Abstract
We present a survey of recent advancements in the emerging field of patient-specific modeling (PSM). Researchers in this field are currently simulating a wide variety of tissue and organ dynamics to address challenges in various clinical domains. The majority of this research employs three-dimensional, image-based modeling techniques. Recent PSM publications mostly represent feasibility or preliminary validation studies on modeling technologies, and these systems will require further clinical validation and usability testing before they can become a standard of care. We anticipate that with further testing and research, PSM-derived technologies will eventually become valuable, versatile clinical tools.
Keywords: computer simulation, clinical decision support techniques, computer-assisted three dimensional imaging
INTRODUCTION
Patient-specific modeling (PSM) is the development of computational models of human pathophysiology that are individualized to patient-specific data. PSM is gaining more attention from research groups around the world because of its potential to improve diagnosis, optimize clinical treatment by predicting outcomes of therapies and surgical interventions, and inform the design of surgical training platforms. Most current medical diagnostic practices lead to rough estimates of outcomes for a particular treatment plan [1], and treatments and their outcomes usually find their basis in the results of clinical trials. However, these results might not apply directly to individual patients [2] because they are based on averages. As an alternative, PSM can be used as a theranostic tool to tailor treatment and optimize an individual's therapy. PSM is also valuable for obtaining information on physical properties that cannot be measured, such as tissue stress, which, for example, might serve as a better predictor of the likelihood of rupture in vascular aneurysms [3].
Recently, PSM has drawn attention from government funding agencies. In 2006, the European Union initiated a consortium ‘Structuring the Europhysiome’, which has led to the Virtual Physiological Human (VPH) project [4, 5]. This project aims to stimulate research in the field of PSM for personalized and predictive healthcare, and encompasses a number of more specific subprojects [6]. In November 2007, the National Institutes of Health (NIH) posted a Funding Opportunity Announcement regarding PSM (Predictive Multiscale Models of the Physiome in Health and Disease [7]). One of the major goals of this grant is to stimulate the design of realistic computational models to make predictions about clinical outcomes. More recently, as a direct result of the American Recovery and Reinvestment Act of 2009, the National Institute of Biomedical Imaging and Bioengineering announced a challenge grant ‘Towards the Virtual Patient’, with the same goal as the prior NIH announcement.
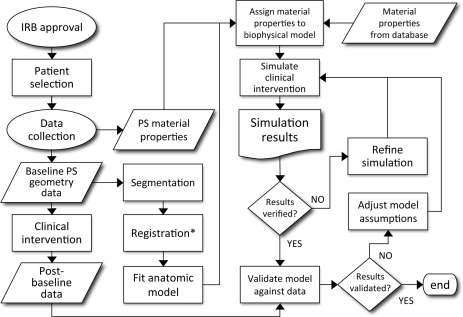
Despite the attention, PSM has not yet become a standard of care in clinical practice because evaluation of the predictive capability of these models has not yet been performed on a large scale [8]. A possible explanation for the lack of such studies could be related to the many manual steps in the workflow from data acquisition to model result [8]. Therefore, one of the challenges is to automate particular steps in this workflow as much as possible [8–14]. Figure 1 shows the typical aspects of this workflow with a specific focus on three-dimensional (3D) model creation and validation.
Figure 1:
Typical workflow for creating and validating a 3D, patient-specific model. Assignment of patient-specific material properties could include additional steps in which material parameters are estimated using a numerical-experimental approach (when properties cannot be measured directly or obtained from external sources). PS, patient-specific; Asterisks, if required.
The goal of this article is to more broadly illuminate recent work in PSM by providing a survey of current publications in the field, to discuss these advancements, and describe the challenges that modelers currently face. We selected the publications included in this survey from a PubMed search on ‘patient-specific modeling’ performed in May 2009. This search was limited to articles published between January 2008 and May 2009. We have organized the article according to the tissues and organs modeled in these publications: blood vessels, heart, bones, brain, skeletal muscle and teeth/periodontia. We also include a section on modeling tumors because it has received much attention in recent literature. We hope that this survey will introduce modelers and clinicians to new modeling methods and technologies, and encourage the cross-pollination of modeling techniques among the various research communities working in PSM.
BLOOD VESSELS
Appearing in the late 1990s, PSM of blood vessels aims to investigate the effects of cardiovascular devices on physiologic function and to predict outcomes of therapies for individual patients [15]. This field uses imaging techniques [e.g. magnetic resonance imaging (MRI), computed tomography (CT), ultrasound (US)] and image processing (acquisition, segmentation and registration techniques) to generate meshes on which mathematical models of fluid flow (e.g. the Navier–Stokes equation) are solved computationally [8, 15]. These are referred to as models of computational fluid dynamics (CFD). In models that incorporate fluid-solid interactions, the mechanics (deformation and stresses) of the vessel wall are also taken into account. In CFD models, fluid velocity vectors and pressures are calculated throughout the domain of interest, constrained by boundary conditions. The latter typically involve no-slip boundary conditions at the vessel wall and in- and outflow boundary conditions at the in- and outlet of the vessel. Computational methods to solve these problems include the finite volume method, finite element (FE) method and finite difference method. The popular FE method is used to solve partial differential equations (PDEs) on complex-shaped domains for a variety of physical problems, including CFD, solid mechanics, electric and magnetic fields and electrophysiology. For the FE method, a continuous domain is discretized into elements and nodes and the solution is approximated on the domain by linear or higher order functions, which are defined in the elements. For an in-depth explanation of the FE method, see Finite Element procedures by Bathe [16].
Of all the modeling subfields discussed here, PSM of blood vessels had the highest number of recent publications. Additionally, several recent reviews focus specifically on PSM of blood vessels. Ricotta et al. [1] reviewed methods regarding carotid and coronary stenoses, aortic aneurysms and aortic dissection. Taylor and Figueroa [15] focused on methods for obtaining anatomic and physiologic data, computational methods and applications regarding disease and intervention prediction and listed important future developments that are necessary for advancing the field.
The majority of papers regarding PSM of blood vessels focus on aneurysms, which are pathological dilations of the artery walls. These dilations are associated with high mortality and morbidity rates due to hemorrhage following rupture. In recent computational studies, two types of aneurysms have gained much attention: those of the abdominal aorta and arteries in the brain.
Abdominal aortic aneurysm
Abdominal aortic aneurysms (AAA) occur in 2–4% of males over the age of 65 in the West [17]. Rupture of an AAA is associated with a mortality rate of 50–75%.
The main objective for modeling of AAA is to improve the prediction of rupture so that an intervention can be performed [3, 18]. Currently, aneurysms are repaired when the maximal diameter exceeds 5.0–5.5 cm, but other factors such as thrombus formation make it impossible to obtain an absolute correlation between size and risk of rupture. Recently, it has been suggested that asymmetry should be taken into account in rupture risk assessment [3] because of a redistribution of wall stress. In assessing wall stress, most studies have assumed isotropic material properties of the cardiac wall. However, researchers have shown that anisotropic properties yield higher peak stresses [19, 20]. This more realistic implementation demonstrates that it is likely more important that material properties be obtained on a patient-specific basis [19].
Prediction of rupture might be greatly improved when vessel growth and remodeling is taken into account [21, 22]. To this end, Figueroa et al. [21] recently presented a new computational framework of fluid-solid mechanics in which their coupled momentum method [23] is combined with stress-mediated growth and remodeling algorithms.
Cerebral aneurysm
Cerebral aneurysms occur in ∼2% of the population [24]. The 1-month case fatality rate of rupture of a cerebral aneurysm (leading to subarachnoid hemorrhage) is estimated to be between 30 and 40% [25]. Two-thirds of survivors of cerebral aneurysm rupture experience a reduction of quality of life.
As with AAAs, it is believed that wall stress is a better predictor of rupture than, for example, aneurysm diameter [26]. Regions of rupture might also be influenced by concentrated jets impinging on the walls [27].
Most computational studies of cerebral aneurysms have focused on assessing the effects of interventions, such as clipping of arteries [28] and cardiovascular devices such as stents [29], on hemodynamics. The main goal is to use computational models to optimize treatments and devices, taking into account factors that play a role, such as thrombus formation [28, 30, 31]. In stent design, Kim et al. [29] recently demonstrated that flow rate and wall shear stress are influenced by stent porosity, strut design and mesh hole shape.
Material properties
Most CFD studies assume Newtonian fluid behavior (constant fluid viscosity, as with water). Rayz et al. [30] showed that in two out of three patient models, the use of non-Newtonian fluid behavior (where viscosity depends on the fluid shear rate, as with blood) improved the relation between regions of slow flow and thrombus formation. Because it has been shown that wall shear stress is increased in the presence of turbulent flow, Tan et al. [32] employed newly developed turbulence models and transitional variants in CFD simulations. They concluded that the results from the transitional models are closer to reality, but that further validation is necessary.
Interventions, cardiovascular devices and surgical planning
Marsden et al. [33] tested the performance of two Y-graft designs used in the conduit Fontan operation with CFD. The Y-grafts improved the distribution of blood flow to both lungs as opposed to other employed methods. The authors conclude that this design is promising and should be evaluated clinically, but more studies are needed to verify improvements.
