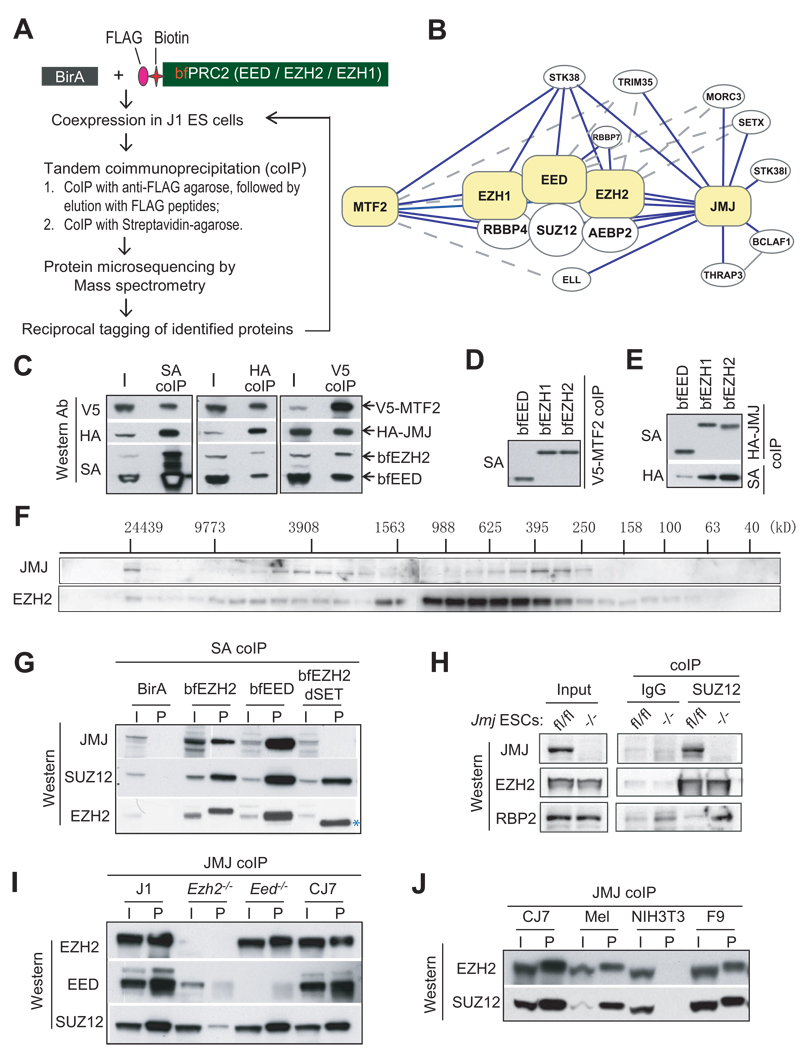
To identify proteins associated with PRC2, we affinity-purified the complex using a previously described in vivo biotinylation strategy (Wang et al., 2006). Nuclear extracts from ESCs coexpressing bacterial biotin ligase (BirA) and subendogenous levels of biotin- and FLAG-tagged EED, EZH1 or EZH2 referred to as bfEED, bfEZH1, bfEZH2 (Shen et al., 2008) were subjected to streptavidin-mediated coimmunoprecipitation or tandem coimmunoprecipitation mediated by anti-FLAG antibody and streptavidin. Interacting proteins were then identified by mass spectrometry sequencing (Figure 1A; Table S1–S4).
Figure 1. JMJ interacts with the PRC2 complex.
(A) The scheme to identify PRC2 interacting proteins.
(B) A PRC2 interactome. Proteins highlighted in yellow are tagged baits for affinity purification. Solid and dotted lines indicate interactions identified by multiple or few replicates, respectively. The interaction between BCLAF1 and THRAP3 was reported previously (Bracken et al., 2008).
(C) – (E) Coimmunoprecipitation (coIP) analysis in 293T cells. V5-tagged MTF2, HA-tagged JMJ, bfEED and bfEZH2 were coexpressed (C). V5-MTF2 (D) or HA-JMJ (E) was coexpressed with bfEED, bfEZH1, or bfEZH2 as indicated.
(F) Gel filtration analysis of nuclear extracts from CJ7 ESCs. Migration of molecular markers is indicated above the panels, and the antibodies for Western blotting are shown on the left.
(G) Streptavidin (SA) mediated coIP in ESCs expressing birA alone, bfEED, bfEZH2 or mutant EZH2 with SET domain deleted (bfEZH2dSET). bfEZH2dSET is indicated by *.
(H) Anti-SUZ12 coIP in Jmfl/fl and Jmj−/− ESCs.
(I) – (J) Anti-JMJ antibody-mediated coIP in ESCs and non-pluripotent cells. In (C) – (J), “I” indicates inputs and “P” indicates coIP fractions. About 5% of nuclear extracts used for coIP were loaded as the inputs.
In addition to known PRC2 core components including EZH1/EZH2, EED, SUZ12, RBBP4 and AEBP2, we identified several novel interacting partners of PRC2 including JUMONJI (JMJ), metal response element-binding transcription factor 2 (MTF2) and serine/threonine kinase 38 (STK38). To confirm the putative interacting proteins and to expand the PRC2 interaction network, we then tagged MTF2 and JMJ with a biotin tag and FLAG epitope (herein referred to as bfMTF2 and bfJMJ) in ESCs. The reverse tagging analysis confirmed association of PRC2 with MTF2 and JMJ (Table S5 and S6). We did not detect peptides representing the H3K9 methyltransferases G9a and GLP that were reported to interact with JMJ when overexpressed in NIH3T3 cells (Shirato et al., 2009).
Based on proteomic results, we constructed an "interactome" surrounding PRC2 (Figure 1B), which includes the core PRC2 components, MTF2, JMJ, STK38, RBBP7 and additional JMJ interacting proteins that may indirectly or transiently associate with PRC2. Interestingly, bfJMJ pulled down few peptides representing MTF2, and bfMTF2 did not capture JMJ (Table S1). Thus, these two PRC2-associated proteins may not coexist in the same PRC2 complex. To validate the above interactions, we co-expressed V5-tagged MTF2, HA-tagged JMJ, bfEED and bfEZH2 in 293T cells. Coimmunoprecipitation of MTF2, JMJ or EED / EZH2 pulled down the other three components (Figure 1C), suggesting their coexistence in one protein complex. To verify binary interactions, we coexpressed V5-tagged MTF2 or HA-tagged JMJ with one of bfEED, bfEZH1 and bfEZH2 in 293T cells. Both MTF2 and JMJ bind to EED, EZH1 or EZH2 (Figure 1D and 1E).
MTF2 (also called PCL2) is a member of the Polycomblike / PHF1 (PCL1) family proteins, which associate with PRC2 in fly and in mammal (Cao et al., 2008; Nekrasov et al., 2007; Sarma et al., 2008; Savla et al., 2008). Because of the recognized importance of Jumonji family proteins in regulating chromatin structure, cellular function and development (Cloos et al., 2008; Nottke et al., 2009), we focused on the interaction of JMJ with PRC2. Fractionation of nuclear extracts from CJ7 ESCs by gel filtration shows that JMJ comigrates with EZH2 with two peaks centered at 3908 kD and 395 kD (Figure 1F), indicating that JMJ is possibly associated with a fraction of PRC2 in ESCs. Streptavidin capture of bfEZH2 and bfEED in ESCs precipitates endogenous JMJ (Figure 1G). An anti-SUZ12 antibody mediates coimmunoprecipitation of JMJ but not RBP2, the H3K4 demethylase, in wild-type ESCs (Figure 1H and S1). Reciprocal coimmunoprecipitation mediated by an anti-JMJ antibody captures endogenous EZH2, EED and SUZ12 (Figure 1I and 1J). These results demonstrate that JMJ interacts with PRC2 in a physiological setting.
In Ezh2−/− mutant ESCs JMJ fails to coimmunoprecipitate with EED, and captures significantly less SUZ12 compared to that in wild-type cells (Figure 1I). In addition, an EZH2 mutant lacking the catalytic SET domain (bfEZH2dSET) (Shen et al., 2008) coimmunoprecipitates with SUZ12 but not JMJ (Figure 1G). These results implicate EZH2 and its SET domain in mediating in vivo association of JMJ and PRC2. EED appears dispensable for this interaction as JMJ coimmunoprecipitates with EZH2 and SUZ12 in Eed−/− ESCs. Endogenous JMJ pulls down PRC2 not only in ESCs, but also in mouse erythroleukemia (MEL) cells and F9 embryonic carcinoma cells (Figure 1J). Therefore, the interaction between JMJ and PRC2 is not unique to pluripotent ESCs.
