Abstract
Background
Mutations in the alpha catalytic subunit of phosphoinositol-3-kinase (PIK3CA) occur in ~30% of ER positive breast cancers. We therefore sought to determine the impact of PIK3CA mutation on response to neoadjuvant endocrine therapy.
Methods
Exon 9 (helical domain - HD) and Exon 20 (kinase domain- KD) mutations in PIK3CA were determined samples from four neoadjuvant endocrine therapy trials. Interactions with clinical, pathological and biomarker response parameters were examined.
Results
A weak negative interaction between PIK3CA mutation status and clinical response to neoadjuvant endocrine treatment was detected (N=235 P=<0.05), but not with treatment-induced changes in Ki67-based proliferation index (N=418). Despite these findings, PIK3CA KD mutation was a favorable prognostic factor for relapse-free survival (RFS log rank P=0.02) in the P024 trial (N=153). The favorable prognostic effect was maintained in a multivariable analysis (N=125) that included the preoperative prognostic index (PEPI), an approach to predicting RFS based on post neoadjuvant endocrine therapy pathological stage, ER and Ki67 levels (HR for no PIK3CA KD mutation, 14, CI 1.9–105 P=0.01).
Conclusion
PIK3CA mutation status did not strongly interact with neoadjuvant endocrine therapy responsiveness in estrogen receptor positive breast cancer. Nonetheless, as with other recent studies, a favorable interaction between PIK3CA kinase domain mutation and prognosis was detected. The mechanism for the favorable prognostic impact of PIK3CA mutation status therefore remains unexplained.
Introduction
Gain-of-function mutations in the catalytic subunit of phosphoinositide-3-kinase occur in approximately 30% of estrogen receptor positive (ER+) breast cancers. Mutations occur predominately in two “hot spots” in exons 9 and 20, encoding the helical (HD) and kinase domains (KD) respectively (1). The association of PIK3CA mutation and ER expression was first reported by Saal et al (2) and is a consistent finding (3, 4). Studies on the effect of PIK3CA mutations on prognosis have been mixed; however recent data suggests that in postmenopausal women with ER+ disease, there may be a favorable effect (5, 6). Based on these findings, we postulated that PIK3CA mutation may be a positive predictive biomarker for endocrine therapy responsiveness. Consist with this hypothesis, the most commonly studied breast cancer cell lines that exhibit estrogen dependence in vitro, MCF7 (HD mutant) and T47D (KD mutant), both harbor PIK3CA mutations (7). Of relevance, in both these cell lines, inhibition of PI3 kinase with RNAi, or with a small molecule inhibitor, in combination with ER inhibition, induces apoptosis in a synthetic lethal manner, raising the possibility that combined inhibition of PI3 kinase and ER could emerge as a new cytotoxic combination for PI3 kinase mutant estrogen dependent breast cancer (8). To further understand the interaction between PIK3CA and breast cancer outcomes in ER+ disease we examined the impact of PIK3CA mutation status on clinical and biomarker outcomes in four neoadjuvant endocrine therapy clinical trials: P024, a randomized trial that compared tamoxifen and letrozole (9), the letrozole alone arm of the RAD2222 study (10), a Phase 2 trial of preoperative letrozole (POL) (11) and the Z1031 trial, a randomized comparison of three aromatase inhibitors, which, while still incomplete, provided additional samples for exploring interactions with the proliferation biomarker Ki67.
Methods
Study Population and tumor bank
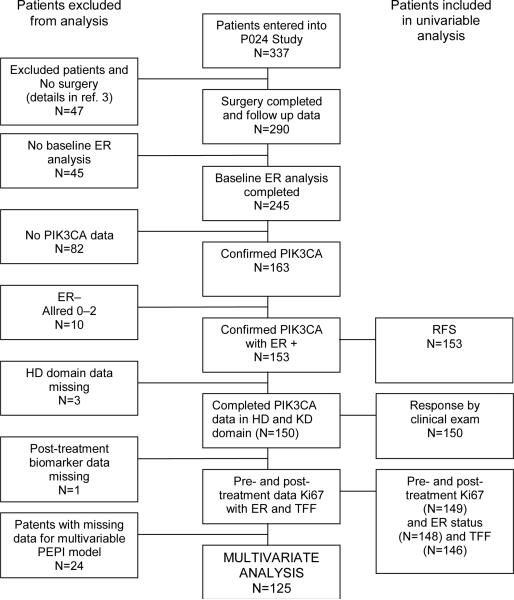
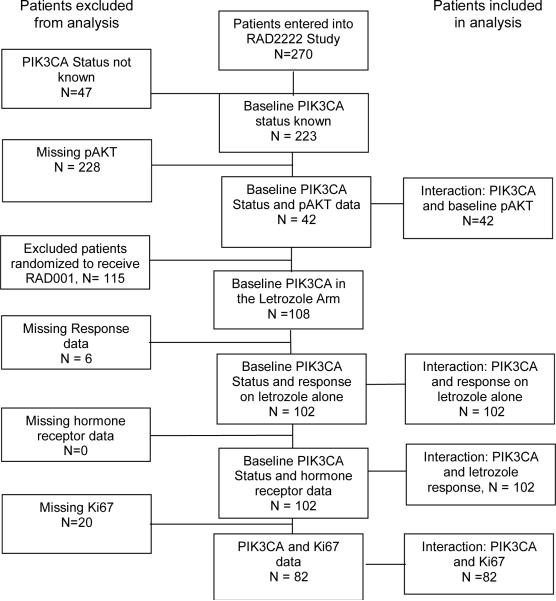
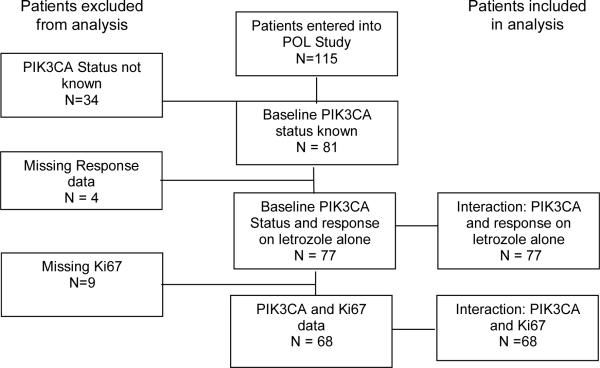
The clinical findings, tumor bank characteristics and biomarker measurements of the P024 trial have been described previously (9, 12, 13). The RAD2222 trial was a randomized study of letrozole versus letrozole in combination with everolimus (10). The samples utilized for this study were from the letrozole monotherapy arm. The POL trial was a Phase 2 study of preoperative letrozole for 4 to 6 months (11). The REMARK diagrams for sample acquisition are provided in Figure 1A–C. To create an adequately sized third data set to examine the interaction between treatment-induced Ki67 changes and PIK3CA status, the POL study was supplemented with 69 samples from an ongoing Z1031 neoadjuvant endocrine therapy study that is comparing letrozole, anastrozole and exemestane. Since Z1031 has not been completed, a REMARK diagram is not provided. Local ethics committees approved all trials and informed consent was obtained from all study participants.