To treat patients who were born with one cardiac ventricle and persistent left superior vena cava (LSVC), the procedure of bilateral bidirectional Glenn (BBDG) is usually applied. Sun et al. [34] investigated the power losses and flow patterns of a patient-specific BBDG connection at different levels of pulmonary flow splits and found this to be dependent on the flow ratio between left and right pulmonary arteries. Such surgeries on congenital heart defects require a prolonged cardiopulmonary bypass (CPB), affecting 20,000 children annually. However, during commonly performed CPB, major disturbances in flow and perfusion were observed through modeling, demonstrating the need for optimized CPB configurations, waveforms and cannula tip designs [35].
Two other recent articles describe systems concerned with planning cardiovascular surgeries, one for peripheral vascular bypass [36], and one for correcting congenital heart defects [37]. The former system uses a one-dimensional (1D) model to simulate blood dynamics in the peripheral vasculature. The latter allows users to manipulate and deform 3D PSM of cardiovascular anatomy like sculpting clay, then recompute the alterations in blood flow through the modified vessels.
Validation
For both aortic and cerebral aneurysms, researchers have focused on validation of their methods. In many cases though, studies were focused on validating newer measurement techniques with CFD simulations. The main findings of these studies were that (i) phase-contrast magnetic resonance angiography (PC-MRA) is accurate in determining velocities, but cannot accurately quantify wall shear stress [38], (ii) that it should only be used for inlet and outlet boundary conditions in numerical simulations [39, 40] and (iii) that inlet boundary conditions may have important implications for aneurysm growth [41].
Future challenges
Investigators are faced with several challenges related to the accurate prediction of vessel wall stress. First, wall thickness is an important determinant of wall stress. Hence, in order to improve predictions of aneurysm rupture, research methods for the assessment of patient-specific vessel wall thickness are necessary [42]. Currently, CT and MRI are not accurate enough to obtain this in vivo. Secondly, growth and remodeling algorithms are expected to improve model predictions in the long term, but experiments need to be performed to validate these types of algorithms [21, 22]. Finally, methods need to be applied that will obtain unloaded vessel geometries [43, 44]. Most current studies generate anatomical models from vessel geometries and define these as the undeformed state in FE models. This will lead to an over-estimation of wall stress, because in reality, vessels in patients are subject to (varying) pressures and are therefore in a deformed state.
BONES
Examples of recent PSM studies on bones focus on vertebrae [13, 45–49], the pelvis [14, 50, 51], the femur [52–54], the mandible [55], glenoid bone [56], the index finger [10], knee joint [57] and skull [58]. Bone is typically assumed to be physically and geometrically linear; hence the linear FE method is often used to model this tissue. Cartilage, however, is much softer, necessitating non-linear FE methods with neo-Hookean material behavior assigned to cartilage [50].
Vertebrae
Drevelle et al. [45] published results on PSM of children's spines, and demonstrated that their model could potentially predict the progression of scoliosis (a condition in which the spine is pathologically curved). In the following year, this group simulated all the steps involved in these specific surgeries and demonstrated that their modeling methods predicted the outcome of corrective surgery for scoliosis in 20 patients [46]. These simulations may help in optimizing scoliosis corrective surgery [46, 49] or scoliosis brace design [48] because it has been shown that techniques employed by different surgeons result in different outcomes [47].
Mesh generation can be a limiting step with respect to speed in the workflow of generating PSM, thus O’Reilly and Whyne [13] proposed a method in which meshes of vertebrae are created from an existing parametric mesh using morphing techniques. This method results in a rapid generation of patient-specific vertebrae geometries. Additionally, in performing a simulation of imposing load on the vertebrae, patient-specific material properties were assigned to the cortical and trabecular bone, based on the density from the CT images [59, 60].
Pelvis
3D modeling of the pelvis has applications in treating osteoarthritis patients and in anticipating the effects of total hip arthroplasty. For example, Anderson et al. [50] developed a new FE model that simulates cartilage contact pressure in the hip joint and provides a non-invasive means of characterizing patient-specific hip joint mechanics. In another recent study, Barrat et al. [51] developed a 3D modeling method based on US data, principle component analysis and statistical shape models (SSM) in order to avoid the risks associated with CT scans performed before orthopedic surgery. They showed that their method could be used to generate accurate models of the pelvis and femur surfaces during total hip replacement surgery without a CT scan. Both of these hip modeling studies used a small number of cadavers for validation data, illustrating the need for more comprehensive clinical validation of these pelvic models. Shim et al. [14] recognized this need and therefore developed a new, faster FE modeling process that may accelerate wider model validation. They show that larger FEs can be used in their models to speed up computations without sacrificing model accuracy and efficacy. They also demonstrated that sparse, or incomplete sections of patient-specific hip models can be supplemented with generic data from the Visible Human [61] to make more complete models. This development is important in determining a ‘minimum PSM dataset’ that is necessary in order to design a reliable predictive model.
Femur
In order to reduce risk of a mal-aligned component and notching with subsequent femoral neck fracture, Bailey et al. [52] validated a computer navigation system for setting the femoral component stem shaft angle during hip resurfacing surgery. Using this imageless method, surgeons identified anatomical landmarks on the shaft, and the computer created a PSM for setting the shaft angle. The study shows that surgeons were able to accurately and consistently place the femoral component at the planned target angle using the computer navigation system. The system was validated using a relatively high number of human patients (35) undergoing hip resurfacing surgery.
Zheng and Schumann [54] provide another example of a study aimed at minimizing the radiation exposure necessary to construct 3D surface models of bone. They tested a method using point distribution models for creating 3D representations of the proximal femur from 2D calibrated X-rays. Their study showed clinically accurate validation against data from 22 cadaver bones.
Finally, Pahr and Zysset [53] published a study aimed at creating numerically efficient FE models of femoral and vertebral bone. The authors detail a modeling technique that includes a new self-correcting cortical shell thickness evaluation algorithm for generating bone iso-surface meshes.
Other bones
Researchers have recently used PSM to model a variety of other bones. de Zee et al. [55] used PSM to determine that differences between the left and right articular eminence inclinations in the mandible of a single patient were consistent with a minimization of joint loads. Their model predictions of the final shape of the eminence following distraction osteogenesis were consistent with measurements made 6.5 years after the procedure.
Diedrichs et al. [56] used PSM to quantify glenoid bone defect sizes and generate bone graft shapes from models of non-pathological contralateral bone. Their method was validated using five cadaveric specimens and may ultimately help surgeons create bone grafts that more anatomically fit these defects.
To further automate the generation of 3D bone models, Gasmann et al. [10] created an artificial neural network (ANN) to automatically segment the bones of the human index finger from CT scans. The results of the neural network showed good agreement with results from two trained human technicians, and were computed in less than one-tenth of the time needed by the technicians. Subburaj et al. [57] also sought to automate model generation further by creating a system that automatically localizes and labels anatomical landmarks on knee joint models. Their system analyzes the surface curvature of the model and labels regions using a ‘spatial adjacency relationship matrix’. The automated program performed as well as or better when compared to labeling performed by three experienced orthopedic surgeons.
Lastly, a study by D’Ambrosio et al. [58] used PSM to locate the least-invasive surgical paths through the skull and into the brain during a craniotomy procedure. This work represents the first patient-specific objective cranial base approach assessment model in the literature
Future challenges
The high contrast that bone tissue provides during CT and US imaging has helped facilitate the automated segmentation techniques mentioned above. However, to further validate these methods, they must be tested using other bones and other imaging modalities [10]. Also, several authors have stressed the need for more patient data for performing further validation studies on bone models [54, 55, 62] and for expanding training sets used for SSM [51] and ANN processing [10].
HEART
Recent studies of patient-specific cardiac modeling other than coronaries (see ‘Blood vessels’ section) focused on ventricular fluid flow [63, 64], electrophysiology [65, 66], growth and remodeling [67], valve mechanical behavior [68, 69], circulation hemodynamics [70] and electromechanics [71, 72]. Since the beginning of this millennium [73, 74], more groups around the world are recognizing that the combination of whole heart electrophysiology and mechanics—electromechanics—is important for a good understanding of cardiac regional and global function. Recently, more efforts are being directed into the development of tightly coupled models of cardiac electromechanics at or above the tissue level [75–80]. Because electromechanical events take place at the (sub-) cellular level, these models are generally multiscale in nature. For a thorough recent review on multiscale modeling that focuses on the heart we refer to Southern et al. [81]. Cardiac tissue is typically assumed to be physically and geometrically non-linear (strain magnitudes of 50% are normal). Hence, non-linear FE methods are used to solve these modeling problems.
Interventions and cardiovascular devices
Doenst et al. [63] investigated the effects of left ventricular reconstructive surgery, which involves removal of diseased cardiac tissue, on left ventricular blood flow and showed that ejection fraction and blood washout increased slightly. For the right ventricle, Tang et al. [64] demonstrated that right ventricular performance and wall stress can be optimized by adjusting the amount of scar removal and patch design.
Although implantable cardiac defibrillator (ICD) systems are routinely implanted in adult patients using a transvenous approach, this is not possible in children due to their small size. Jolley et al. [65] used models of the torso and heart in children to investigate the effects of ICD lead placement on electric fields. Relatively small changes in electrode position resulted in significant changes in predicted defibrillation thresholds (DFT) and lengthening the electrode decreased DFT and distribution of the voltage gradient.