JMJ colocalizes with PRC2 on chromatin
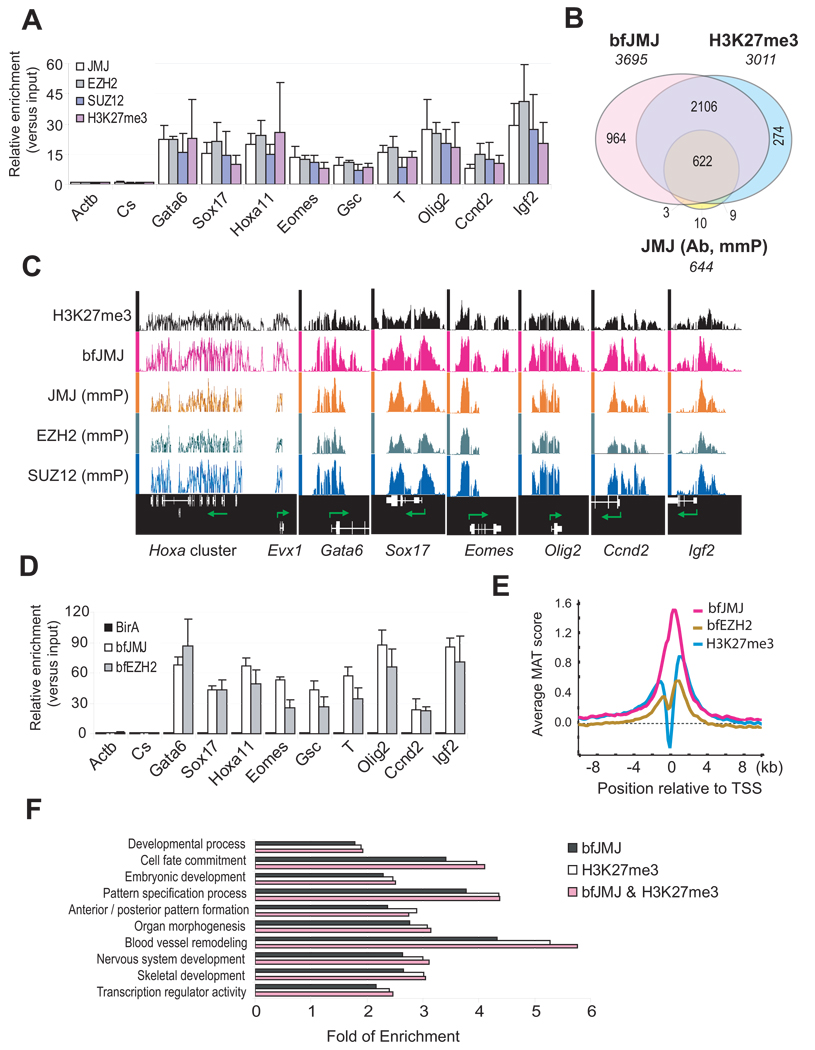
JMJ contains a conserved ARID / BRIGHT domain, which can bind DNA (Kim et al., 2003). We sought to identify DNA targets of JMJ. Chromatin immunoprecipitation (ChIP) in ESCs using antibody to JMJ shows that endogenous JMJ is significantly enriched at promoters of PRC2 target genes (Figure 2A). To identify gene targets of JMJ in a global fashion, we performed ChIP-chip on Affymetrix promoter tiling arrays. Out of 644 target genes bound by JMJ (Figure 2B; Table S7, S8 and S13), 632 (98%) genes overlap with genes marked by H3K27me3 (Shen et al., 2008). Moreover, 81% ~ 98% of JMJ target genes overlap with targets of EZH2, SUZ12 and bfEED (Figure 2C; Table S7A and S10–S13). The extent of overlap between JMJ and PRC2 / H3K27me3 targets greatly exceeds the reported colocalization between RBP2 and H3K27me3, in which only 195 out of 606 RBP2 target genes overlap with H3K27me3 targets (Pasini et al., 2008).
Figure 2. JMJ colocalizes with the PRC2 and H3K27me3 mark on chromatin.
(A) ChIP-qPCR analysis in ESCs. The Actb promoter and an intergenic region (Cs) serve as negative controls. Error bars represent standard deviations of relative enrichments to inputs.
(B) Venn diagram of genome-wide co-localization of JMJ, bfJMJ and the H3K27me3 mark.
(C) Representative view of ChIP-chip peaks at various genomic loci in the Affymetrix Integrated Genome Browser. Arrows indicate transcription start and direction. Unless indicated by “mmP” (Affymetrix mouse promoter tiling array), chip hybridization was performed on mouse whole genome tiling array sets.
(D) BioChIP-qPCR analysis in ESCs expressing birA control only, bfJMJ or bfEZH2. Error bars represent standard deviations of relative enrichments to inputs.
(E) ChIP-chip signals around transcription start sites (TSS). Average enrichment ratios within +/− 10kb regions relative to TSS were calculated based on MAT scores.
(F) Gene ontology (GO) analysis of H3K27me3 target genes (p < 1e-5).
Results with the conventional ChIP method are highly dependent on the quality of available antibodies to the target protein, whereas bioChIP obviates the need for antibodies and takes advantage of the strong interaction between biotin and streptavidin (Kim et al., 2008; Shen et al., 2008). To refine the genome-wide targets of JMJ, we analyzed ESCs expressing subendogenous level of bfJMJ (data not shown) and confirmed the enrichment of bfJMJ on PRC2 targets (Figure 2D). BioChIP-chip of bfJMJ on Affymetrix whole genome tiling arrays revealed many more targets (3695 genes) than JMJ antibody-mediated ChIP-chip (Figure 2B and Table S9). As 97% of JMJ ChIP-chip targets overlap bioChIP-chip targets (Table S7), bioChIP-chip faithfully maps comprehensive coverage of chromatin binding of endogenous JMJ. Over 80% of H3K27me3, bfEZH1 or bfEZH2 targets overlap those of bfJMJ (Table S7), confirming genome-wide colocalization of JMJ with PRC2 and H3K27me3. JMJ occupies genomic regions in close proximity to transcription start sites (TSS) (Figure 2E). Similar to H3K27me3 target genes, JMJ target genes are enriched in development-related functions (Figure 2F). Thus, JMJ interacts with PRC2 at both protein and chromatin levels.
JMJ-containing protein complex shows methyltransferase activity on H3K27
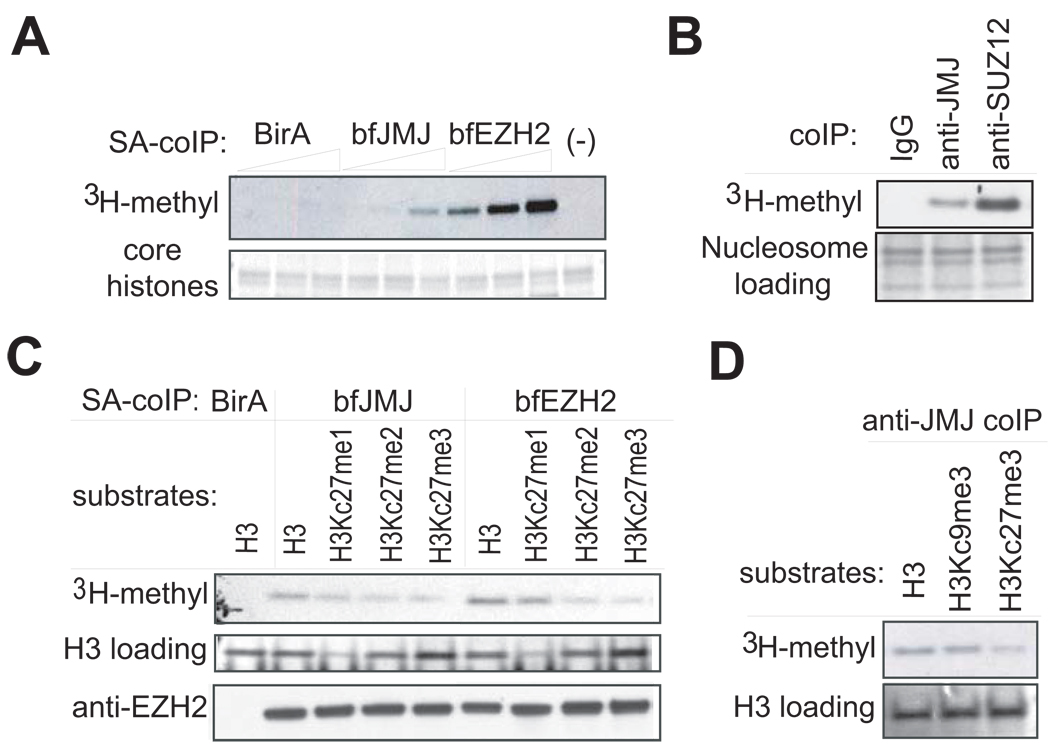
To address functional implication of the interaction of JMJ with PRC2, we assayed JMJ-associated proteins for lysine methyltransferase (KMT) activity. Indeed, proteins that coprecipitate with bfJMJ in ESCs demonstrate KMT activity towards core histones in a dose-dependent manner similar to the bfEZH2-containing complex captured by streptavidin (Figure 3A). In addition, the endogenous JMJ-containing complex precipitated by an anti-JMJ antibody shows KMT activity towards nucleosomes, which are the natural substrates in vivo (Figure 3B).
Figure 3. Histone methyltransferase assays show intrinsic KMT activity within the JMJ-containing complex.
(A) Addition of increasing amounts of the JMJ-containing complex captured by streptavidin (SA)-mediated coIP from ESCs expressing bfJMJ exhibits increasing KMT activity on chicken core histones. BirA and bfEZH2 serve as the negative and positive control, respectively.
(B) Proteins captured by anti-JMJ and anti-SUZ12 antibodies show KMT activity toward nucleosomes from Hela cells. Nucleosomes were pretreated at 50°C for 10 minutes to inactivate copurified, endogenous KMTs.