Figure 1A, B, C.
Remark diagrams for the P024 trial (A), the RAD2222 trial (B) and POL (C) indicating the distribution of specimens in the various analyses reported in the paper.
Mutation screening
For the RAD2222 study, 8uM tumor biopsy sections were macro-dissected and the Picopure kit (Arcturus) was used for DNA recovery. Mutations were analyzed by direct sequencing and by the Surveyor-Wave system (Transgenomic). PCR was performed using the Advantage HF2 PCR kit (Clontech). Primer sequences are TGCTTTTTCTGTAAATCATCTGTG (forward) and TATGGTAAAAACATGCTGAGATCA (reverse) for PIK3CA exon 9 and AGCTATTCGACAGCATGCCAATC (forward) and ATCGGTCTTTGCCTGCTGAGAG (reverse) for PIK3CA exon 20. For direct sequencing, PCR products were purified on a Biomek FX liquid handling robotic workstation using AMPure magnetic beads (Agencourt) and sequenced with an ABI BigDye Terminator v3.1 Cycle Sequencing kit on an ABI 3730xl DNA Analyzer. Sequence variation was analyzed using Polyphread software. For the Surveyor-Wave analysis, PCR products were digested directly using the Surveyor Nuclease kit (Transgenomic) according to the manufacturer's instructions. Digested PCR products were analyzed with the Transgenomic Wave 3500HT system under non-denaturing conditions. Samples showing distinct elution profiles when compared with normal control sample were identified as mutants and confirmed by direct sequencing.
For the P024 trial, laser capture microdissection (LCM, Arcturus /MDS, Sunnyvale, CA) was employed to isolate tumor cells from 10 micron formalin fixed sections. For the Z1031 and POL studies DNA was extracted from frozen biopsies that contained in excess of 70% cancer. DNA was isolated and purified using the DNeasy Tissue Kit (Qiagen, Valencia, CA) with over-night proteinase K digestion at 60C. For RNA isolation, macrodissection was conducted to obtain material with tumor cellularity higher than 50% based on H&E staining of an adjacent section. RNA was isolated with either Optimum FFPE RNA isolation kits (Applied Biosystems/Ambion, Foster City, CA ) or RecoverALL Total Nucleic Acid Isolation kits (Ambion). PCR products were generated with primers designed to span mutation hotspots in the HD (exon 9) and the KD (exon20). Initially, genomic DNA was screened by PCR using Taq LA DNA polymerase (Sigma-Aldrich, St. Louis, MO). If no product was detected, RT-PCR of RNA was conducted using the iScript cDNA synthesis Kit (Bio-Rad, Hercules, CA) followed by HotStart-IT Taq DNA polymerase (USB, Cleveland, Ohio). 7.5–10ng DNA or cDNA were used as the template for the PCR reactions in 15ul or 25 ul respectively. A “Step down” in the iCycler (Bio-Rad) thermal cycler was used for optimum annealing. The primers used for genomic PCR are TGAAAATGTATTTGCTTTTTCTGT (forward) and TCATGTAAATTCTGCTTTATTTATTCC (reverse) for amplification of exon 9; and CTATTCGACAGCATGCCAATC (forward) and AATTGTGTGGAAGATCCAATCC (reverse) for amplification of exon 20. The primers used for RT-PCR were GCAGGACTGAGTAACAGACTAGCTAGAGA (forward) and GTTACACAATAGTGTCTGTGACTCCATAGA (reverse) for exon 9; And CGAAAG ACCCTAGCCTTAG (forward) and CAGTTCAATGCATGCTGT (reverse) for exon 20. All of the forward and reverse primers were tailed with M13 universal primers TGTAAAACGACGGCCAGT (forward) and CAGGAAACAGCTATGACC (reverse) for facilitating downstream sequencing. All products were gel-purified (QIAquick gel extraction kit, Qiagen) before sequencing using the BigDye Terminator v3.1 sequencing kit (Applied Biosystems). Sequences were obtained using an ABI 3731 capillary sequencing machine. For mutational analysis, Mutation Surveyor, v 3.01software (Softgenetics) was employed along with manual reviewing of chromatogram trace files with Finch TV v. 1.4 (Geospiza, Seattle, WA).