Starfinger et al. [70] proposed a method to identify septic shock with a minimum set of measurements to obtain patient-specific parameters for the circulation and tested this successfully in pigs. The model accurately captured hemodynamics throughout the course of sepsis. The investigators proposed that the estimation of systemic resistance could be used as an indicator for admission of medicine in critical care.
Cardiac electromechanics
Le Rolle et al. [71] proposed a model-based approach to analyze Tissue Doppler Imaging (TDI). TDI is used to evaluate regional contraction in the myocardium of the heart and has been useful in differentiating healthy from ischemic tissue. Model parameters for the left ventricle were estimated by minimizing strain signals between the computational model and strain signals obtained with TDI in several myocardial segments. The model was able to identify failing segments in a patient with dilated cardiomyopathy.
A set of simplified models of cardiac electromechanics and a workflow for obtaining patient-specific data was proposed by Kerckhoffs et al. [82] and Sermesant et al. [72]. Their workflow includes obtaining ventricular geometry with MRI, electrical parameter values by electroanatomical mapping and mechanical parameter values by matching simulated and measured LV volumes. A similar strategy regarding electrophysiology was proposed by Pfeifer et al. [66] in which the torso was also included.
Future challenges
Most future challenges of PSM of blood vessels, such as experiments needed to validate growth and remodeling algorithms [83] and determination of unloaded geometries, also apply to cardiac modeling. Southern et al. [81] noted the challenge of designing computational models that can be used for long-QT drug trials. To date, many (non-cardiovascular) drugs do not pass the trial phase, because they have the side effect of prolonging the QT phase in the ECG, which may lead to arrhythmias. Indeed, computer-aided drug design is now a widely recognized field that facilitates the design and discovery of new drugs [84, 85]. The preDiCT project, part of the VPH [86], aims to predict the impact of drugs on the heart's rhythm using computer models. Multi-scale computational cardiovascular models could reduce the costs associated with clinical trials and could even lead to drug design tailored to the patient.
BRAIN
PSM of the brain has recently focused on simulating deep brain stimulation (DBS) for patients with Parkinson's Disease and other movement disorders [87–89], brain deformations as a result of craniotomy procedures [90, 91], needle-tissue brain interactions [92], and intracranial pressure dynamics in traumatic brain injury patients [93]. All these studies employed the FE method to obtain model solutions except the publication by Wakeland et al., in which a compartmental fluid dynamics model was used.
Deep brain stimulation
DBS is an established treatment procedure for patients with Parkinson's disease (PD) where either the globus pallidus or subthalamic nuclei are stimulated using surgically implanted electrodes. However, researchers do not completely understand the mechanisms whereby this treatment imparts benefits to the patient. To illuminate these mechanisms two research groups [87, 88] developed 3D PSM from either MRI or CT scans that simulated the electrical stimulation procedure. Astrom et al. [87] describe their general FE model-generation method and its use in identifying brain regions affected during DBS in one patient studied retrospectively. Their simulations help explain why the patient showed negative side effects following DBS: the simulated electrical field generated during stimulation covered brain regions implicated in those side effects.
Maks et al. [88] also created 3D patient-specific brain models to help understand the positive effects of DBS in PD patients. The authors simulated DBS using models created from scans of 10 different PD patients. They found that in the five patients that responded best to DBS, all but one had the majority of their activated tissue volume outside the subthalamic nucleus. This finding suggests that the stimulation of tissue outside the subthalamic nucleus may be an important contributor to the therapeutic benefits of DBS.
Brain deformation during craniotomy
Two recent papers describe PSM studies aimed at predicting brain deformation following the removal of part of the skull. To find an optimally detailed model for predicting deformation, Wittek et al. [91] produced FE brain models from a single patient at three different complexity levels. They found that the linear elastic model, the least complex, performed just as well as hyperviscoelastic and hyperelastic alternatives in predicting intra-operative positions of brain landmarks. They recommend using the simpler model because it affords a 29% savings in computational time, but note that, while applicable to a wide variety of neurosurgical situations, their method does not extend to surgeries involving cutting and/or tissue removal.
Shiavone et al. [90] also considered the problem of predicting brain deformation during craniotomy. In their recent article they describe a new method for extracting a constitutive law of brain elasticity using an elasticity-measuring device applied in vivo. Clinicians may be able to better tune patient-specific FE brain models using data from this device and thus better predict deformation during surgery.
Needle-tissue brain interactions and intracranial pressure dynamics
In developing technology for modeling/planning surgeries, Wittek et al. [92] validated a new FE model that predicts force dynamics during needle insertion into the brain. Their model accounts for material and geometric non-linearities and showed good agreement with empirical validation measurements from pigs. However, their method depends on measuring patient-specific material properties of the brain and meninges. The authors stress the need for technology, such as that described above by Shiavone et al. [90] that would provide such measurements in humans.
Using a compartmental fluid dynamics model, Wakeland et al. [93] developed a method for predicting intracranial pressure (ICP) dynamics in patients with traumatic brain injury. Their system predicted changes in ICP for nine patients during respiratory rate and head of bed elevation challenge sessions. The system show modest success in predicting ICP dynamics given an initial parameterization phase at the start of a session, but less success when this parameterization was used to predict dynamics in subsequent sessions.
Future challenges
Recent authors have stressed the need for more accurate segmentation and registration methods for brain structures [87, 89]. These methods may improve if the resolution of diffusion tensor imaging for modeling heterogeneous brain tissue can be increased [89]. There are also future challenges for integrating in vivo brain elasticity measurements with other published models [90] and for more precisely characterizing the physical properties of brain tissues such as gray and white matter [87], the pia mater and meninges [92].
SKELETAL MUSCLE
Recent studies regarding musculoskeletal PSM have focused on shoulder muscle forces during wheel chair propulsion [94], treatment of walking patterns [95] with multibody dynamical models, and treatment of deep tissue injury with non-linear large-deformation FE models [96, 97].
Muscle dynamics
Many wheelchair users may experience shoulder strain injuries. The design and analysis of patient-specific musculoskeletal models of the upper body might help to optimize wheel chair design to reduce shoulder pain [94]. Results from a 3D musculoskeletal model of the upper body showed a large dependence of shoulder joint forces on seat height and position, demonstrating the possibility of wheel chair design optimization.
Reinbolt et al. [95] presented a computational framework to predict post-treatment changes in walking patterns from pre-treatment measured data for specific patients. First, model parameters are optimized to the patient's movement data and a 3D full body gait model is then used to predict gait after an intervention. The model successfully predicted motion after two interventions: gait modification (in which the patient is instructed to change his or her walking pattern) and high tibial osteotomy surgery.
Deep tissue injury
Deep tissue injury is a muscle lesion under intact skin. Knowledge of deep tissue injury etiology is yet to be established; however, causative factors include pressure-related ischemia together with excessive soft tissue deformation [98, 99]. Linder-Ganz et al. [96] created a musculoskeletal FE model of the thigh region of a 30-year-old male who was found unconscious after 3 days. The person was lying on his cell phone and MRI revealed deep tissue injury between the cell phone and the thighbone. Different material properties were assigned to different tissue types. Using a damage law [100] and muscle cell-death threshold, injured cells were made stiffer linearly with the magnitude of compression stress and time. Stress served as a somewhat better predictor of deep tissue injury, for regions of predicted injury more closely matched those obtained with MRI measurements.
In another study of deep tissue injury, a patient-specific FE model was created of a residual limb bearing the load from a prosthesis [97]. In this study, the goal was to characterize the mechanics of muscle tissue at the end of the limb using pressure and MRI measurements and isotropic, non-linearly viscoelastic large-deformation material models. The authors found strain and stress concentrations in the flap under the tibial end, exposing the patient to relative higher risk to develop deep tissue injury.
Future challenges
An important future advancement in skeletal muscle modeling will be to obtain injury thresholds for muscles [101] in order to predict injury and optimize prosthetic designs to minimize muscle stress and strain concentrations.
TEETH/PERIODONTIA
Researchers in orthodontics have also recently published PSM studies using FE-based models. Cattaneo et al. [102] created an FE model of the left segment of a single human mandible to investigate the type of tooth movements associated with different orthodontic forces (as in the application of braces). Their findings suggest that tooth movement based on a given moment-to-force ratio varies among individuals, and therefore may best be predicted using PSM rather than the more generalized, classically prescribed theory.
In a subsequent study [103], these same authors used patient-specific FE modeling to examine orthodontic forces on the tissues surrounding teeth. Again, their findings did not confirm canonical orthodontic theory, suggesting that orthodontic loading regimes can be better optimized using PSM.
A third orthodontic modeling study by Kondo et al. [104] used patient-specific FE models to determine whether molar loss increases stress and strain in the periodontium. The authors found significantly higher stress and strains in periodontal tissues of the second premolars in patients with bilateral molar loss versus those with unilateral or no molar loss. They assert that this stress could not be prevented by wearing a denture and advocate future studies aimed at improving the usability of their modeling system by clinicians.
Future challenges
Future directions for modeling teeth and surrounding tissues include validating models against in vivo loading data from patients, incorporating the viscoelastic properties of the periodontal ligament into simulations, and obtaining patient-specific material properties for model parameterization [102, 103]. Kondo et al. [104] also emphasized their intent to enhance the usability of their simulation systems for clinical investigators.