(C) KMT activity of bfJMJ and bfEZH2 complexes on recombinant histone H3 substrates. The bottom panel shows Western blotting of EZH2 to ensure equal addition of coIP’ed protein complexes.
(D) KMT activity of endogenous JMJ protein complex captured by an anti-JMJ antibody on recombinant histone H3 substrates. The lower panels in (A), (B) and (D) and the middle panel in (C) are protein gels stained by Colloidal blue to show equal addition of substrates.
To study substrate specificity, recombinant histones bearing methyl lysine analogues (MLAs) were used to examine desired methylation patterns on histone H3. These MLAs (indicated as Kc) behave similarly to their natural counterparts (Simon et al., 2007). The bfEZH2-containing complex shows decreasing activity on the H3 substrates with increasing numbers of methyl groups on residue 27 (Figure 3C). This result is consistent with the report that mono-methylation activity of PRC2 is dominant to its tri-methylation activities in vitro (Sarma et al., 2008). A weak methylation signal observed on H3Kc27me3, which contains saturated methyl groups, may reflect non-specific activity of PRC2 on lysine 9 in vitro. Although PRC2 specifically acts on H3K27 in vivo, recombinant human PRC2 exhibits weak KMT activity on H3K9 in vitro (Kuzmichev et al., 2002).
The bfJMJ complex exhibits similar substrate specificity as the bfEZH2 complex with the highest and lowest activity towards unmethylated H3 and H3Kc27me3, respectively, supporting physical existence of JMJ and PRC2 in one protein complex. Moreover, the endogenous JMJ-containing complex shows similar activity on unmethylated H3 and H3Kc9me3 substrates, but significantly weaker activity on the H3Kc27me3 substrate (Figure 3D), implying that the JMJ-PRC2 complex specifically acts on H3K27, but not H3K9.
JMJ inhibits PRC2 activity in vitro
Mammalian JMJ contains several conserved motifs including JmjN, ARID/BRIGHT, JmjC and C5HC2 zinc finger domains (Figure S2). Mutation of the critical residues involved in cofactor binding in the JmjC domain predicts that JMJ is inactive as a demethylase, a function distinguishing many Jumonji family members (Cloos et al., 2008). Consistent with the prediction, we failed to detect lysine demethylase (KDM) activity of JMJ regardless of the presence of EZH2 (Figure S3). Moreover, addition of JMJ had no obvious effect on known KDMs (Figure S3).
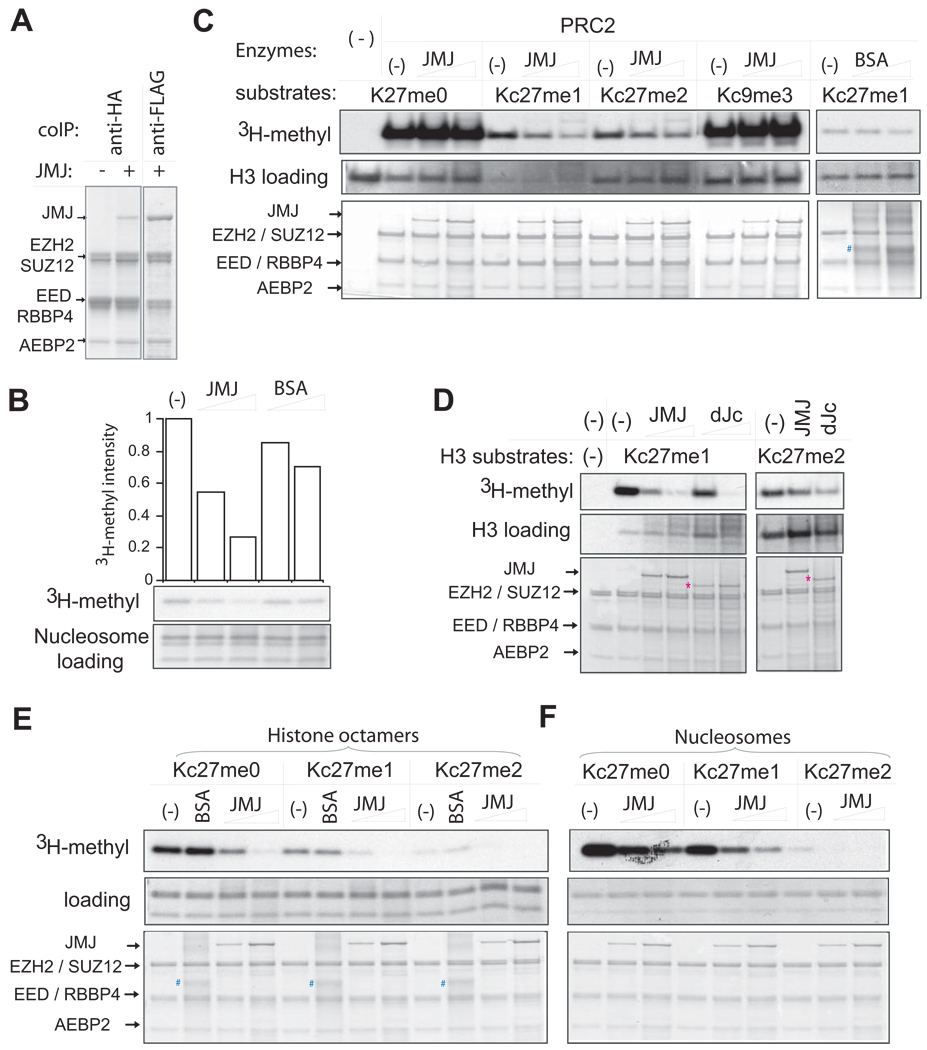
To ask whether JMJ assembles as a stoichiometric component of PRC2, we reconstituted JMJPRC2 through baculoviral expression. Anti-HA coimmunoprecipitation targeted to EED captures JMJ, which is present at one-fourth the amount of EZH2. Therefore, JMJ is a substoichiometric component of PRC2 (Figure 4A). This result suggests that PRC2 may exist in two forms: JMJ-containing and JMJ-free PRC2 complexes.
Figure 4. JMJ inhibits methyltransferase activity of PRC2.
(A) Reconstitution of JMJ with PRC2 in vitro. Except for the first lane in which JMJ-expressing baculovirus is omitted, FLAG-tagged JMJ was coexpressed with FLAG-tagged EZH2, HA-tagged EED, SUZ12, RBBP4 and AEBP2 in Sf21 cells. Captured proteins were separated on the gel and stained by Colloidal blue. While equal amounts of JMJ and EZH2 were captured by anti-FLAG antibody, JMJ captured with HA-EED is one-fourth the amount of EZH2, indicating JMJ as a substoichiometric component of PRC2.
(B) JMJ inhibits intrinsic methyltransferase activity associated with crude nucleosomes purified from Hela cells. The top plot shows the quantification of the autoradiography (in the middle). The lower panel shows equal loading of nucleosomes (5 µg). About 0.5 – 2 µg of JMJ and 4 – 10 µg of bovine serum albumin (BSA) were added as indicated.
(C) JMJ inhibits di- and tri-methyltransferase activities of PRC2 on recombinant histones (1 µg per reaction). About 2 – 10 µg of BSA (indicated by #) was added as indicated.
(D) The JmjC and C5HC2 domains of JMJ are dispensable for the inhibitory function of JMJ on PRC2. The protein band corresponding to the JMJ mutant (dJc) is indicated by *.
(E) – (F) JMJ inhibits all methylation activities of PRC2 on MLA octamers (E) and nucleosomes (F). About 2 µg of BSA was added. In (C) – (F), 5 µg of PRC2 and 0.5 µg – 2 µg of JMJ or dJc were added as indicated. The upper panels show autoradiography. The middle and bottom panels are protein gels stained by Colloidal blue to show additions of substrates and enzymes, respectively.
A chromatin fraction mainly comprised of oligonucleosomes from Hela cells contains endogenous KMT activity (Figure 4B). Addition of increasing amounts of JMJ but not bovine serum albumin (BSA) markedly inhibits this activity. Close interaction between JMJ and PRC2 suggests that JMJ may directly regulate the KMT activity of PRC2. To test this possibility, we reconstituted KMT reactions with recombinant PRC2 and histone H3 MLAs with the defined methylation pattern at residues Kc27 and Kc9.
Addition of JMJ did not affect PRC2-mediated mono-methylation on H3 and H3Kc9me3 substrates (Figure 4C). JMJ dramatically down-modulates PRC2 activity on H3Kc27me1 and me2 substrates in a dose-dependent manner (Figure 4C). BSA has no effect on PRC2, indicating the specificity of the assay. JMJ failed to inhibit PRC2 on chicken core histones (data not shown), presumably due to dominant monomethylation catalyzed by PRC2 in vitro, which may mask the inhibitory effect of JMJ on di- and tri-methylation. The JMJ mutant (dJc) lacking the C-terminal JmjC and C5HC2 domains also inhibits di- and tri-methylation activity of PRC2 (Figure 4D), suggesting that the JmjC and C5HC2 domains are dispensable for JMJ activity on PRC2.