Phospho AKT, Ki67, ER and PgR immunohistochemistry
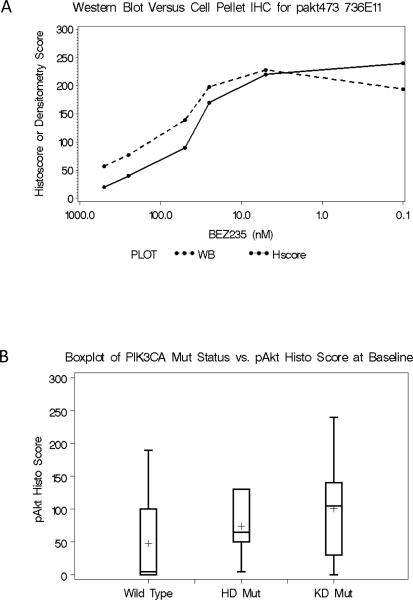
4uM sections were cut fresh from paraffin-embedded core needle biopsies and stained for phospho-Akt S473 with the rabbit mAb clone 736E11 in a subset of samples from the RAD2222 trial (Cell Signaling Technologies) on a Ventana XT immunostainer using CC1 buffer, mild, no heat, for antigen retrieval and Omni-UltraMap HRP for visualization. Staining was recorded as a histoscore, calculated as the percentage of positive cells times an intensity scale of either 1+, 2+ or 3+. To validate IHC quantitation of pAkt473, Jurkat cells were treated for 1 hour with increasing concentrations of BEZ235, a PIK3CA inhibitor in Phase 1 clinical development (14), and both lysates and cell pellets were prepared from the treated cells. The latter were mounted as a tissue microarray and subject to IHC using the protocol described above. Histoscores were generated by one blinded observer. These were compared to western blots of the lysates, using the same antibody, assessed by densitometry. The correlation between IHC and western blot quantification was acceptable (Figure 2A). Pretreatment of the TMA slide with lambda phosphatase abolished all staining, indicating specificity for phospoproteins. The Ki67, ER and PgR methodology and results for the P024 samples have already been published (9, 12) . In the RAD2222 trial standard reagents from Ventana were used according to manufacturer's recommendations (ER: clone SP1; PR: clone 1E2; Ki67: clone 30–9). In the Z1031 and POL trials, the Ki67 assay was done using the SP6 antibody (Neomarkers) on a Shandon Sequenza® Immunostainer. Envision HRP secondary antibodies and hematoxylin counter staining were used for visualization. For Ki67 scoring, photomicrographs were taken under 40X magnification and projected on a background grid. Ki67 staining was quantified independently by two observers using point counting and the average percent of Ki67 positive cells was determined. Methodology used to assign HER2 status in the P024 and the RAD2222 trials has been described previously (9, 10).
Figure 2A and 2B.
A. The effect of BEZ235 on pAKT levels in Jurkat cells prepared as either as a lysate and estimated by immunoblotting, or as a cell block and estimated by the semi-quantitative immunohistochemical technique used for the breast tumor analysis. B. A comparison of pAKT levels according to PIK3CA mutation status in samples from the letrozole alone arm of the RAD2222 trial. The Kruskal-Wallis test was used to compare the difference of cytoplasmic pAKT histology score amongst the three groups degrees of freedom (DF)=2 P=0.0453. The Mann-Whitley test was applied to compare PIK3CA wt group to PIK3CA mutant group when KD and HD mutation groups are combined DF=1, P=0.0135.