TUMORS
Recent applications of PSM in oncology have focused on optimizing radiotherapy [105–107], chemotherapy [108] and thermal treatment [109] in cancer patients, researching the mechanics of brain tumor growth [110, 111], and imaging cancerous breast tissue [112]. For further review of current advancements in modeling tumors, please see Juffer et al. [113] and Deisboeck et al. [114].
Optimization of cancer treatment
Treatment of cancer often requires the clinician to find a means of destroying cancerous targets while minimizing damage to surrounding tissues. As an optimization problem, this challenge lends itself to computer-based solutions, and several recent articles have used PSM as a way of optimizing cancer therapy. Campbell et al. [105] used patient-specific single photon emission CT data to compute radiation doses for treating liver tumors with yttrium-90 microspheres. They found that their PSM was better than more generic alternatives at predicting outcomes of 14 cancer patients studied retrospectively. They also infer that the model is better at predicting the response to therapy, especially for smaller tumors.
Gorelik et al. [108] also present a study on optimizing cancer treatment for rare tumors based on PSM. They used a computer algorithm in combination with patient-specific chemosensitivity tests on xenografted tumors to devise an optimal treatment plan for a single patient. After being treated using the optimal therapy method predicted by the model, the patient experienced relief of pancytopenia and a stabilization of the metastatic disease. The patient retained a good quality of life for 6 months until succumbing.
Titz and Jeraj [107] recently published a multiscale model that accounts for oxygenation levels in simulating head and neck tumor growth. They used this model to investigate the influence of oxygenation on tumor responses to radiation therapy. If validated against clinical cases, this type of model could also be used to anticipate patient-specific responses to radiotherapy.
Taking a more general approach to the application of modeling for theranostic anti-cancer treatment, South et al. [106] present a ‘theoretical framework for optimizing radiotherapy based on patient-specific radiobiological parameters derived from a series of images’. This more basic approach is not constrained to one type of model. Rather, it is a generally applicable tool for assessing the validity of a range of clinically useful cancer models.
Simulating brain tumors
Hogea et al. [110] published a model that simulates the spatio-temporal spread of gliomas, and also accounts for mechanical deformations of the surrounding brain tissue caused by tumor growth. Given that patient-specific data on tumor growth is generally sparse, their publication is concerned with the inverse problem of identifying parameters that control tumor dynamics.
Szeto et al. [111] also modeled glioma growth to characterize the link between tumor kinetics and the tumor's hypoxic burden. They used data from MRI and 18F-fluoromisonidazole PET to tune PSM of tumors in 11 different human subjects. Their results imply that more abnormally shaped tumors tend to have a greater hypoxic burden, and also support the theory that more aggressive cell lines produce less spherical tumors.
Other tumor modeling
In developing a new computer-assisted method of therapy for prostate cancer, Fuentes et al. [109] created a complex, high-speed ‘cyberinfrastructure’ between two research sites in Texas. This integrated system includes heat-transfer models of the prostate and tumor, 3D tissue imaging, laser optics and surgical control systems. The authors used their cyberinfrastucture to develop and test a dynamic data driven application system (DDDAS) that guides laser therapy of prostate cancer. They tested their system on two in vivo canine prostates and in both cases obtained encouraging results.
Finally, Pathamanathan et al. [112] created 3D FE models of the breast in order to help radiologists and surgeons more precisely locate cancerous breast tissue. By introducing artificial tumors into their models, they were able to simulate deformations of normal and pathological tissue. Their deformable models can simulate breast shape during a variety of clinical procedures, such as surgery or mammography.
Future challenges
While there has been much recent progress in modeling cancerous tissues, researchers require more data for performing additional preclinical and clinical model validation [107–109, 111, 112]. Other authors [106, 111] have emphasized the need for validated imaging methods that measure tumor burden and hypoxia in vivo. Such methods can accelerate clinical trials that test model-predicted tumor response to therapy.
STATE OF THE ART AND FUTURE CHALLENGES
In this survey, we have found that PSM has recently been employed to model the dynamics of a wide variety of anatomical entities. Aside from the canonical and pathological entities we have discussed above, we found PSM examples for lung [115, 116], larynx [117] and kidneys [118] as well. We chose to omit these studies from our discussion above in order to provide ample room for addressing more heavily researched areas. We have also found that the vast majority of recent publications on PSM are concerned with developing 3D mechanistic models versus models of lower dimension and that most articles represent feasibility or small-n validation studies where models simulate a very particular clinical condition. Given these findings, we foresee large-n clinical validation studies, especially those of 3D models, as the major challenge facing patient-specific modelers in the near future. Such studies will also be necessary for FDA approval of medical modeling systems. We see several rate-limiting factors that impede progress towards these validation studies: manual steps in the workflow between data acquisition and model simulation, the identification of patient-specific material properties, and the need for model standardization. Eliminating these bottlenecks will help accelerate PSM research and will be vital for performing the studies required for PSM to become a clinical standard of care.
Patient-specific material properties
With current imaging modalities, a geometry of a tissue or organ is relatively easy to obtain, and many of the 3D PSM reviewed here have incorporated patient-specific geometry. Assigning material properties to these geometries using previously quantified values (by in vitro measurements, for example) may work in certain instances, but are not ideal due to biological variation. Such assumptions will cause even greater problems when material properties are altered by disease. Hence, in order to create accurate predictive PSM, obtaining patient-specific material properties may well be as important as obtaining geometries. For example, methods have been proposed and tested to obtain the Young's modulus of bone by quantifying radio-density in CT images [59, 60]. Mixed numerical-experimental methods to obtain mechanical properties of biological tissue in vitro [119] can also be used to estimate material properties in patients [120].
In 2007, the National Heart, Lung and Blood Institute convened a workshop on research directions needed to develop methods to elucidate human cardiac function [121]. One of the workshop's recommendations related to PSM was that material properties obtained through molecular and cellular investigations of explanted tissues, including histological and/or immunohistochemical characterization, should be combined with clinical data and high-resolution, non-invasive structural, functional (MR tagging, MR elastography, 3D US speckle tracking), and electrophysiological imaging. We extend this recommendation to include fields other than cardiac research.
Translation to the clinic
Before a PSM can become standard of care, the workflow involving modeling methods and equations has to be approved by the Food and Drug Administration (FDA) and by similar institutions in other countries. We anticipate that the model would have to undergo the same approval procedures as for medical devices. Currently, software involved in cardiac electrophysiological mapping is treated and approved by the FDA in this manner.
In anticipating the process of translating PSM modeling research into a clinical standard of care, we see the need for more standardized criteria for evaluating published models. We anticipate that as PSM moves further into the applied clinical domain, clinicians will require metrics and standards for evaluating and comparing competing models. PSM tools will likely need to be flexible and accommodating of a spectrum of clinical cases, given the variability among patients’ anatomy, physiology and clinical situations. Model users must know, therefore, the assumptions that are inherent in the models available to them, since these assumptions may invalidate their use for some patient cases. Given multiple models to choose from, clinicians will need to know how well each candidate model matches empirical data for a given patient population. This kind of validation is common among biological modeling studies, and thus we encourage clinicians interested in PSM to become familiar with metrics often used by modelers to assess model accuracy (e.g. the root mean square test). Conversely, we also encourage model developers to anticipate how they will test and communicate the clinical efficacy of their simulation systems to medical personnel. While a model may be well validated against empirical data, its use may not change clinical decision-making or improve patient outcomes. PSM model developers will be challenged to demonstrate not only their systems’ predictive capabilities, but also their enhancement of patient care.
The NIBIB's Interagency Modeling and Analysis Group (IMAG) [122] was created, in part, to address some of the issues surrounding modeling standards for PSM, and other efforts to standardize information about model content and results are being actively researched [123–128]. For example, members of the systems biology community have developed the Systems Biology Markup Language (SBML), a standardized biochemical pathway model description format that allows semantic links to concepts in reference ontologies [124], the Minimum Information Required in the Annotation of Biochemical Models (MIRIAM) [125], a set of model curation standards and the Minimum Information About a Simulation Experiment (MIASE) [123], a standard for reproducing numerical simulation results. By facilitating model sharing and reuse, these kinds of standards can help advance PSM research and ease its adoption as a clinical tool. However, because the publications returned by our literature search focused on modeling at the tissue and organ level, we foresee a demand for model description and annotation standards that scale to higher levels of biological organization and extend beyond chemical networks into other physical domains.
Finally, because certain medical cases are more time-sensitive than others, clinicians will need to know the computational efficiency of the model they are using. For example, the monitoring of a patient in an intensive care unit requires faster-than-real-time solutions as provided by ODE models [70], but FE-based predictions for identifying cardiac resynchronization therapy candidates [129] are less time-sensitive. In the latter case, a model that takes days to compute may still be clinically viable. However, model users under more strict time constraints must know approximately how long a simulation will take to solve, and what tradeoffs exist between a model's accuracy and its computational timeliness. Likewise, model developers addressing clinical problems must be sensitive to the time constraints of those who will be applying their models in a clinical setting. Although computational power remains a rate-limiting step in some areas of 3D PSM, we anticipate that High Performance Computing (HPC) using parallelization techniques will widen this bottleneck. Recently there has been much scientific interest in applying Graphical Processor Units (GPUs) [130] to FE models. When compared to conventional CPU clusters, GPU computing has several significant advantages related to performance, cost and accessibility and can easily be applied in a clinical setting.