To test whether JMJ specifically inhibits PRC2 activity on more physiological substrates, we assembled recombinant histone octamers and nucleosomes containing H3Kc27me0, me1 or me2. Consistent with histone substrates, JMJ inhibited PRC2 mediated di- and tri-methylation in a dose-dependent manner on MLA octamers and nucleosomes (Figure 4E and 4F). JMJ also inhibited monomethylation activity of PRC2 on H3Kc27me0 octamers and nucleosomes, indicating that JMJ-PRC2 may exhibit different enzymatic activities towards H3 histone, octamers, core histones (which contain octamers plus the linker histone H1) and nucleosomal substrates in vitro. Nevertheless, these results demonstrate that JMJ inhibits di- and tri-methylation activity of PRC2.
JMJ fine-tunes the H3K27me3 level in vivo
To investigate how JMJ regulates PRC2 function in vivo, we isolated Jmj conditional knockout (fl/fl) ESCs. Deletion of exon 3 creates a frame shift and subsequent termination mutation (Figure S2A), resulting in a mutant peptide of 15 amino acids that contains none of the known functional motifs of JMJ (Mysliwiec et al., 2006). By Cre recombinase-mediated excision of the fl alleles in vitro, we established Jmj−/− ESCs, in which JMJ expression is completely abolished and JMJ is no longer detected at PRC2 target loci by ChIP (Figure 5A – 5C). Anti-SUZ12 coimmunoprecipitation in Jmj−/− cells fails to detect a protein band representing JMJ (Figure 1H), confirming the specificity of the interaction between JMJ and PRC2. Jmj−/− ESCs show normal expression of pluripotency markers (Table S14) and proliferate at a similar rate as wild-type ESCs under standard conditions (data not shown). Thus, JMJ is dispensable for ESC self-renewal and maintenance.
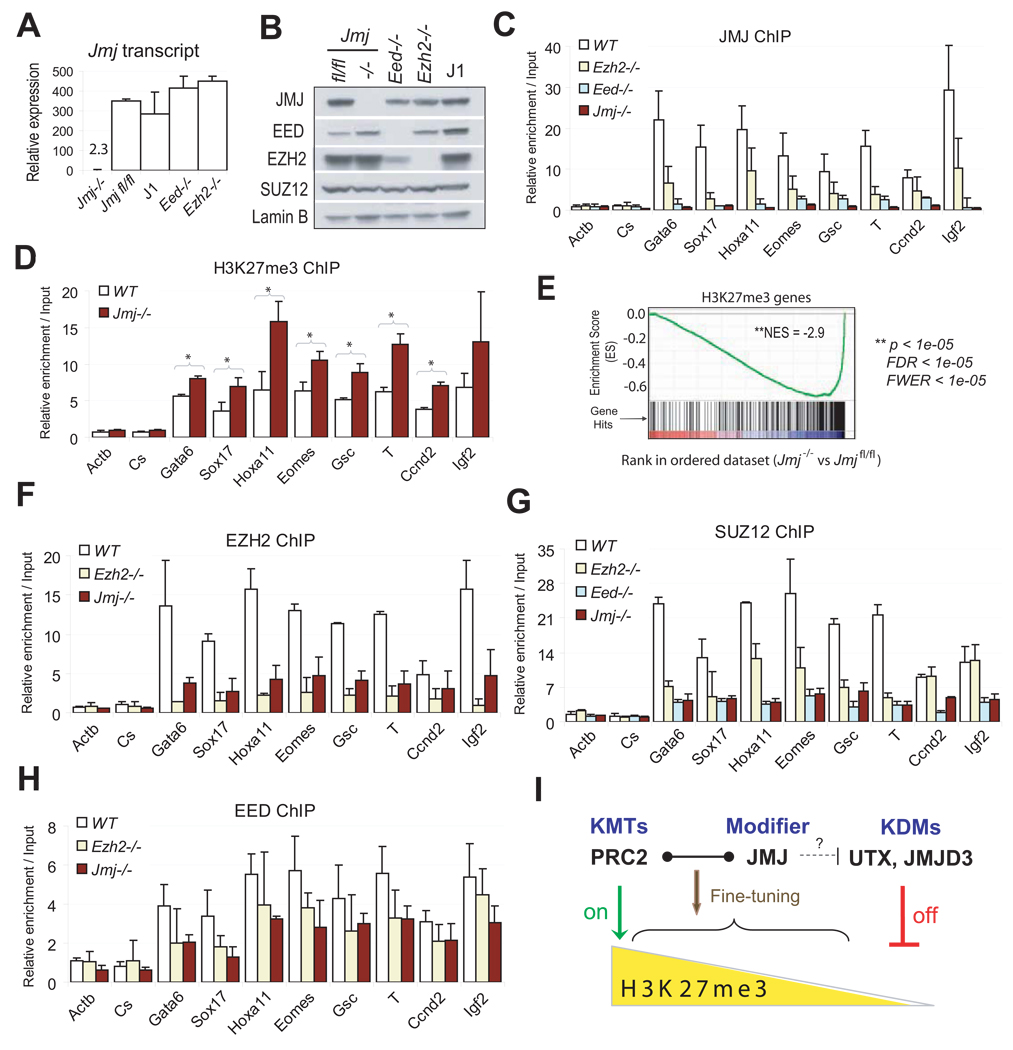
Figure 5. Characterization of Jmj−/− ESCs.
(A) Reverse transcription (RT) and qPCR analysis of Jmj transcripts in wild-type and mutant ESCs. Error bars are standard deviations of relative expression to GADPH.
(B) Western blotting analysis of wild-type and mutant ESCs.
(C) ChIP-qPCR analysis of JMJ in wild-type (WT) and mutant ESCs.
(D) ChIP-qPCR analysis of the H3K27me3 mark in Jmj−/− and wild-type ESCs. * indicates p < 0.05 by a Student's paired t-Test with a two-tailed distribution.
(E) GSEA profile of the set of H3K27me3 target genes. Genes are ranked into an ordered list based on the correlation between their expression levels in Jmj−/− and Jmjfl/fl ESCs. Genes which are upregulated in Jmj−/− ESCs are ranked at the top of the list (left, red), while downregulated genes are ranked toward the bottom (right, blue). The middle portion of the plot shows where the members of the gene set (indicated by vertical lines) locate in the ranked list shown in the bottom portion. The green curve in the top portion shows the running enrichment score (ES) for the gene set as the analysis walks down the ranked list. The distribution of H3K27me3 target genes appear toward the bottom of the rank list with a normalized enrichment score (NES) of −2.9. Statistic significance is indicated by nominal p value, familywise-error rate (FWER) and false discovery rate (FDR).
(F) – (H) ChIP-qPCR analysis of EZH2 (F), SUZ12 (G) and EED (H) at PRC2 targets in wild-type and mutant ESCs. In (C), (D) and (F)–(H), error bars are standard deviations of relative enrichments based on at least three biological repeats. The Actb promoter and an intergenic region (Cs) serve as negative controls.
(I) A model shows dynamic regulation of H3K27me3. PRC2, the H3K27 methyltransferase complex (KMT), and UTX and JMJD3, the H3K27me3 demethylases (KDM), represent on-off controls on H3K27me3. While PRC2 catalyzes the formation of H3K27me3, UTX and JMJD3 demethylate H3K27me3. In contrast to KMTs and KDMs, JMJ, as a modifier of PRC2, fine-tunes the levels of H3K27me3 by regulating the activity and recruitment of PRC2.
As JMJ inhibits PRC2 tri-methyltransferase activity in vitro, we investigated whether loss of JMJ affects H3K27me3 levels in vivo. Global levels of H3K27 methylation, as well as other histone marks, are not affected in Jmj−/− ESCs (Figure S4), consistent with a role of JMJ as a modifier of PRC2. However, moderate, but consistent, enhancement of H3K27me3 binding at individual PRC2 target loci is observed in Jmj−/− cells (Figure 5D).
To study the effect of Jmj loss on gene expression of PRC2 and H3K27me3 targets, we performed microarray profiling of Jmj−/− and Jmjfl/fl ESCs (Table S14). By Gene Set Enrichment Analysis (GSEA), we ranked genes from high to low based on the correlation between their expression levels in these two cell types (Figure 5E). Genes that are upregulated in Jmj−/− ESCs compared to Jmjfl/fl ESCs are ranked at the top of the list, while genes that are downregulated are ranked toward the bottom of the list. We previously identified a subset of H3K27me3 target genes referred to as ‘H3K27me3-(WT)-Day6 UP’ genes that are repressed in undifferentiated ESCs but upregulated at day 6 of ESC differentiation (Shen et al., 2008). We then asked where members of this H3K27me3 target gene set are distributed in the ranked dataset. We find that H3K27me3 target genes distribute toward the bottom of the rank list with a significant normalized enrichment score (NES) of −2.9 (Figure 5E), indicating reduced expression in Jmj−/− ESCs. Thus, accentuated repression of H3K27me3 genes is consistent with ChIP analysis, indicating that increased presence of H3K27me3 on chromatin may further repress PRC2 targets that normally exhibit low levels of transcription.