Statistical methodology
All P values reported were two sided; P<=0.05 were considered to be statistically significant. The Bonferroni correction for multiple testing was used when multiple comparisons were made. Fisher's exact and Chi squared tests were used to assess associations between PIK3CA mutation status and clinical trial and biomarker data. The effect of PIK3CA wild type and HD and/or KD mutants and biomarker changes were evaluated by Mann-Whitney test or Kruskal-Wallis test. The Mann-Whitney test was also applied to compare differences in Ki67 changes between PIK3CA wild type and HD and/or KD mutation status. The 95% confidence interval of the geometric Ki67 mean was calculated to show the size of effects in pair-wise comparisons. Relapse free survival (RFS) was defined as the interval between randomization and the earliest subsequent breast cancer event. For univariate analysis, survival curves were estimated by the Kaplan–Meier method, a P-value from a two-sided log-rank test was calculated. A multivariable Cox proportional hazard regression model was applied to evaluate the independent prognostic relevance of PIK3CA KD mutation status within the context of the prognostic variables in the PEPI score obtained upon pathological analysis of the surgical specimen: i.e. tumor size, lymph node status, ER and Ki67 levels (15). All statistical analyses were performed using SAS 9.1.3 (SAS Institute Inc., Cary NC USA).
Results
PIK3CA mutation and baseline pathological characteristics, HER2 gene amplification/over-expression, phospho-AKT levels and treatment induced changes in hormone receptor expression
Targeted sequencing of the PIK3CA HD and KD domains (Table 1) revealed that the frequency of PIK3CA mutations was 45/153 (29.4%) in the P024 trial, 38/97 (39.2%) in the RAD2222 trial and (25/79 (31.7%) in the POL trial and 19/69 (27.5%) for the samples from the Z1031 trial. The distribution of mutations between KD and HD domains (approximately twice as many KD as HD mutations) was similar to that reported in previous studies (2, 4). The KD mutation H1047R was the most prevalent anomaly observed in all study cohorts, followed by the HD mutation, E545K. Rarer alleles were also present. Thus, although different techniques and primer sets were used to assign PIK3CA mutation status in each data set, the similar distribution of mutations indicates the analytical approaches were comparable. In the individual trials (data not shown) and combined data set created from the P024 and RAD2222 trials, relationships between baseline HER2 status and histological grade and pathological stage after neoadjuvant therapy (pathological tumor size, node status) and PIK3CA mutation status were explored. However, no significant interactions emerged (Table 2A). The data sets could not be easily merged for an analysis of hormone receptor status, because different scoring approaches were used. However, PIK3CA mutation status had no impact on baseline or treatment induced changes in ER and PgR expression on the letrozole arms in either study (Table 2B, RAD2222 study and Table 2C, P024 study). Expression of the estrogen-regulated gene TFF1 was also examined in the letrozole arm of the P024 trial. TFF1 baseline levels and estrogen deprivation-induced changes were also not affected by the presence of a PIK3CA mutation, although a trend was observed for KD domain-expressing tumors to have higher baseline TFF1 expression that was also seen for ER in the P024 study (the P value was corrected for multiple comparisons). The tamoxifen arm in the P024 trial was not included in the pre and post treatment pair-wise analysis since tamoxifen had very inconsistent effects on the expression of estrogen-regulated genes that was likely due to the mixed agonist/antagonist properties of this agent (12). PhosphoAKT levels were examined in a subset of RAD2222 samples (N=42) to determine if PIK3CA mutation was associated with increased phosphorylation of AKT, a key downstream mediator of signal transduction (Figure 2B). A Kruskal-Wallis test indicated a significant difference in cytoplasmic pAKT amongst the three PI3KCA groups (wt, PIK3CA HD domain mutant and PIK3CA KD domain mutant (P=0.045) and the Mann-Whitney test was also significant when pAKT levels in the PIK3CA wild-type group were compared to a combined group of all samples harboring either class of PIK3CA mutation (P=0.01).
Table 1. PIK3CA Mutation Frequency.
Results from exon 9 and exon 20 mutational screens from the P024, RAD2222 POL and Z1031 trials. For the P024 and RAD2222 trials, total sample numbers are from the cases in which mutation status of both PIK3CA exons were determined. PIK3CA KD and HD mutation status was determined in all POL and Z1031 cases.