As PSM tools become more available to clinicians, it will also be important to research how PSM fits into clinical workflows. The introduction of new computer-based technology in the medical industry can be difficult and slow, and we hope to see more studies in the near future that examine how PSM, once put into clinical use, can be optimized as a medical tool. Researchers have much work ahead of them before PSM becomes a standard in clinical practice; however, we believe that eventually, PSM will prove to be a valuable and versatile technology that improves medical care in a myriad of disciplines.
Key Points.
PSM is being applied to model the dynamics of a wide variety of tissues and organs within a wide variety of clinical domains.
Most of the recent work in PSM employs 3D models versus models of lower dimension.
Most of the recent work in PSM represents feasibility and/or preliminary validation studies.
Major challenges in PSM include the need for more model validation, further automation of common tasks in the modeling workflow (such as image segmentation), and model description and annotation standards that scale to higher levels of biological organization.
PSM model developers will be challenged to demonstrate not only their systems’ predictive capabilities, but also their enhancement of patient care.
FUNDING
National Institutes of Health grants T15 LM007442-06, R01 HL96544, American Heart Association Award 09PRE210064 and the National Biomedical Computation Resource (National Institutes of Health grant P41 RR08605).
Acknowledgments
The authors thank Dr Russ Altman for inviting to submit this article, Fred Lionetti and Dr Jazmin Aguado-Sierra for their assistance on the section on High Performance Computing, and Dr Owen Faris for a discussion on FDA approval of PSM systems.
Biographies
Maxwell Lewis Neal earned his BS in Biology from Case Western Reserve University in 1999. He has experience modeling brain, bones and the cardiovascular system. He is presently a PhD candidate in the Division of Biomedical and Health Informatics at the University of Washington, and his current research focuses on creating informatics tools to support biosimulation modeling.
Roy Kerckhoffs earned his PhD degree in biomedical engineering in 2003 at Eindhoven University of Technology in the Netherlands. He is working as a Project Scientist at the Cardiac Mechanics Research Group at the University of California San Diego. His research interests include multiscale modeling of cardiovascular mechanics and electrophysiology. Currently, his main focus lies on the development of multiscale (animal- and patient-specific) computational models of cardiac electromechanics.
References
*Papers describing some advancement in automating or accelerating modeling tasks and papers that used some patient-specific data for validation (n < 10)
**Papers that used a high number of patient-specific data sets for validation (n > 10), papes that described a major advancement in automating modeling tasks and all PSM reviews
- Ricotta JJ, Pagan J, Xenos M, et al. Cardiovascular disease management: the need for better diagnostics. Med Biol Eng Comput. 2008;46:1059–68. doi: 10.1007/s11517-008-0416-x. [**] [DOI] [PubMed] [Google Scholar]
- Kent DM, Hayward RA. Limitations of applying summary results of clinical trials to individual patients: the need for risk stratification. JAMA. 2007;298:1209–12. doi: 10.1001/jama.298.10.1209. [DOI] [PubMed] [Google Scholar]
- Doyle BJ, Callanan A, Burke PE, et al. Vessel asymmetry as an additional diagnostic tool in the assessment of abdominal aortic aneurysms. J Vasc Surg. 2009;49:443–54. doi: 10.1016/j.jvs.2008.08.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virtual Physiological Human—Latest News. [(10 October 2009, date last accessed)]. http://www.vph-noe.eu/home 2009. [Google Scholar]
- Hunter P, Robbins P, Noble D. The IUPS human physiome project. Pflügers Archiv Eur J Physiol. 2002;445:1–9. doi: 10.1007/s00424-002-0890-1. [DOI] [PubMed] [Google Scholar]
- VPH Projects. [(10 October 2009, date last accessed)]. http://www.vph-noe.eu/vph-projects 2009.
- NIH. Predictive Multiscale Models of the Physiome in Health and Disease (PAR-08-023) [(10 October 2009, date last accessed)]. http://grants.nih.gov/grants/guide/pa-files/PAR-08-023.html 2009.
- Antiga L, Piccinelli M, Botti L, et al. An image-based modeling framework for patient-specific computational hemodynamics. Med Biol Engin Comput. 2008;46:1097–112. doi: 10.1007/s11517-008-0420-1. [*] [DOI] [PubMed] [Google Scholar]
- Bekkers EJ, Taylor CA. Multiscale vascular surface model generation from medical imaging data using hierarchical features. IEEE Trans Med Imag. 2008;27:331–41. doi: 10.1109/TMI.2007.905081. [DOI] [PubMed] [Google Scholar]
- Gassman EE, Powell SM, Kallemeyn NA, et al. Automated bony region identification using artificial neural networks: reliability and validation measurements. Skeletal Radiol. 2008;37:313–19. doi: 10.1007/s00256-007-0434-z. [*] [DOI] [PubMed] [Google Scholar]
- Goel VR, Greenberg RK, Greenberg DP. Mathematical analysis of DICOM CT datasets: Can endograft sizing be automated for complex anatomy? J Vascular Surg. 2008;47:1306–12. doi: 10.1016/j.jvs.2007.12.046. [DOI] [PubMed] [Google Scholar]
- Grosland NM, Bafna R, Magnotta VA. Automated hexahedral meshing of anatomic structures using deformable registration. Comput Methods Biomech Biomed Engin. 2009;12:35–43. doi: 10.1080/10255840903065134. [DOI] [PubMed] [Google Scholar]
- O'Reilly MA, Whyne CM. Comparison of computed tomography based parametric and patient-specific finite element models of the healthy and metastatic spine using a mesh-morphing algorithm. Spine. 2008;33:1876–81. doi: 10.1097/BRS.0b013e31817d9ce5. [DOI] [PubMed] [Google Scholar]
- Shim VB, Pitto RP, Streicher RM, et al. Development and validation of patient-specific finite element models of the hemipelvis generated from a sparse CT data set. J Biomech Eng. 2008;130:051010. doi: 10.1115/1.2960368. [DOI] [PubMed] [Google Scholar]
- Taylor CA, Figueroa CA. Patient-specific modeling of cardiovascular mechanics. Annu Rev Biomed Eng. 2009;11:109–34. doi: 10.1146/annurev.bioeng.10.061807.160521. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bathe KJ. Finite Element Procedures. New Jersey: Englewood Cliffs; 1996. [Google Scholar]
- Zankl AR, Schumacher H, Krumsdorf U, et al. Pathology, natural history and treatment of abdominal aortic aneurysms. Clin Res Cardiol. 2007;96:140–51. doi: 10.1007/s00392-007-0472-5. [DOI] [PubMed] [Google Scholar]
- Martufi G, Di Martino ES, Amon CH, et al. Three-dimensional geometrical characterization of abdominal aortic aneurysms: image-based wall thickness distribution. J Biomech Eng. 2009;131:061015. doi: 10.1115/1.3127256. [DOI] [PubMed] [Google Scholar]
- Rissland P, Alemu Y, Einav S, et al. Abdominal aortic aneurysm risk of rupture: patient-specific FSI simulations using anisotropic model. J Biomech Eng. 2009;131:031001. doi: 10.1115/1.3005200. [DOI] [PubMed] [Google Scholar]
- Vande Geest J, Schmidt D, Sacks M, et al. The effects of anisotropy on the stress analyses of patient-specific abdominal aortic aneurysms. Ann Biomed Engin. 2008;36:921–32. doi: 10.1007/s10439-008-9490-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa AC, Baek S, Taylor CA, et al. A computational framework for fluid-solid-growth modeling in cardiovascular simulations. Comput Meth Appl Mech Eng. 2009;198:3583–602. doi: 10.1016/j.cma.2008.09.013. [*] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helderman F, Manoch I, Breeuwer M, et al. A numerical model to predict abdominal aortic aneurysm expansion based on local wall stress and stiffness. Med Biol Eng Comput. 2008;46:1121–7. doi: 10.1007/s11517-008-0358-3. [*] [DOI] [PubMed] [Google Scholar]
- Figueroa CA, Vignon-Clementel IE, Jansen KE, et al. A coupled momentum method for modeling blood flow in three-dimensional deformable arteries. Comp Meth Appl Mech Eng. 2006;195:5685–706. [Google Scholar]
- Rinkel GJ, Djibuti M, Algra A, et al. Prevalence and risk of rupture of intracranial aneurysms: a systematic review. Stroke. 1998;29:251–6. doi: 10.1161/01.str.29.1.251. [DOI] [PubMed] [Google Scholar]
- Burns J, Brown R. Treatment of unruptured intracranial aneurysms: surgery, coiling, or nothing? Curr Neurol Neurosci Rep. 2009;9:6–12. doi: 10.1007/s11910-009-0002-0. [DOI] [PubMed] [Google Scholar]