The enrichment of JMJ at PRC2 targets is markedly reduced in Ezh2−/− and Eed−/− ESCs (Figure 5C), indicating a requirement of EZH2 and EED in targeting JMJ to chromatin. This result is consistent with the observation that JMJ lacks specific binding affinity to methylated histones, as revealed by histone peptide binding assays (Figure S5). On the other hand, enrichment of SUZ12, EZH2 and EED at PRC2 targets is significantly decreased in Jmj−/− ESCs (Figure 5F – 5H), pointing to a positive role of JMJ in facilitating PRC2 binding to chromatin. Thus, JMJ modulates PRC2 function in opposing ways. As PRC2 and UTX / JMJD3 function as lysine methyltransferases (KMTs) and demethylases (KDMs), respectively, to control “on” and “off” switches of H3K27me3, JMJ functions as a modifier of PRC2 to fine-tune the level of H3K27me3 by promoting chromatin binding, while also inhibiting the KMT activity of PRC2 (Figure 5I).
JMJ is required for proper ESC differentiation
The core components of PRC2 including EZH2, EED and SUZ12 appear to be more important in executing than maintaining pluripotency (Pasini et al., 2007; Shen et al., 2008). To investigate the role of JMJ in regulating ESC pluripotency, we differentiated Jmj−/− and Jmjfl/fl ESCs by withdrawal of leukemia inhibitory factor (LIF) and performed microarray expression profiling.
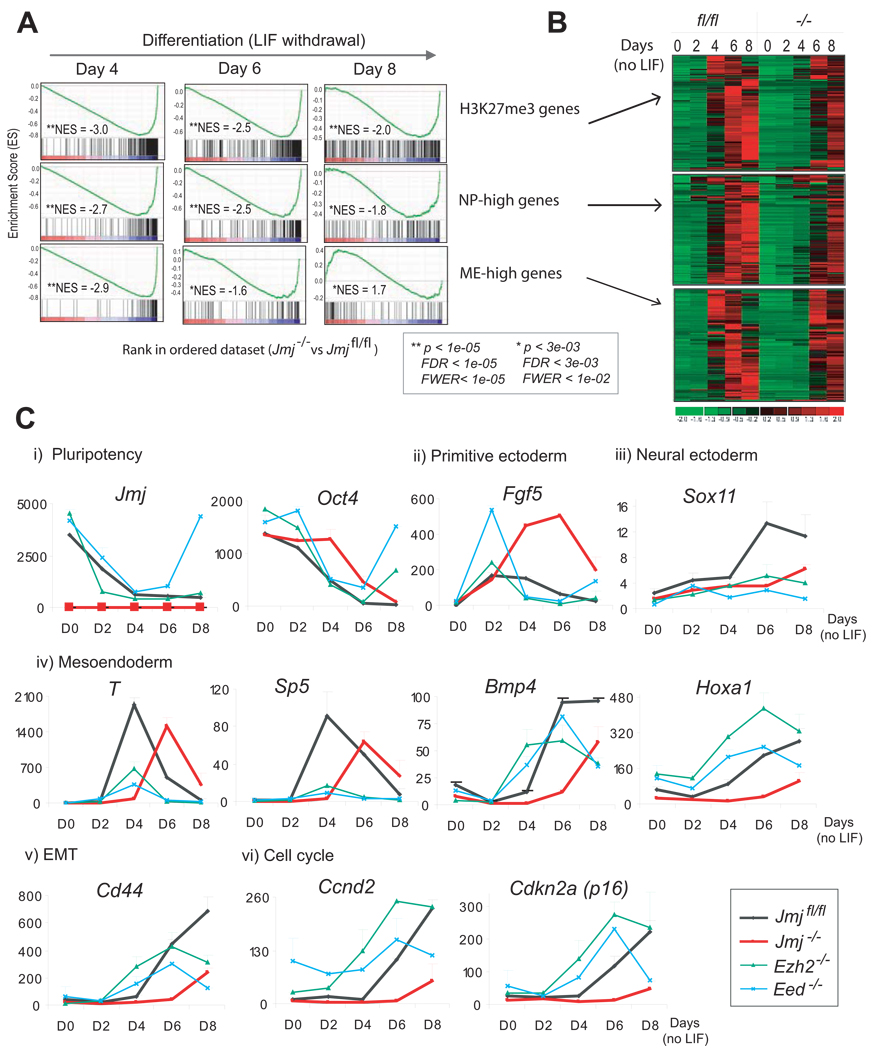
Global gene expression in Jmj−/− ESCs was compared to that in Jmjfl/fl ESCs by GSEA. H3K27me3 targets are distributed primarily toward the bottom of the ranked list in their enrichment profiles with the lowest NES of −3.0 at day 4 compared to −2.5 and −2.0 at day 6 and 8 of differentiation, respectively (Figure 6A). Consistent with GSEA which provides a statistical comparison of a prior defined gene set between two cell types, heatmap analysis shows that the activation of H3K27me3 target genes in Jmj−/− ESCs is delayed and not activated to the same extent as in Jmjfl/fl cells during differentiation (Figure 6B).
Figure 6. Jmj is required for ESC differentiation.
(A) GSEA profiles of the sets of H3K27me3 target genes, NP-high (enriched in neuronal progenitors) and ME-high genes (enriched in mesoendodermal lineage).
(B) Heatmaps show expression of genes in three gene sets indicated in (A) in Jmjfl/fl and Jmj−/− cells during ESC differentiation.
(C) Time-course analysis of marker genes during differentiation by RT-qPCR. Error bars are standard deviations of relative expression shown in the y-axis.
Upon LIF withdrawal pluripotent ESCs differentiate into three germ layers including neuroectoderm, mesoderm and endoderm. Mesoendoderm (ME) is a transient cell state prior to further differentiation into mesoderm and endoderm. Both JMJ and H3K27me3 occupy a set of genes critical for development. We studied the effect of JMJ loss on lineage differentiation, particularly genes enriched in neuronal progenitors (NP) and mesoendodermal (ME) lineages, which were previously defined as “NP-high” genes and “ME-high” genes, respectively (Shen et al., 2008).
GSEA and heatmap analysis indicate significant underrepresentation of NP-high genes in Jmj−/− cells during differentiation (Figure 6A and 6B). ME-high genes in Jmj−/− cells are also significantly downregulated at day 4 and day 6 of differentiation, but eventually become overrepresented with a positive NES of 1.7 at day 8 of differentiation (Figure 6A and 6B). Thus, loss of JMJ results in compromised and delayed differentiation of ESCs. Imposition of excessive H3K27me3 marks in the absence of JMJ may render a less permissive chromatin conformation on target genes and attenuate rapid reconfiguration of chromatin states in response to differentiation signals.
To confirm microarray analysis, we analyzed expression of marker genes in wild-type (Jmjfl/fl) and polycomb mutant ESCs including Jmj−/−, Ezh2−/− and Eed−/− cells (Figure 6C). Jmj transcripts are downregulated 6-fold and 8-fold, respectively, at day 4 and day 8 of differentiation, implying early involvement of JMJ in regulation of ESC differentiation. Upon differentiation higher expression of ESC specific genes, including the pluripotency marker Oct4, is observed in Jmj−/− cells as compared to Jmjfl/fl cells (Figure 6C and S6). Delayed attenuation of pluripotency genes confirms a delay in differentiation.
Fgf5 is a marker for primitive ectoderm, a transient cell state that pluripotent ESCs must proceed through before differentiating to three germ layers. In wild-type ESCs, the expression of Fgf5 reaches its peak at day 2 of differentiation and is gradually downregulated thereafter (Figure 6C). This pattern of expression is consistent with a transitory existence of primitive ectoderm during development. However, Ezh2−/− and Eed−/− cells show abnormal activation of Fgf5 at day 2 but an abrupt drop to a significantly lower level at day 4, suggesting rapid entrance into and exit from the primitive ectoderm state. Despite normal activation at day 2 in Jmj−/− cells, Fgf5 expression progressively rises to nearly 8-fold higher than that in wild-type cells at day 6. Persistent, high level expression of Fgf5 suggests that Jmj−/− cells differentiate into primitive ectoderm but further development is stalled at this stage.
Consistent with prolongation of the primitive ectodermal state, Jmj−/− cells show delayed commitment to the three somatic lineages (Figure 6C). For example, neuronal gene Sox11 fails to be upregulated upon differentiation in mutant cells. Brachyury (T) – an early marker for mesoendoderm reaches its peak expression at day 4 in wild-type cells. However, Jmj−/− cells weakly express T at day 4 and show peak activation at day 6, at a time when T expression in wild-type cells has already been downregulated. Additional mesoderm markers, such as Sp5, Bmp4 and Hoxa1, also show delayed and compromised activation. In Ezh2−/− and Eed−/− cells, both T and Sp5 show a similar time-course of activation as wild-type ESCs, but the extent of activation is reduced, reflecting induction but failure to sustain mesoendoderm in these mutant cells.
Expression of CD44, a cell surface marker in mesenchymal stem cells important for epithelial-mesenchymal transition (EMT), is not activated appropriately in Jmj−/− cells (Figure 6C). In contrast, CD44 expression is activated but not sustained in Ezh2−/− and Eed−/− cells. Lastly, cell cycle regulators, including cyclin D2 (Ccnd2) and Cdkn2a (p16), which are highly expressed in all three germ layers, are expressed at abnormally high levels in Ezh2−/− and Eed−/− cells, suggesting that differentiation may be initiated but cannot be sustained due to cell-cycle arrest. In contrast, Ccnd2 and Cdkn2a are not activated until day 8 of differentiation in Jmj−/− cells with expression levels significantly lower that those in Jmjfl/fl cells (Figure 6C). Therefore, global and individual gene analysis demonstrate that Jmj−/− ESCs fail to respond to differentiation signals promptly and exhibit delayed, compromised lineage commitment.