| PIK3CA Mutations in P024 | Cases | Domain | Exon |
|---|---|---|---|
| G1624A:E542K | 8 | Helical | 9 |
| G1633A:E545K | 8 | Helical | 9 |
| T1568C:L523S | 1 | Helical | 9 |
| G1573A:E525K | 1 | Helical | 9 |
| T1592C:L531P | 1 | Helical | 9 |
| Total in HD (%) | 18/152 (11.8%) | ||
| A3140G:H1047R | 29 | Kinase | 20 |
| Total in KD (%) | 29/163 (17.8%) | ||
| Total | 45/153 (29.4%) | ||
| PIK3CA Mutations in RAD2222 | Cases | Domain | Exon |
|---|---|---|---|
| G1633A:E545K | 9 | Helical | 9 |
| G1624A:E542K | 2 | Helical | 9 |
| C1636A:Q546K | 2 | Helical | 9 |
| A1634G:E545G | 1 | Helical | 9 |
| Total in HD (%) | 14/102 (13.7 %) | ||
| A3140G:H1047R | 19 | Kinase | 20 |
| A3140T:H1047R | 2 | Kinase | 20 |
| A3127G:M1043V | 1 | Kinase | 20 |
| C3139T:H1047Y | 1 | Kinase | 20 |
| C3145G:G1049R | 1 | Kinase | 20 |
| C3145T:G1049A | 1 | Kinase | 20 |
| 3204_3205ins A:NSTOP>KLKRSTOP | 1 | Kinase | 20 |
| Total in KD (%) | 26/102 (25.5%) | ||
| Total | 38/97 (39.2%) | ||
| PIK3CA Mutations in POL | Cases | Domain | Exon |
|---|---|---|---|
| G1624A:E542K | 2 | Helical | 9 |
| G1633A:E545K | 4 | Helical | 9 |
| C1370G:N457K | 1 | C2 | 7 |
| Total in HD (%) | 7/79 (10%) | ||
| A3140G:H1047R | 13 | Kinase | 20 |
| A3140T:H1047L | 4 | Kinase | 20 |
| G3068T:R1023L | 1 | Kinase | 20 |
| Total in KD (%) | 18/79 (22.8%) | ||
| Total | 25/79 (31.7%) | ||
| PIK3CA Mutations in Z1031 | Cases | Domain | Exon |
|---|---|---|---|
| G1624A:E542K | 3 | Helical | 9 |
| G1633A:E545K | 4 | Helical | 9 |
| Total in HD (%) | 7/69 (10.1%) | ||
| A3140G:H1047R | 12 | Kinase | 20 |
| Total in KD (%) | 12/69 (17.4%) | ||
| Total | 19/69 (27.5%) | ||
Table 2.
Interactions between PIK3CA mutation status and pathological and biomarker characteristics. A. A pooled analysis of pathological parameters for the P024 and RAD2222 trial. B. An analysis of ER and PgR expression at baseline and upon treatment in the letrozole alone arm of the RAD2222 trial. C. An analysis of ER, PgR and trefoil factor 1 (TFF1) in the P024 trial. Changes with treatment were also examined but only the letrozole arm.
| A | |||
|---|---|---|---|
| Characteristics (Combined Data¶) | PIK3CA HD | PIK3CA KD | PIK3CA WT |
| Histological Grade | |||
| I | 4 (15%) | 8 (18%) | 16 (10%) |
| II/III | 23 | 36 | 141 |
| Path tumor status | |||
| <=20mm | 11 (39%) | 15 (35%) | 62 (38%) |
| >20mm | 17 | 28 | 100 |
| Path node status | |||
| Negative | 12 (43%) | 22 (51%) | 85 (54%) |
| Positive | 16 | 21 | 72 |
| Histology | |||
| Lobular | 1 (4%) | 3 (7%) | 17 (11%) |
| Ductal | 24 | 43 | 142 |
| HER2 status | |||
| Negative | 26 (93%) | 44 (88%) | 173 (90%) |
| Positive | 2 | 6 | 19 |
| B | |||||
|---|---|---|---|---|---|
| Mutation Status RAD 2222 Trial* | PIK3CA HD (mean) | PIK3CA KD (mean) | PIK3CA WT (mean) | Comparison | P Value† |
| ER | 234 | 234 | 260 | mut vs wt | 0.19 |
| Baseline | KD vs wt | 0.28 | |||
| Histoscore | HD vs wt | 0.34 | |||
| PgR | 160 | 174 | 154 | mut vs wt | 0.36 |
| Baseline | KD vs wt | 0.37 | |||
| Histoscore | HD vs wt | 0.71 | |||
| ER | 4.5 | −14.5 | −7.8 | mut vs wt | 0.93 |
| Change | KD vs wt | 0.47 | |||
| Histoscore | HD vs wt | 0.36 | |||
| PgR | −82.7 | −75.5 | −85.9 | mut vs wt | 0.86 |
| Change | KD vs wt | 0.84 | |||
| Histoscore | HD vs wt | 0.89 | |||
| C | |||||
|---|---|---|---|---|---|
| Mutation Status P024 Trial | PIK3CA HD (mean) | PIK3CA KD (mean) | PIK3CA WT (mean) | Comparison | P value† |
| ER | 8.0 | 8.0 | 7.0 | mut vs wt | 0.1* |
| Allred | KD vs wt | 0.06* | |||
| Baseline | HD vs wt | 0.53 | |||
| PgR | 1.5 | 6.5 | 5.0 | mut vs wt | 0.82 |
| Allred | KD vs wt | 0.13 | |||
| Baseline | HD vs wt | 0.23 | |||
| TFF1 | 4.5 | 6.0 | 4.0 | mut vs wt | 0.1* |
| Allred | KD vs wt | 0.06* | |||
| Baseline | HD vs wt | 0.72 | |||
| ER | 0 | −0.5 | 0 | mut vs wt | 0.92 |
| Allred | KD vs wt | 0.58 | |||
| Change | HD vs wt | 0.26 | |||
| PgR | −1.5 | −5 | −4 | mut vs wt | 0.64 |
| Allred | KD vs wt | 0.1 | |||
| Change | HD vs wt | 0.52 | |||
| TFF1 | −3.5 | −2 | −1 | mut vs wt | 0.12* |
| Allred | KD vs wt | 0.35 | |||
| Change | HD vs wt | 0.12* | |||
P=NS, chi Square and Fishers exact tests.