- Isaksen JG, Bazilevs Y, Kvamsdal T, et al. Determination of wall tension in cerebral artery aneurysms by numerical simulation. Stroke. 2008;39:3172–8. doi: 10.1161/STROKEAHA.107.503698. [DOI] [PubMed] [Google Scholar]
- Cebral JR, Hendrickson S, Putman CM. Hemodynamics in a lethal basilar artery aneurysm just before its rupture. AJNR Am J Neuroradiol. 2009;30:95–8. doi: 10.3174/ajnr.A1312. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayz VL, Lawton MT, Martin AJ, et al. Numerical simulation of pre- and postsurgical flow in a giant basilar aneurysm. J Biomech Eng. 2008;130:021004. doi: 10.1115/1.2898833. [**] [DOI] [PubMed] [Google Scholar]
- Kim M, Taulbee DB, Tremmel M, et al. Comparison of two stents in modifying cerebral aneurysm hemodynamics. Ann Biomed Eng. 2008;36:726–41. doi: 10.1007/s10439-008-9449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayz VL, Boussel L, Lawton MT, et al. Numerical modeling of the flow in intracranial aneurysms: prediction of regions prone to thrombus formation. Ann Biomed Eng. 2008;36:1793–804. doi: 10.1007/s10439-008-9561-5. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olgac U, Poulikakos D, Saur SC, et al. Patient-specific three-dimensional simulation of LDL accumulation in a human left coronary artery in its healthy and atherosclerotic states. Am J Physiol Heart Circ Physiol. 2009;296:1969–82. doi: 10.1152/ajpheart.01182.2008. [DOI] [PubMed] [Google Scholar]
- Tan FP, Soloperto G, Bashford S, et al. Analysis of flow disturbance in a stenosed carotid artery bifurcation using two-equation transitional and turbulence models. J Biomech Eng. 2008;130:061008. doi: 10.1115/1.2978992. [DOI] [PubMed] [Google Scholar]
- Marsden AL, Bernstein AJ, Reddy VM, et al. Evaluation of a novel Y-shaped extracardiac Fontan baffle using computational fluid dynamics. J Thoracic Cardiovasc Surg. 2009;137:394–403. doi: 10.1016/j.jtcvs.2008.06.043. e392. [DOI] [PubMed] [Google Scholar]
- Sun Q, Wan D, Liu J, et al. Patient-specific computational fluid dynamic simulation of a bilateral bidirectional Glenn connection. Med Biol Engin Comput. 2008;46:1153–9. doi: 10.1007/s11517-008-0376-1. [DOI] [PubMed] [Google Scholar]
- Pekkan K, Dur O, Sundareswaran K, et al. Neonatal aortic arch hemodynamics and perfusion during cardiopulmonary bypass. J Biomech Eng. 2008;130:061012. doi: 10.1115/1.2978988. [DOI] [PubMed] [Google Scholar]
- Marchandise E, Willemet M, Lacroix V. A numerical hemodynamic tool for predictive vascular surgery. Med Engin Phys. 2009;31:131–44. doi: 10.1016/j.medengphy.2008.04.015. [DOI] [PubMed] [Google Scholar]
- Pekkan K, Whited B, Kanter K, et al. Patient-specific surgical planning and hemodynamic computational fluid dynamics optimization through free-form haptic anatomy editing tool (SURGEM) Med Biol Engin Comput. 2008;46:1139–52. doi: 10.1007/s11517-008-0377-0. [DOI] [PubMed] [Google Scholar]
- Boussel L, Rayz V, Martin A, et al. Phase-contrast magnetic resonance imaging measurements in intracranial aneurysms in vivo of flow patterns, velocity fields, and wall shear stress: comparison with computational fluid dynamics. Magn Reson Med. 2009;61:409–17. doi: 10.1002/mrm.21861. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollnagel DI, Summers PE, Poulikakos D, et al. Comparative velocity investigations in cerebral arteries and aneurysms: 3D phase-contrast MR angiography, laser Doppler velocimetry and computational fluid dynamics. NMR Biomed. 2009;22:795–808. doi: 10.1002/nbm.1389. [**] [DOI] [PubMed] [Google Scholar]
- Canstein C, Cachot P, Faust A, et al. 3D MR flow analysis in realistic rapid-prototyping model systems of the thoracic aorta: Comparison with in vivo data and computational fluid dynamics in identical vessel geometries. Magn Resonance Med. 2008;59:535–46. doi: 10.1002/mrm.21331. [**] [DOI] [PubMed] [Google Scholar]
- Rayz VL, Boussel L, Acevedo-Bolton G, et al. Numerical simulations of flow in cerebral aneurysms: comparison of CFD results and in vivo MRI measurements. J Biomech Eng. 2008;130:051011. doi: 10.1115/1.2970056. [**] [DOI] [PubMed] [Google Scholar]
- Breeuwer M, de Putter S, Kose U, et al. Towards patient-specific risk assessment of abdominal aortic aneurysm. Med Biol Eng Comput. 2008;46:1085–95. doi: 10.1007/s11517-008-0393-0. [*] [DOI] [PubMed] [Google Scholar]
- de Putter S, Wolters BJBM, Rutten MCM, et al. Patient-specific initial wall stress in abdominal aortic aneurysms with a backward incremental method. J Biomech. 2007;40:1081–90. doi: 10.1016/j.jbiomech.2006.04.019. [*] [DOI] [PubMed] [Google Scholar]
- Huang X, Yang C, Yuan C, et al. Patient-specific artery shrinkage and 3D zero-stress state in multi-component 3D FSI models for carotid atherosclerotic plaques based on in vivo MRI data. Mol Cell Biomech. 2009;6:121–34. [*] [PMC free article] [PubMed] [Google Scholar]
- Drevelle X, Dubousset J, Lafon Y, et al. Analysis of the mechanisms of idiopathic scoliosis progression using finite element simulation. Stud Health Technol Inform. 2008;140:85–9. [PubMed] [Google Scholar]
- Lafon Y, Lafage V, Dubousset J, et al. Intraoperative three-dimensional correction during rod rotation technique. Spine. 2009;34:512–9. doi: 10.1097/BRS.0b013e31819413ec. [**] [DOI] [PubMed] [Google Scholar]
- Majdouline Y, Aubin CE, Labelle H. Influence of correction objectives on the optimal scoliosis instrumentation strategy: a preliminary study. Stud Health Technol Inform. 2008;140:116–20. [PubMed] [Google Scholar]
- Nie WZ, Ye M, Wang ZY. Infinite models in scoliosis: a review of the literature and analysis of personal experience. Biomed Tech. 2008;53:174–80. doi: 10.1515/BMT.2008.029. [DOI] [PubMed] [Google Scholar]
- Wang X, Aubin CE, Labelle H, et al. Biomechanical modelling of a direct vertebral translation instrumentation system: preliminary results. Stud Health Technol Inform. 2008;140:128–32. [PubMed] [Google Scholar]
- Anderson AE, Ellis BJ, Maas SA, et al. Validation of finite element predictions of cartilage contact pressure in the human hip joint. J Biomech Eng. 2008;130:051008. doi: 10.1115/1.2953472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barratt DC, Chan CSK, Edwards PJ, et al. Instantiation and registration of statistical shape models of the femur and pelvis using 3D ultrasound imaging. Med Image Anal. 2008;12:358–74. doi: 10.1016/j.media.2007.12.006. [*] [DOI] [PubMed] [Google Scholar]
- Bailey C, Gul R, Falworth M, et al. Component alignment in hip resurfacing using computer navigation, clinical orthopaedics and related research. 2009;467:917–22. doi: 10.1007/s11999-008-0584-x. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahr DH, Zysset PK. From high-resolution CT data to finite element models: development of an integrated modular framework. Comp Meth Biomech Biomed Engin. 2009;12:45–57. doi: 10.1080/10255840903065399. [DOI] [PubMed] [Google Scholar]
- Zheng G, Schumann S. 3D reconstruction of a patient-specific surface model of the proximal femur from calibrated x-ray radiographs: a validation study. Med Phys. 2009;36:1155–66. doi: 10.1118/1.3089423. [*] [DOI] [PubMed] [Google Scholar]
- de Zee M, Cattaneo PM, Svensson P, et al. Prediction of the articular eminence shape in a patient with unilateral hypoplasia of the right mandibular ramus before and after distraction osteogenesis—a simulation study. J Biomech. 2009;42:1049–53. doi: 10.1016/j.jbiomech.2009.02.027. [*] [DOI] [PubMed] [Google Scholar]
- Diederichs G, Seim H, Meyer H, et al. CT-based patient-specific modeling of glenoid rim defects: a feasibility study. AJR Am J Roentgenol. 2008;191:1406–11. doi: 10.2214/AJR.08.1091. [*] [DOI] [PubMed] [Google Scholar]
- Subburaj K, Ravi B, Agarwal M. Automated identification of anatomical landmarks on 3D bone models reconstructed from CT scan images. Comp Med Imag Graph. 2009;33:359–68. doi: 10.1016/j.compmedimag.2009.03.001. [DOI] [PubMed] [Google Scholar]
- D'Ambrosio AL, Mocco J, Hankinson TC, et al. Quantification of the frontotemporal orbitozygomatic approach using a three-dimensional visualization and modeling application. Neurosurgery. 2008;62:251–60. doi: 10.1227/01.neu.0000317401.38960.f6. discussion 260–51. [DOI] [PubMed] [Google Scholar]