DISCUSSION
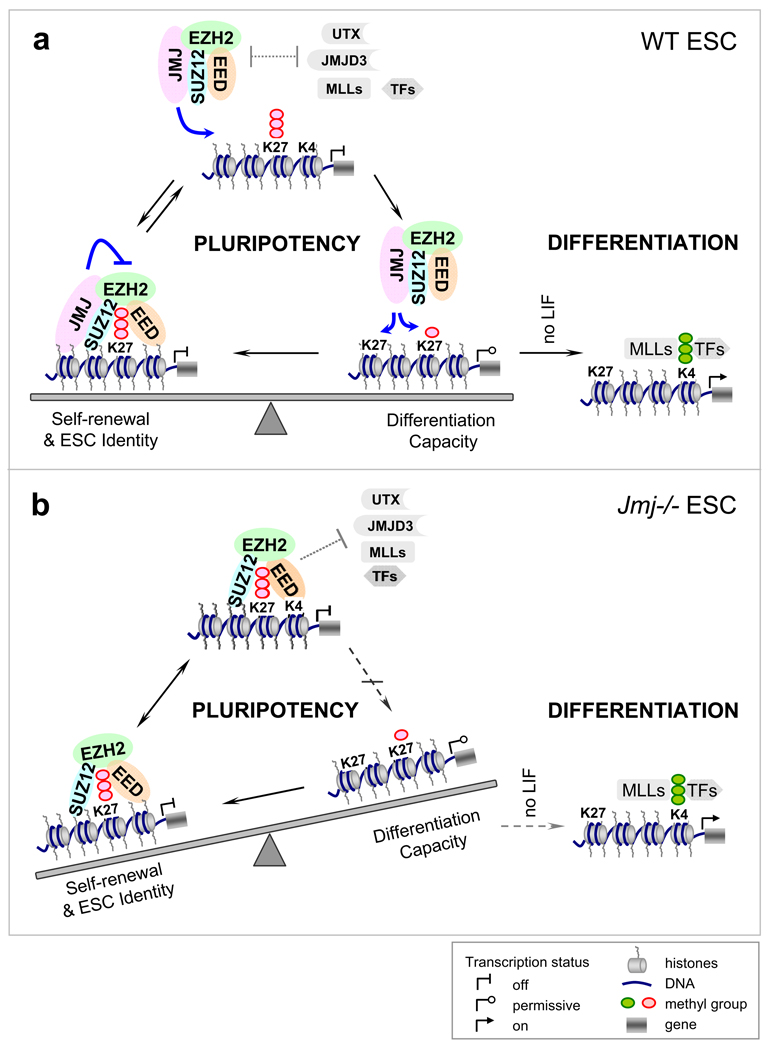
We have identified JMJ as a novel associated protein of the PRC2 complex. JMJ colocalizes with PRC2 and the H3K27me3 mark in chromatin. The endogenous JMJ-containing complex exhibits specific methyltransferase activity on H3K27, indicating that JMJ serves as a subunit of the active PRC2 complex. Despite its lack of KDM activity, JMJ modulates PRC2 function in opposing ways. While promoting PRC2 recruitment to DNA targets, JMJ inhibits methyltransferase activity of PRC2. Jmj−/− ESCs show increased levels of H3K27me3 at target loci and fail to activate lineage differentiation in a proper time-course and to the same extent as in wild-type cells. Thus, loss of Jmj shifts the balance of pluripotency towards maintenance of the ESC phenotype and compromises differentiation (Figure 7). These consequences of JMJ loss establish its critical functions in modulating H3K27me3 levels and orchestrating developmental transitions. Our study reveals a new dimension in the dynamic epigenetic regulation of gene expression.
Figure 7. A model for the interplay between JMJ and PRC2 in regulating ESC pluripotency.
(A) In wild-type ESC, the inhibitory role of JMJ on PRC2 activity disfavors formation of H3K27me3 and may facilitate attacks by opposing enzyme or factors, such as UTX, JMJD3 and MLLs, resulting in transient H3K27me3 demethylation and a permissive chromatin structure for transcription. Paradoxically, the positive role of JMJ in promoting PRC2 binding may redirect PRC2 to chromatin to establish the H3K27me3 mark. Dynamic control of the H3K27me3 mark defines the fine balance between the maintenance of the ESC state and differentiation capacity, and is required for rapid relief of repression and subsequent gene activation upon differentiation signals.
(B) In Jmj−/− ESCs, H3K27me3 may be more rigidly positioned at target loci. The balance tips towards maintenance of the stem cell phenotype. Genes marked and repressed by H3K27me3 become refractory to activation on differentiation signals.
JMJ interacts with PRC2
Both JMJ and RBP2 belong to the JARID family. It was reported previously that RBP2 interacts with PRC2 to maintain transcriptional repression by coordinating H3K4me3 demethylation and H3K27 methylation (Pasini et al., 2008). However, we have not been able to confirm the association of RBP2 with PRC2 in ESCs. Sequencing of PRC2 protein complexes failed to reveal RBP2-derived peptides (Table S1–S4), and RBP2 did not precipitate with EZH2 and SUZ12 (Figure 1H and S1). Coimmunoprecipitation against RBP2 in ESCs did not capture EZH2 (Q. Yan and W. Kaelin’s unpublished data). In addition, RBP2 was barely detected when RBP2 and PRC2 components were reconstituted in vitro (Figure S1). RBP2 colocalizes with H3K4me3 but not H3K27me3 in human promonocytic U937 cells (Lopez-Bigas et al., 2008). Depletion of RBP2 does not affect PRC2 binding to chromatin (Pasini et al., 2008). Thus, we believe that JMJ but not RBP2 is the bona fide JARID family protein interacting with PRC2 in ESCs. However, other JMJ-like, Jumonji family proteins may associate with PRC2 in other developmental contexts. For instance, FBXL10 (KDM2B), which is found to interact with EZH2 in 293T cells, promotes H3K4me3 and H3K36me2 demethylation, but facilitates PRC2 binding and H3K27me3 formation at the Ink4a/Arf locus (Tzatsos et al., 2009).
PRC2 regulates multiple aspects of stem cell proliferation and differentiation, and is overexpressed in diverse cancers. Considering that the association of JMJ and PRC2 is not unique to pluripotent ESCs, we speculate JMJ may modulate PRC2 functions in other stem cells. As a portion of PRC2 appears free of JMJ, it will be of interest to determine how the interaction is regulated. We find that JMJ is highly prone to protein degradation during isolation of nuclear extracts (X. Shen and S.H. Orkin’s unpublished data). Regulation of JMJ stability through posttranslational modification or by other protein factors may be a possible means to modulate its function.
JMJ regulates PRC2 activity
Cooperation between SET domain-containing KMTs and Jumonji family KDMs appears to be a consistent theme in epigenetic regulation of transcription. The association of the H3K27me2/3 demethylases UTX and JMJD3 with the H3K4 methyltransferase complexes (MLLs) provides an example of coupling removal of a repressive mark with establishment of an active mark (De Santa et al., 2007; Issaeva et al., 2007; Lee et al., 2007). JMJ promotes chromatin targeting of PRC2, while it inhibits the enzymatic activity of PRC2. This interplay reveals a novel mechanism by which an epigenetic modifier regulates gene expression by modulating its interacting KMT.
Despite compromised recruitment of PRC2 in Jmj−/− ESCs, the level of H3K27me3 on PRC2 target loci is paradoxically increased. ChIP analyses of PRC2 and the H3K27me3 mark may reflect different kinetics: transient recruitment of enzymatic activity versus steady-state levels of a modification, respectively. H3K27me3 occupancy on chromatin integrates PRC2 binding and activity, while PRC2 association on chromatin is a snapshot of protein binding. With respect to H3K27me3 levels, our model suggests increased activity of PRC2 gained by loss of JMJ inhibition appears to outweigh the diminished recruitment of PRC2 to chromatin in Jmj−/− ESCs. Alternately, loss of JMJ allows other modifiers to interact with and modulate PRC2. For example, sequence analysis implies that JMJ-containing PRC2 and MTF2-containing PRC2 may be distinct protein complexes. Interestingly, PCL1, a homolog of MTF2, in fly and mammals is required to generate high levels of H3K27me3 at PcG target sites by directly stimulating PRC2 activity and possibly stabilizing PRC2 binding to chromatin (Cao et al., 2008; Nekrasov et al., 2007; Sarma et al., 2008; Savla et al., 2008). Loss of JMJ may augment MTF2 binding to PRC2, hence increasing PRC2 activity and H3K27me3 levels in vivo. In addition, we cannot rule out a possible role of EZH1 in establishing high levels of H3K27me3 in Jmj−/− ESCs. Despite decreased chromatin association of SUZ12 in Ezh2−/− ESCs, H3K27me3 levels on a subset of developmental genes are paradoxically upregulated due to the non-canonical EZH1-containing PRC2 (Shen et al., 2008). Whether JMJ regulates EZH1-PRC2 activity awaits further analysis.