Mann-Whitney test
indicates results that were significant without Bonferroni correction.
Analysis of interactions between PIK3CA mutation status, clinical response and Ki67 changes associated with neoadjuvant endocrine treatment
A series of contingency tables were examined to identify interactions between clinical response and PIK3CA mutation status (Table 3). The possibility that PIK3CA HD mutations may be associated with a poorer clinical response to letrozole was raised in the P024 data set, due to only one of six PIK3CA HD mutation positive tumors exhibiting a clinical response (17%) compared to 35 out of 46 (76%) of the PIK3CA wild-type tumors P=0.01 (Table 3). We therefore combined data from the P024, RAD2222 and POL trials (N=235) and detected a weak adverse effect on response with a response rate for both HD (56% response rate) and KD (57% response rate) in comparison with PIK3CA wild-type cases (70% response rate P=<0.05). To further explore the impact of PIK3CA mutation and the efficacy of endocrine therapy, endocrine treatment-induced changes in the proliferation marker Ki67 were studied within groups defined by the presence of PIK3CA KD and HD domain mutation (Table 4). All PIK3CA mutation-defined subgroups showed evidence for a significant decline in the Ki67 levels within each group (i.e. no evidence of resistance in a subgroup). Multiple exploratory cross group comparisons (mutant versus wild-type) confirmed equivalent falls in Ki67 expression levels in all groups defined by PIK3CA mutation status, even when the POL data set was expanded to include an additional 69 cases from the Z1031 trial (Table 4).
Table 3.
Relationships between PIK3CA mutation status and clinical response to neoadjuvant endocrine therapy.
| Clinical Responses† | PIK3CA Status [n (% response within subset)]* |
p-value§ | |||
|---|---|---|---|---|---|
| HD Mut | KD Mut | Double Mut | Wild Type | ||
| P024 Letrozole only | |||||
| Response/total | 1/6 (17%) | 7/10 (70%) | 0/1 (0%) | 35/46 (76%) | 0.0161 |
| P024 Tamoxifen only | |||||
| Response/total | 5/10 (50%) | 4/16 (25%) | 0/1 (0%) | 27/60 (45%) | 0.3025 |
| P024 Fused | |||||
| Response/total | 6/16 (38%) | 11/26 (42%) | 0/2 (0%) | 62/106 (58%) | 0.0525 |
| RAD 2222 trial Letrozole alone arm | |||||
| Response/total | 7/12 (58%) | 14/24 (58%) | 1/2 (50%) | 41/62 (66%) | 0.8428 |
| POL trial | |||||
| Response/total | 6/7 (86%) | 8/17 (47%) | 35/51 (69%) | 0.3822 | |
| Fused (P024 letrozole alone arm/RAD2222 letrozole alone arm/POL) | |||||
| Response/total | 14/25 (56%) | 29/51 (57%) | 111/159 (70%) | 0.0459 | |
Clinical response, defined as CR and PR versus SD and PD.
Rare double mutations (KD and HD mutant) was excluded from the analysis.
Fisher exact test Chi square test or Fisher exact test was used to evaluate associations between PIK3CA mutation status and clinical response.
Table 4.
Relationships between PIK3CA mutation status and endocrine therapy associated changes in Ki67 expression.