- Cattaneo PM, Dalstra M, Frich LH. A three-dimensional finite element model from computed tomography data: a semi-automated method. Proceedings of the Institution of Mechanical Engineers. J Engin Med. 2001;215:203–213. doi: 10.1243/0954411011533760. [DOI] [PubMed] [Google Scholar]
- Diamant I, Shahar R, Masharawi Y, et al. A method for patient-specific evaluation of vertebral cancellous bone strength: in vitro validation. Clin Biomech. 2007;22:282–91. doi: 10.1016/j.clinbiomech.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Ackerman MJ. The visible human project. Proc IEEE. 1998;86:504–11. [Google Scholar]
- Aubin CE, Labelle H, Chevrefils C, et al. Preoperative planning simulator for spinal deformity surgeries. Spine. 2008;33:2143–52. doi: 10.1097/BRS.0b013e31817bd89f. [*] [DOI] [PubMed] [Google Scholar]
- Doenst T, Spiegel K, Reik M, et al. Fluid-dynamic modeling of the human left ventricle: methodology and application to surgical ventricular reconstruction. Ann Thorac Surg. 2009;87:1187–95. doi: 10.1016/j.athoracsur.2009.01.036. [DOI] [PubMed] [Google Scholar]
- Tang D, Yang C, Geva T, et al. Patient-specific MRI-based 3D FSI RV/LV/patch models for pulmonary valve replacement surgery and patch optimization. J Biomech Eng. 2008;130:041010. doi: 10.1115/1.2913339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley M, Stinstra J, Pieper S, et al. A computer modeling tool for comparing novel ICD electrode orientations in children and adults. Heart Rhythm. 2008;5:565–72. doi: 10.1016/j.hrthm.2008.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer B, Hanser F, Seger M, et al. Patient-specific volume conductor modeling for non-invasive imaging of cardiac electrophysiology. Open Med Inform J. 2008;2:32–41. doi: 10.2174/1874431100802010032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon W, Delhaas T, Arts T, et al. Computational modeling of volumetric soft tissue growth: application to the cardiac left ventricle. Biomech Model Mechanobiol. 2009;8:301–9. doi: 10.1007/s10237-008-0136-z. [*] [DOI] [PubMed] [Google Scholar]
- Ionasec RI, Georgescu B, Gassner E, et al. Dynamic model-driven quantitative and visual evaluation of the aortic valve from 4D CT. Med Image Comput Comput Assist Interv Int Conf Med Image Comput Comput Assist Interv. 2008;11:686–94. doi: 10.1007/978-3-540-85988-8_82. [DOI] [PubMed] [Google Scholar]
- Votta E, Caiani E, Veronesi F, et al. Mitral valve finite-element modelling from ultrasound data: a pilot study for a new approach to understand mitral function and clinical scenarios. Phil Trans Royal Soc A: Math, Phys and Engin Sci. 2008;366:3411–34. doi: 10.1098/rsta.2008.0095. [DOI] [PubMed] [Google Scholar]
- Starfinger C, Chase JG, Hann CE, et al. Model-based identification and diagnosis of a porcine model of induced endotoxic shock with hemofiltration. Math Biosci. 2008;216:132–9. doi: 10.1016/j.mbs.2008.08.014. [DOI] [PubMed] [Google Scholar]
- Le Rolle V, Hern·ndez AI, Richard P-Y, et al. Model-based analysis of myocardial strain data acquired by tissue Doppler imaging. Artificial Intell Med. 2008;44:201–19. doi: 10.1016/j.artmed.2008.06.001. [*] [DOI] [PubMed] [Google Scholar]
- Sermesant M, Peyrat JM, Chinchapatnam P, et al. Toward patient-specific myocardial models of the heart. Heart Fail Clin. 2008;4:289–301. doi: 10.1016/j.hfc.2008.02.014. [*] [DOI] [PubMed] [Google Scholar]
- Kerckhoffs RC, Bovendeerd PH, Kotte JC, et al. Homogeneity of cardiac contraction despite physiological asynchrony of depolarization: a model study. Ann Biomed Eng. 2003;31:536–47. doi: 10.1114/1.1566447. [DOI] [PubMed] [Google Scholar]
- Usyk TP, LeGrice IJ, McCulloch AD. Computational model of three dimensional cardiac electromechanics. Comput Visual Sci. 2002;4:249–57. [Google Scholar]
- Campbell SG, Howard E, Aguado-Sierra J, et al. Effect of transmurally heterogeneous myocyte excitation-contraction coupling on canine left ventricular electromechanics. Exp Physiol. 2009;94:541–52. doi: 10.1113/expphysiol.2008.044057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash MP, Panfilov AV. Electromechanical model of excitable tissue to study reentrant cardiac arrhythmias. Progr Biophys Mol Biol. 2004;85:501–22. doi: 10.1016/j.pbiomolbio.2004.01.016. [DOI] [PubMed] [Google Scholar]
- Nickerson D, Smith N, Hunter P. New developments in a strongly coupled cardiac electromechanical model. Europace. 2005;7(Suppl 2):118–27. doi: 10.1016/j.eupc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Niederer SA, Smith NP. An improved numerical method for strong coupling of excitation and contraction models in the heart. Prog Biophys Mol Biol. 2008;96:90–111. doi: 10.1016/j.pbiomolbio.2007.08.001. [DOI] [PubMed] [Google Scholar]
- Sermesant M, Moireau P, Camara O, et al. Cardiac function estimation from MRI using a heart model and data assimilation: advances and difficulties. Med Image Anal. 2006;10:642–56. doi: 10.1016/j.media.2006.04.002. [*] [DOI] [PubMed] [Google Scholar]
- Smith NP, Buist ML, Pullan AJ. Altered T wave dynamics in a contracting cardiac model. J Cardiovascular Electrophysiol. 2003;14:203–9. doi: 10.1046/j.1540.8167.90312.x. [DOI] [PubMed] [Google Scholar]
- Southern J, Pitt-Francis J, Whiteley J, et al. Multi-scale computational modelling in biology and physiology. Prog Biophys Mol Biol. 2008;96:60–89. doi: 10.1016/j.pbiomolbio.2007.07.019. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckhoffs RC, Narayan SM, Omens JH, et al. Computational modeling for bedside application. Heart Fail Clin. 2008;4:371–8. doi: 10.1016/j.hfc.2008.02.009. [*] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh AL, Wang Y, Ross RS, et al. Mechanotransduction in single cardiac myocyte studied using laser tweezers and FRET. Biophys J. 2009;96:316a. [Google Scholar]
- Amaro R, Baron R, McCammon J. An improved relaxed complex scheme for receptor flexibility in computer-aided drug design. J Comp-Aid Mol Design. 2008;22:693–705. doi: 10.1007/s10822-007-9159-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song CM, Lim SJ, Tong JC. Brief Bioinform. 2009. Recent advances in computer-aided drug design. Advance Access published online on May 11, 2009; doi:10.1093/bib/bbp023. [DOI] [PubMed] [Google Scholar]
- VPH—preDiCT. [(10 October 2009, date last accessed)]. http://www.vph-predict.eu/ 2009. [Google Scholar]
- Astrom M, Zrinzo LU, Tisch S, et al. Method for patient-specific finite element modeling and simulation of deep brain stimulation. Med Biol Eng Comput. 2009;47:21–8. doi: 10.1007/s11517-008-0411-2. [*] [DOI] [PubMed] [Google Scholar]
- Maks CB, Butson CR, Walter BL, et al. Deep brain stimulation activation volumes and their association with neurophysiological mapping and therapeutic outcomes. J Neurol Neurosurg Psychiatry. 2009;80:659–66. doi: 10.1136/jnnp.2007.126219. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasques X, Cif L, Hess O, et al. Stereotactic model of the electrical distribution within the internal globus pallidus during deep brain stimulation. J Comput Neurosci. 2009;26:109–18. doi: 10.1007/s10827-008-0101-y. [DOI] [PubMed] [Google Scholar]
- Schiavone P, Chassat F, Boudou T, et al. In vivo measurement of human brain elasticity using a light aspiration device. Med Image Anal. 2009;13:673–8. doi: 10.1016/j.media.2009.04.001. [*] [DOI] [PubMed] [Google Scholar]
- Wittek A, Hawkins T, Miller K. On the unimportance of constitutive models in computing brain deformation for image-guided surgery. Biomech Model Mechanobiol. 2009;8:77–84. doi: 10.1007/s10237-008-0118-1. [*] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittek A, Dutta-Roy T, Taylor Z, et al. Subject-specific non-linear biomechanical model of needle insertion into brain. Comput Methods Biomech Biomed Engin. 2008;11:135–46. doi: 10.1080/10255840802296665. [*] [DOI] [PubMed] [Google Scholar]
- Wakeland W, Agbeko R, Vinecore K, et al. Assessing the prediction potential of an in silico computer model of intracranial pressure dynamics. Crit Care Med. 2009;37:1079–89. doi: 10.1097/CCM.0b013e31819b629d. [*] [DOI] [PubMed] [Google Scholar]
- Dubowsky SR, Rasmussen J, Sisto SA, et al. Validation of a musculoskeletal model of wheelchair propulsion and its application to minimizing shoulder joint forces. J Biomech. 2008;41:2981–8. doi: 10.1016/j.jbiomech.2008.07.032. [*] [DOI] [PubMed] [Google Scholar]
- Reinbolt JA, Haftka RT, Chmielewski TL, et al. A computational framework to predict post-treatment outcome for gait-related disorders. Med Engin Phys. 2008;30:434–43. doi: 10.1016/j.medengphy.2007.05.005. [*] [DOI] [PubMed] [Google Scholar]