Of 27 Jumonji family proteins in human genome, only 15 have been reported to contain demethylase activity (Cloos et al., 2008). Many Jumonji family proteins, including Hairless (HR), PHF2 and Epe1, lack demethylase activity but remain important in development. For example, HR is a transcriptional corepressor essential for hair growth, but how HR regulates the balance of proliferation and differentiation in epidermis cells is elusive (Zarach et al., 2004). JMJ function within the PRC2 complex may provide a paradigm for regulation of KMT activity by other enzymatically-inactive Jumonji family proteins.
JMJ in stem cell differentiation and development
Similar to Ezh2 and Eed, Jmj serves an important role in regulating ESC differentiation. However, Jmj−/− ESCs exhibit a distinct differentiation time course from Ezh2−/− and Eed−/− cells. Whereas Jmj−/− cells show abnormal, prolonged activation of primitive ectoderm and delayed entrance into mesoendoderm, Ezh2−/− and Eed−/− cells initiate but fail to sustain primitive ectoderm and mesoendoderm. These phenotypic differences may reflect intrinsic differences in how the H3K27me3 level at specific target genes is altered in these cells. Jmj−/− mutants contain excessive H3K27me3 at targets, which may lead to augmented repression and prevent efficient gene activation required for differentiation. On the contrary, a global decrease or absence of H3K27me3 in Ezh2−/− and Eed−/− cells may facilitate initial lineage induction but fail to orchestrate a normal sequence of events and epigenetically establish new cell fates that require de novo establishment of H3K27me3 on chromatin. Despite compromised and delayed differentiation of Jmj−/− ESCs in culture, Jmj-null mouse embryos gastrulate and form three germ layers, suggesting that additional compensatory mechanisms exist in vivo.
JMJ has been implicated in regulating development and differentiation. In Drosophila, Jmj is required for metamorphosis (Sasai et al., 2007). Jmj loss results in a delay of pupation and lethality at larval and pupal stages. In mouse, Jmj−/− embryos show hyperproliferating cardiac and neural progenitor cells, but lack differentiating and mature cardiac myocytes and post-mitotic neurons (Jung et al., 2005; Takeuchi et al., 2006). Jmj−/− hepatocytes proliferate normally but fail to differentiate and mature in vivo and in vitro. Taken together with our findings, these observations support a critical role for JMJ in promoting stem / progenitor cell differentiation and mediating cell fate transitions.
Fine-tuning of H3K27me3 in ESC pluripotency
The role of JMJ in execution of pluripotency is consistent with JMJ as a common target of multiple (>5) pluripotency transcription factors (Chen et al., 2008; Kim et al., 2008; Zhou et al., 2007). The expression of JMJ is robust in ESCs and induced pluripotent stem (iPS) cells (Ambrosi et al., 2007; Mikkelsen et al., 2008; Walker et al., 2007), and falls abruptly on differentiation. Levels of EED, EZH2 and SUZ12 also decrease on differentiation (Kuzmichev et al., 2005; Pasini et al., 2007). This series of events presumably permits access of active KDMs to remove the repressive H3K27me3 mark and facilitates rapid switching of expression states in response to differentiation signals.
Global low-level transcriptional activity is a hallmark of the ESC genome, and is necessary for maintenance of the pluripotent state (Efroni et al., 2008). It remains obscure how tissue-specific genes are kept at a low-level expression state in ESCs and readily activated upon differentiation signals. The interplay between PRC2 and JMJ, which enables fine-tuning, rather than on-off control, of methylation states on H3K27, provides a mechanism for preserving expression plasticity.
In wild-type ESCs, the inhibitory role of JMJ on PRC2 activity disfavors formation of H3K27me3 and may facilitate attacks by opposing enzymes or factors, such as UTX, JMJD3 and MLLs, resulting in transient H3K27me3 demethylation and a permissive chromatin structure for transcription (Figure 7). On the other hand, the positive role of JMJ in PRC2 recruitment may redirect PRC2 to chromatin to establish the H3K27me3 mark. Thus, at steady-state the distribution of H3K27me3 at target sites reflects a dynamic equilibrium controlled by JMJ and PRC2, and likely influenced by UTX, MLLs and other factors. Dynamic modulation of H3K27me3 levels allows rapid reprogramming of the epigenome upon developmental or environmental signals, and facilitates rapid relief of repression and subsequent gene activation.
In Jmj−/− ESCs, H3K27me3 may be more rigidly positioned at target loci. Imposition of static H3K27me3 modification may convert a repressive, low expression state, with inherent plasticity that permits transcriptional activation during differentiation, to an "off" state. Developmental genes marked by H3K27me3 may become refractory to gene activation in the absence of JMJ. As the balance in Jmj−/− cells tips towards maintenance of stem cell identity, these cells exhibit delayed and compromised activation of differentiation program in response to developmental signals.
Dynamic control of NANOG, a key pluripotency regulator, provides another example of the fine balance between self-renewal and differentiation. Transitory downregulation of NANOG mediated by TCF3 predisposes ESCs to differentiation but does not mark commitment, while reexpression of NANOG reverses precocious commitment and reestablishes the ESC state (Chambers et al., 2007; Cole et al., 2008). TCF3 deletion, or NANOG overexpression, renders ESCs refractory to differentiation. Thus, the balance between maintenance of stem cell identity and preservation of differentiation capability lies at the heart of pluripotency.
Because reduced or excessive H3K27me3 modification appears equally detrimental to gene regulation during cell fate switches, our finding that JMJ modulates PRC2 functions and ESC differentiation further supports the importance of chromatin dynamics for early lineage decisions.
EXPERIMENTAL PROCEDURES
Cells and culture
Mouse ESCs expressing bfEED, bfEZH1 and bfEZH2 have been described previously (Shen et al., 2008). ESCs expressing bfJMJ or bfMTF2 were generated by introducing cDNA expressing biotinand FLAG-tagged JMJ or MTF2 into J1 ESCs expressing bacterial biotin ligase birA and stable clones were selected by puromycin. Jmjfl/fl ESCs were isolated from mouse blastocysts with the same genotype and treated with GFPCre (Gagneten et al., 1997) to establish Jmj−/− ESCs. Ezh2−/− and Eed−/− ESCs were described previously (Montgomery et al., 2005; Shen et al., 2008). ESC culture and differentiation were described previously (Shen et al., 2008).
Gel filtration chromatography, affinity purification, coimmunoprecipitation and mass spectrometry
These procedures were performed as described (Shen et al., 2008; Wang et al., 2006). Details and antibodies are described in the Supplemental Data.
ChIP and bioChIP analysis
Antibody-mediated ChIP or streptavidin-mediated bioChIP were performed as described previously (Shen et al., 2008). ChIP-chip analysis of endogenous JMJ, EZH2 and SUZ12 were performed on Affymetrix mouse promoter tiling arrays. BioChIP-chip of bfJMJ was performed on Affymetrix mouse tiling 2.0R array sets. Model-based analysis of tiling array (MAT) (Johnson et al., 2006) was used to predict and map significant peaks (p < 1e-7) to the genome based on mouse mm8 genome assembly. For ChIP and quantitative qPCR analysis, DNA levels at various chromatin loci were first normalized to an internal control region located in the first intron of Actb. Relative enrichments were calculated by dividing the normalized level of ChIP DNA to that of input DNA at corresponding locus. Error bars represent standard deviations of relative enrichments from at least three biological replicates of ChIP. Primers were listed previously (Shen et al., 2008).
Microarray expression profiling and RNA analysis
Total RNA from undifferentiated (day 0) and differentiated ESCs (day 2, 4, 6 and 8) was isolated by Trizol (Invitrogen), reversely transcribed by SuperScript III (Invitrogen), labeled and hybridized onto Affymetrix mouse genome 430 2.0 arrays. The GenePattern software was used for processing raw data and GSEA (Reich et al., 2006). Heatmap comparison was generated by dChip (Li and Wong, 2001). Gene ontology (GO) analysis was performed by DAVID tools (Dennis et al., 2003). For individual RNA transcripts analysis, gene expression was normalized to GADPH. Error bars represent standard deviations of mean expression based on at least three cell and RNA isolations. Primers sequences are listed in the Supplemental Data.
Baculoviral expression and protein purification
Recombinant PRC2 was described previously (Shen et al., 2008). FLAG-tagged JMJ and the dJc mutant were expressed by the Bac-to-Bac system (Invitrogen) and purified as previously described (Shen et al., 2008).
Histone methyltransferase assay
Histone methyltransferase assays were performed as described previously (Shen et al., 2008). Crude oligonucleosomes (S2 chromatin fraction) were isolated from Hela cells as described (Umlauf et al., 2004). Recombinant histones including H3 (#31207), H3Kc9me3 (#31213) and H3Kc27me1/2/3 (#31214, #31215, #31216) were from Active Motif. MLA histone octamers and nucleosomes were prepared as described (Simon et al., 2007). About 5 µg of native nucleosomes from Hela cells, 1 µg of recombinant histones, 0.5 µg of MLA octamers or nucleosomes were used as substrates per reaction. Bovine serum albumin (BSA, Sigma) was used as a negative control for the assays.