| Ki67 [Geometric mean(95% CI)] | PIK3CA mutation Status |
||
|---|---|---|---|
| HD mutation | HD Wild type | mut vs wt, P-value† | |
| P024 | |||
| Letrozole | |||
| Pre | 2.69(0.8–9.0) | 3.74(2.38–5.89) | 0.9085 |
| Surgery | 0.23(0.1–0.53) | 0.48(0.29–0.8) | |
| P-value - pre vs post within genotype* | 0.0313 | 0.0001 | |
| Tamoxifen | |||
| Pre | 4.72(2.9–7.66) | 6.29(4.48–8.83) | 0.2251 |
| Surgery | 2.65(1.24–5.65) | 1.43(0.92–2.25) | |
| P-value - pre vs post within genotype* | 0.0840 | 0.0001 | |
| RAD 2222 Letrozole alone arm | |||
| Pre | 38.52(26.85–55.26) | 19.44(13.78–27.44) | 0.2680 |
| Surgery | 3.72(0.5–27.51) | 0.88(0.42–1.85) | |
| P-value - pre vs post within genotype* | 0.002 | 0.0001 | |
| POL & Z1031 | |||
| Pre | 18.18(10.23–32.31) | 15.76(12.46–19.93) | 0.1153 |
| Surgery | 0.63(0.11–3.42) | 2.66(1.76–4.03) | |
| P-value - pre vs post within genotype* | 0.0012 | 0.0001 | |
| Ki67 [Geometric mean(95% CI)] | PIK3CA mutation Status |
||
|---|---|---|---|
| KD mutation | KD Wild type | mut vs wt, P-value† | |
| P024 | |||
| Letrozole | |||
| Pre | 6.21(2.37–16.29) | 3.23(2.02–5.16) | 0.7890 |
| Surgery | 0.74(0.15–3.62) | 0.4(0.24–0.65) | |
| P-value - pre vs post within genotype* | 0.0273 | 0.0001 | |
| Tamoxifen | |||
| Pre | 6.34(3.2–12.58) | 5.93(4.31–8.16) | 1.0 |
| Surgery | 1.57(0.55–4.5) | 1.5(0.98–2.28) | |
| P-value - pre vs post within genotype* | 0.0052 | 0.0001 | |
| RAD 2222 Letrozole alone arm | |||
| Pre | 19.99(13.26–30.14) | 20.76(14.15–30.48) | 0.2316 |
| Surgery | 1.39(0.31–6.16) | 0.96(0.44–2.11) | |
| P-value - pre vs post within genotype* | 0.0001 | 0.0001 | |
| POL & Z1031 | |||
| Pre | 17.88(13.15~24.31) | 15.49(11.88–20.2) | 0.3117 |
| Surgery | 3.64(1.62~8.18) | 2.0(1.24–3.23) | |
| P-value - pre vs post within genotype* | 0.0002 | 0.0001 | |
The Wilcoxon rank rank test was used to compare Ki67 changes within each subgroup defined by PIK3CA mutation status.
In comparison of Ki67 changes between groups the Mann-Whitney test was applied.
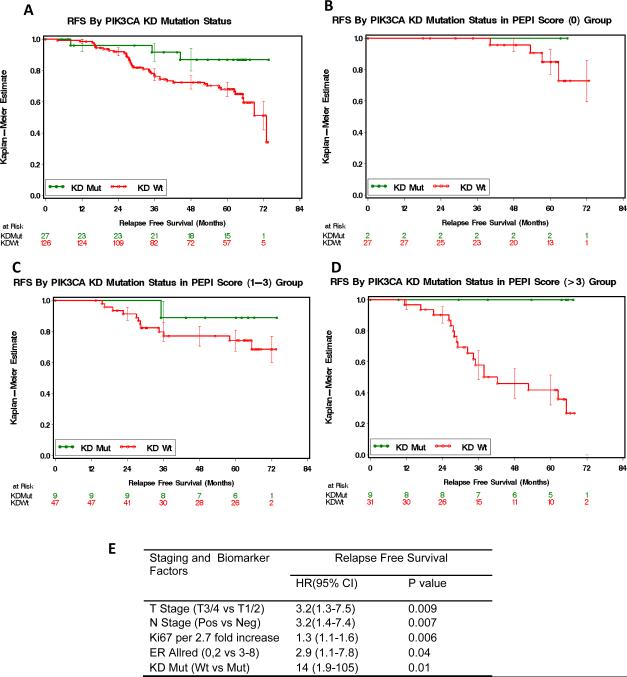
Analysis of the effect of PIK3CA mutation status and RFS and interactions with the preoperative endocrine prognostic index (PEPI)
Univariable analysis for the P024 cohort (N=153) indicated a modest association between PIK3CA KD domain mutation and more favorable RFS (PIK3CA KD mutant versus all others log rank 0.025 – Figure 3A). In contrast, we did not detect any interaction between PIK3CA HD mutation positive cases and RFS (data not shown). However PIK3CA HD domain mutations are less common, consequently this small data set does not rule out an interaction. Long-term outcomes associated with the P024 neoadjuvant endocrine therapy trial have been analyzed to develop a preoperative endocrine therapy index (PEPI), which provides a prognostic score based on a pathological staging and biomarker analysis of the tumor at surgery post neoadjuvant endocrine therapy (15). Cox proportional hazards were therefore used to weight the prognostic effects of PIK3CA KD domain mutation against the other four components of the PEPI (pathological tumor stage, pathological nodal stage, estrogen receptor status and the level of Ki67 expression, N=125). The PEPI model defines three broad risk groups. Group 1 tumors with a PEPI score of 0 (good prognosis), group 2 tumors with a PEPI of 1–3 (intermediate prognosis) and group 3 tumors with a PEPI of 4 or more (worst prognosis). PIK3CA KD mutation status did not have a significant effect on RFS in groups 1 and 2 (Figure 3B and 3C). However, in group 3, where most of the relapses occurred, the presence of a PIK3CA KD mutation was associated with more favorable RFS (log rank P=0.007, Figure 3D). When PIK3CA KD mutation status was added to the standard factors in the PEPI Cox proportional hazards model (Figure 3E), the absence of PIK3CA KD mutation was found to be a significant and independent adverse prognostic factor (HR 14 CI 1.9–105).
Figure 3A–E.