- Linder-Ganz E, Shabshin N, Gefen A. Patient-specific modeling of deep tissue injury biomechanics in an unconscious patient who developed myonecrosis after prolonged lying. J Tissue Viability. 2009;18:62–71. doi: 10.1016/j.jtv.2009.02.004. [*] [DOI] [PubMed] [Google Scholar]
- Portnoy S, Yizhar Z, Shabshin N, et al. Internal mechanical conditions in the soft tissues of a residual limb of a trans-tibial amputee. J Biomech. 2008;41:1897–909. doi: 10.1016/j.jbiomech.2008.03.035. [*] [DOI] [PubMed] [Google Scholar]
- Bouten CV, Oomens CW, Baaijens FP, et al. The etiology of pressure ulcers: skin deep or muscle bound? Arch Phys Med Rehabil. 2003;84:616–9. doi: 10.1053/apmr.2003.50038. [DOI] [PubMed] [Google Scholar]
- Gefen A. Bioengineering models of deep tissue injury. Adv Skin Wound Care. 2008;21:30–6. doi: 10.1097/01.ASW.0000305403.89737.6c. [DOI] [PubMed] [Google Scholar]
- Linder-Ganz E, Engelberg S, Scheinowitz M, et al. Pressure-time cell death threshold for albino rat skeletal muscles as related to pressure sore biomechanics. J Biomech. 2006;39:2725–32. doi: 10.1016/j.jbiomech.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Stekelenburg A, Strijkers GJ, Parusel H, et al. Role of ischemia and deformation in the onset of compression-induced deep tissue injury: MRI-based studies in a rat model. J Appl Physiol. 2007;102:2002–11. doi: 10.1152/japplphysiol.01115.2006. [*] [DOI] [PubMed] [Google Scholar]
- Cattaneo PM, Dalstra M, Melsen B. Moment-to-force ratio, center of rotation, and force level: A finite element study predicting their interdependency for simulated orthodontic loading regimens. Am J Orthod Dentofac Orthoped. 2008;133:681–9. doi: 10.1016/j.ajodo.2006.05.038. [DOI] [PubMed] [Google Scholar]
- Cattaneo PM, Dalstra M, Melsen B. Strains in periodontal ligament and alveolar bone associated with orthodontic tooth movement analyzed by finite element. Orthod Craniofac Res. 2009;12:120–8. doi: 10.1111/j.1601-6343.2009.01445.x. [DOI] [PubMed] [Google Scholar]
- Kondo T, Wakabayashi N. Influence of molar support loss on stress and strain in premolar periodontium: a patient-specific FEM study. J Dent. 2009;37:541–8. doi: 10.1016/j.jdent.2009.03.015. [**] [DOI] [PubMed] [Google Scholar]
- Campbell JM, Wong CO, Muzik O, et al. Early dose response to yttrium-90 microsphere treatment of metastatic liver cancer by a patient-specific method using single photon emission computed tomography and positron emission tomography. Int J Radiat Oncol Biol Phys. 2009;74:313–20. doi: 10.1016/j.ijrobp.2008.12.058. [**] [DOI] [PubMed] [Google Scholar]
- South CP, Partridge M, Evans PM. A theoretical framework for prescribing radiotherapy dose distributions using patient-specific biological information. Med Phys. 2008;35:4599–611. doi: 10.1118/1.2975229. [*] [DOI] [PubMed] [Google Scholar]
- Titz B, Jeraj R. An imaging-based tumour growth and treatment response model: investigating the effect of tumour oxygenation on radiation therapy response. Phys Med Biol. 2008;53:4471–88. doi: 10.1088/0031-9155/53/17/001. [*] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorelik B, Ziv I, Shohat R, et al. Efficacy of weekly docetaxel and bevacizumab in mesenchymal chondrosarcoma: a new theranostic method combining xenografted biopsies with a mathematical model. Cancer Res. 2008;68:9033–40. doi: 10.1158/0008-5472.CAN-08-1723. [*] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuentes D, Oden JT, Diller KR, et al. Computational modeling and real-time control of patient-specific laser treatment of cancer. Ann Biomed Eng. 2009;37:763–82. doi: 10.1007/s10439-008-9631-8. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogea C, Davatzikos C, Biros G. An image-driven parameter estimation problem for a reaction-diffusion glioma growth model with mass effects. J Math Biol. 2008;56:793–825. doi: 10.1007/s00285-007-0139-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szeto MD, Chakraborty G, Hadley J, et al. Quantitative metrics of net proliferation and invasion link biological aggressiveness assessed by MRI with hypoxia assessed by FMISO-PET in newly diagnosed glioblastomas. Cancer Res. 2009;69:4502–9. doi: 10.1158/0008-5472.CAN-08-3884. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pathmanathan P, Gavaghan DJ, Whiteley JP, et al. Predicting tumor location by modeling the deformation of the breast. IEEE Trans Biomed Eng. 2008;55:2471–80. doi: 10.1109/TBME.2008.925714. [*] [DOI] [PubMed] [Google Scholar]
- Juffer AH, Marin U, Niemitalo O, et al. Computer modeling of brain tumor growth. Mini Rev Med Chem. 2008;8:1494–506. doi: 10.2174/138955708786786471. [**] [DOI] [PubMed] [Google Scholar]
- Deisboeck TS, Zhang L, Yoon J. In silico cancer modeling: is it ready for prime time? Nat Clin Pract Oncol. 2009;6:34–42. doi: 10.1038/ncponc1237. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashwath S, Toshinori Y, Christopher EH, et al. A minimal model of lung mechanics and model-based markers for optimizing ventilator treatment in ARDS patients. Comp Meth Programs Biomed. 2009;95:166–80. doi: 10.1016/j.cmpb.2009.02.008. [DOI] [PubMed] [Google Scholar]
- Shi Y, Qi F, Xue Z, et al. Segmenting lung fields in serial chest radiographs using both population-based and patient-specific shape statistics. IEEE Trans Med Imag. 2008;27:481–94. doi: 10.1109/TMI.2007.908130. [DOI] [PubMed] [Google Scholar]
- Li NY, Verdolini K, Clermont G, et al. A patient-specific in silico model of inflammation and healing tested in acute vocal fold injury. PLoS ONE. 2008;3:e2789. doi: 10.1371/journal.pone.0002789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X, Zhang N, Sha D, et al. A discrete mechanics framework for real time virtual surgical simulations with application to virtual laparoscopic nephrectomy. Stud Health Technol Inform. 2009;142:459–64. [PubMed] [Google Scholar]
- Oomens CWJ, Ratingen MRV, Janssen JD, et al. A numerical-experimental method for a mechanical characterization of biological materials. J Biomech. 1993;26:617. doi: 10.1016/0021-9290(93)90024-9. [DOI] [PubMed] [Google Scholar]
- Hendriks FM, Brokken D, Van Eemeren J, et al. A numerical-experimental method to characterize the non-linear mechanical behaviour of human skin. Skin Res Technol. 2003;9:274–83. doi: 10.1034/j.1600-0846.2003.00019.x. [DOI] [PubMed] [Google Scholar]
- Rudy Y, Ackerman MJ, Bers DM, et al. Systems approach to understanding electromechanical activity in the human heart: a national heart, lung, and blood institute workshop summary. Circulation. 2008;118:1202. doi: 10.1161/CIRCULATIONAHA.108.772715. [**] [DOI] [PMC free article] [PubMed] [Google Scholar]
- NIH. Working Group 10: Model Sharing and the Defining of Modeling Standards. http://www.imagwiki.org/mediawiki/index.php?title=Working_Group_10 2008.
- EMBL-EBI. MIASE—Minimum Information About a Simulation Experiment. http://www.ebi.ac.uk/compneur-srv/miase/2009. [DOI] [PMC free article] [PubMed]
- Hucka M, Finney A, Sauro HM, et al. The Systems Biology Markup Language (SBML): A Medium for Representation and Exchange of Biochemical Network Models. Bioinformatics. 2003;19:524–31. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Le Novere N, Finney A, Hucka M, et al. Minimum information requested in the annotation of biochemical models (MIRIAM) Nat Biotechnol. 2005;23:1509–15. doi: 10.1038/nbt1156. [DOI] [PubMed] [Google Scholar]
- Neal ML, Gennari JH, Arts T, et al. Advances in semantic representation for multiscale biosimulation: a case study in merging models. Pac Symp Biocomput. 2009;14:304–15. [PMC free article] [PubMed] [Google Scholar]
- Nickerson D, Hunter P. Using CellML in computational models of multiscale physiology. Conf Proc IEEE Eng Med Biol Soc. 2005;6:6096–9. doi: 10.1109/IEMBS.2005.1615884. [DOI] [PubMed] [Google Scholar]
- Smith NP, Crampin EJ, Niederer SA, et al. Computational biology of cardiac myocytes: proposed standards for the physiome. J Experiment Biol. 2007;210:1576. doi: 10.1242/jeb.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass DA. Cardiac resynchronization therapy. J Cardiovascular Electrophysiol. 2005;16:S35–41. doi: 10.1111/j.1540-8167.2005.50136.x. [DOI] [PubMed] [Google Scholar]
- Owens JD, Houston M, Luebke D, et al. GPU computing. Proc IEEE. 2008;96:879–99. [Google Scholar]