Supplementary Material
ACKNOWLEDGMENTS
We thank T. Magnuson, K. Helin, Q. Yan, K.H. Hansen, R. Fang for reagents; K.H. Hansen, A. Kuo and O. Gozani for advice on peptide binding assays; R. Tomaino and S. Gygi for performing mass spec sequencing; DFCI Microarray Core for processing microarray samples; T. Liu and X.S. Liu for assistance with MAT; Q. Yan, B. Wilson, J. Xu and J. Perry for criticism. X.S. is supported by the Claudia Adams Barr Program in Cancer Research. M.D.S. is a HHW fellow. S.H.O is an investigator of the HHMI.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray data are available in the Gene Expression Omnibus (GEO) database under accession number GSE19169.
SUPPLEMENTAL DATA
The supplemental Data include Supplemental Experimental Procedures, 6 figures, and 14 tables and can be found with this article online at (http://).
REFERENCES
- Ambrosi DJ, Tanasijevic B, Kaur A, Obergfell C, O'Neill RJ, Krueger W, Rasmussen TP. Genome-wide reprogramming in hybrids of somatic cells and embryonic stem cells. Stem cells (Dayton, Ohio) 2007;25:1104–1113. doi: 10.1634/stemcells.2006-0532. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, et al. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Bracken CP, Wall SJ, Barre B, Panov KI, Ajuh PM, Perkins ND. Regulation of cyclin D1 RNA stability by SNIP1. Cancer research. 2008;68:7621–7628. doi: 10.1158/0008-5472.CAN-08-1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang H, He J, Erdjument-Bromage H, Tempst P, Zhang Y. Role of hPHF1 in H3K27 methylation and Hox gene silencing. Mol Cell Biol. 2008;28:1862–1872. doi: 10.1128/MCB.01589-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers I, Silva J, Colby D, Nichols J, Nijmeijer B, Robertson M, Vrana J, Jones K, Grotewold L, Smith A. Nanog safeguards pluripotency and mediates germline development. Nature. 2007;450:1230–1234. doi: 10.1038/nature06403. [DOI] [PubMed] [Google Scholar]
- Chen X, Xu H, Yuan P, Fang F, Huss M, Vega VB, Wong E, Orlov YL, Zhang W, Jiang J, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- Cloos PA, Christensen J, Agger K, Helin K. Erasing the methyl mark: histone demethylases at the center of cellular differentiation and disease. Genes Dev. 2008;22:1115–1140. doi: 10.1101/gad.1652908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MF, Johnstone SE, Newman JJ, Kagey MH, Young RA. Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 2008;22:746–755. doi: 10.1101/gad.1642408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G. The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell. 2007;130:1083–1094. doi: 10.1016/j.cell.2007.08.019. [DOI] [PubMed] [Google Scholar]
- Dennis G, Jr, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome biology. 2003;4:P3. [PubMed] [Google Scholar]
- Efroni S, Duttagupta R, Cheng J, Dehghani H, Hoeppner DJ, Dash C, Bazett-Jones DP, Le Grice S, McKay RD, Buetow KH, et al. Global transcription in pluripotent embryonic stem cells. Cell stem cell. 2008;2:437–447. doi: 10.1016/j.stem.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagneten S, Le Y, Miller J, Sauer B. Brief expression of a GFP cre fusion gene in embryonic stem cells allows rapid retrieval of site-specific genomic deletions. Nucleic acids research. 1997;25:3326–3331. doi: 10.1093/nar/25.16.3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol. 2007;27:1889–1903. doi: 10.1128/MCB.01506-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS. Model-based analysis of tiling-arrays for ChIP-chip. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung J, Mysliwiec MR, Lee Y. Roles of JUMONJI in mouse embryonic development. Dev Dyn. 2005;232:21–32. doi: 10.1002/dvdy.20204. [DOI] [PubMed] [Google Scholar]
- Kim J, Chu J, Shen X, Wang J, Orkin SH. An extended transcriptional network for pluripotency of embryonic stem cells. Cell. 2008;132:1049–1061. doi: 10.1016/j.cell.2008.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TG, Kraus JC, Chen J, Lee Y. JUMONJI, a critical factor for cardiac development, functions as a transcriptional repressor. J Biol Chem. 2003;278:42247–42255. doi: 10.1074/jbc.M307386200. [DOI] [PubMed] [Google Scholar]
- Klose RJ, Zhang Y. Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol. 2007;8:307–318. doi: 10.1038/nrm2143. [DOI] [PubMed] [Google Scholar]
- Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- Kuzmichev A, Margueron R, Vaquero A, Preissner TS, Scher M, Kirmizis A, Ouyang X, Brockdorff N, Abate-Shen C, Farnham P, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–450. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503–518. doi: 10.1016/j.molcel.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen TS, Hanna J, Zhang X, Ku M, Wernig M, Schorderet P, Bernstein BE, Jaenisch R, Lander ES, Meissner A. Dissecting direct reprogramming through integrative genomic analysis. Nature. 2008;454:49–55. doi: 10.1038/nature07056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery ND, Yee D, Chen A, Kalantry S, Chamberlain SJ, Otte AP, Magnuson T. The murine polycomb group protein Eed is required for global histone H3 lysine-27 methylation. Curr Biol. 2005;15:942–947. doi: 10.1016/j.cub.2005.04.051. [DOI] [PubMed] [Google Scholar]
- Mysliwiec MR, Chen J, Powers PA, Bartley CR, Schneider MD, Lee Y. Generation of a conditional null allele of jumonji. Genesis. 2006;44:407–411. doi: 10.1002/dvg.20221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nekrasov M, Klymenko T, Fraterman S, Papp B, Oktaba K, Kocher T, Cohen A, Stunnenberg HG, Wilm M, Muller J. Pcl-PRC2 is needed to generate high levels of H3-K27 trimethylation at Polycomb target genes. Embo J. 2007;26:4078–4088. doi: 10.1038/sj.emboj.7601837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottke A, Colaiacovo MP, Shi Y. Developmental roles of the histone lysine demethylases. Development. 2009;136:879–889. doi: 10.1242/dev.020966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769–3779. doi: 10.1128/MCB.01432-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Hansen KH, Christensen J, Agger K, Cloos PA, Helin K. Coordinated regulation of transcriptional repression by the RBP2 H3K4 demethylase and Polycomb-Repressive Complex 2. Genes Dev. 2008;22:1345–1355. doi: 10.1101/gad.470008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- Sarma K, Margueron R, Ivanov A, Pirrotta V, Reinberg D. Ezh2 requires PHF1 to efficiently catalyze H3 lysine 27 trimethylation in vivo. Mol Cell Biol. 2008;28:2718–2731. doi: 10.1128/MCB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai N, Kato Y, Kimura G, Takeuchi T, Yamaguchi M. The Drosophila jumonji gene encodes a JmjC-containing nuclear protein that is required for metamorphosis. The FEBS journal. 2007;274:6139–6151. doi: 10.1111/j.1742-4658.2007.06135.x. [DOI] [PubMed] [Google Scholar]
- Savla U, Benes J, Zhang J, Jones RS. Recruitment of Drosophila Polycomb-group proteins by Polycomblike, a component of a novel protein complex in larvae. Development. 2008;135:813–817. doi: 10.1242/dev.016006. [DOI] [PubMed] [Google Scholar]
- Shen X, Liu Y, Hsu YJ, Fujiwara Y, Kim J, Mao X, Yuan GC, Orkin SH. EZH1 Mediates Methylation on Histone H3 Lysine 27 and Complements EZH2 in Maintaining Stem Cell Identity and Executing Pluripotency. Mol Cell. 2008;32:491–502. doi: 10.1016/j.molcel.2008.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirato H, Ogawa S, Nakajima K, Inagawa M, Kojima M, Tachibana M, Shinkai Y, Takeuchi T. A jumonji (Jarid2) protein complex represses cyclin D1 expression by methylation of histone H3-K9. J Biol Chem. 2009;284:733–739. doi: 10.1074/jbc.M804994200. [DOI] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–1012. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi T, Watanabe Y, Takano-Shimizu T, Kondo S. Roles of jumonji and jumonji family genes in chromatin regulation and development. Dev Dyn. 2006;235:2449–2459. doi: 10.1002/dvdy.20851. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Pfau R, Kampranis SC, Tsichlis PN. Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2641–2646. doi: 10.1073/pnas.0813139106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umlauf D, Goto Y, Feil R. Methods in molecular biology. Vol. 287. Clifton, NJ: 2004. Site-specific analysis of histone methylation and acetylation; pp. 99–120. [DOI] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet. 2009;41:521–523. doi: 10.1038/ng.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker E, Ohishi M, Davey RE, Zhang W, Cassar PA, Tanaka TS, Der SD, Morris Q, Hughes TR, Zandstra PW, et al. Prediction and testing of novel transcriptional networks regulating embryonic stem cell self-renewal and commitment. Cell stem cell. 2007;1:71–86. doi: 10.1016/j.stem.2007.04.002. [DOI] [PubMed] [Google Scholar]
- Wang J, Rao S, Chu J, Shen X, Levasseur DN, Theunissen TW, Orkin SH. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Zarach JM, Beaudoin GM, 3rd, Coulombe PA, Thompson CC. The co-repressor hairless has a role in epithelial cell differentiation in the skin. Development. 2004;131:4189–4200. doi: 10.1242/dev.01303. [DOI] [PubMed] [Google Scholar]
- Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.