Interactions between PIK3CA mutations status and RFS in the P024 trial in subgroups defined by the PEPI score (15). Panel A: PIK3CA KD mutation positive cases versus other cases P=0.025 (log rank test). In Panel B–D: effect of PIK3CA status examined within the context of PEPI group 1 (B), group 2 (C) and group 3 (D). In PEPI group 3, the favorable effect of PIK3CA mutation status was significant P=0.007. Panel E: Cox proportional Hazards model that contains the four components of the PEPI model with the addition of PIK3CA KD mutation status.
Discussion
Neoadjuvant endocrine therapy trials are an efficient setting for biomarker studies because of the simultaneous capacity to examine the predictive qualities of a biomarker for endocrine therapy responsiveness and, when the data set matures, the prognostic impact in the setting of a cohort of patients uniformly treated with adjuvant endocrine treatment. However, a general limitation of neoadjuvant endocrine therapy data sets is the typically modest sample size. Thus, when looking at a relatively uncommon parameter, such as subclasses of PI3KCA mutation, it becomes necessary to combine data from several trials to study the biomarker question adequately.
Contrary to our hypothesis we found no evidence that PIK3CA mutation status strongly interacts with endocrine therapy responsiveness in the neoadjuvant setting, using an analysis approach that examined the impact of the two major classes of mutation, PIK3CA KD and PIK3CA HD, on clinical and biomarker outcomes in four independent studies. The weak negative interaction with clinical response (Table 3) is not of sufficient magnitude to be clinically meaningful and is contrary to the favorable effect on prognosis. Since the interaction is modest, these data must be confirmed in another data set, preferably a single study with uniform response criteria and a large sample size. The lack of a significant interaction between PIK3CA mutation status and treatment-induced changes in Ki67 is quite a robust negative finding since this interaction was explored in over 400 samples. In contrast to this negative finding, a similar analysis that examined the impact of HER2 gene amplification on the Ki67-based proliferation index revealed strong evidence that HER2 drives endocrine therapy-resistant proliferation (13). With the HER2 result as a “positive control”, we can therefore be fairly certain that PI3KCA mutations do not strongly perturb the short term (3 or 4 months) cell cycle response to endocrine agents in ER+ breast cancer. The data we report has implications for the design of exploratory neoadjuvant clinical trials that target mutant PIK3CA tumors with pharmacological PI3 kinase inhibitors in combination with an endocrine agent (8, 16), since the endocrine responsiveness of the PIK3CA mutant subgroup is an important consideration.
An overview of the influence of PI3 kinase mutations on prognosis presents a mixed picture but a view point is emerging that in post menopausal women with ER+ disease, PIK3CA mutation may have a favorable prognostic effect (17). Our findings in the context of the P024 trial, the only study of the four with sufficient long term outcome to address the prognosis question, support this hypothesis, at least for PIK3CA KD mutations (5). However we remain circumspect about our findings regarding the interaction with the PEPI score given the small sample size. The potential of PI3KCA mutation status to influence prognosis independently of endocrine response parameters is mechanistically interesting and will be studied further in the Z1031 trial, which will eventually produce a well-powered confirmatory data set to examine the role of PIK3CA mutations further.
Acknowledgments
Research Support This work was supported by a research grant to MJE by Novartis Pharma for the P024 trial, the POL trial was funded by an AVON NCI partners in progress award (3P50 CA68438-07S2) and R01 CA095614, a St Louis Affiliate of the Susan G Komen for the Core CRAFT grant, and the Z1031 analysis was supported by R01 CA095614, a Breast Cancer Research Foundation grant to the American College of Surgeons Oncology Group (NCI U10 CA076001) and grant support from Novartis and Pfizer.
References
- 1.Samuels Y, Wang Z, Bardelli A, et al. High frequency of mutations of the PIK3CA gene in human cancers. Science. 2004;304(5670):554. doi: 10.1126/science.1096502. [DOI] [PubMed] [Google Scholar]
- 2.Saal LH, Holm K, Maurer M, et al. PIK3CA mutations correlate with hormone receptors, node metastasis, and ERBB2, and are mutually exclusive with PTEN loss in human breast carcinoma. Cancer Res. 2005;65(7):2554–9. doi: 10.1158/0008-5472-CAN-04-3913. [DOI] [PubMed] [Google Scholar]
- 3.Perez-Tenorio G, Alkhori L, Olsson B, et al. PIK3CA mutations and PTEN loss correlate with similar prognostic factors and are not mutually exclusive in breast cancer. Clin Cancer Res. 2007;13(12):3577–84. doi: 10.1158/1078-0432.CCR-06-1609. [DOI] [PubMed] [Google Scholar]
- 4.Stemke-Hale K, Gonzalez-Angulo AM, Lluch A, et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008;68(15):6084–91. doi: 10.1158/0008-5472.CAN-07-6854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kalinsky K, Jacks LM, Heguy A, et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin Cancer Res. 2009;15(16):5049–59. doi: 10.1158/1078-0432.CCR-09-0632. [DOI] [PubMed] [Google Scholar]
- 6.Barbareschi M, Buttitta F, Felicioni L, et al. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin Cancer Res. 2007;13(20):6064–9. doi: 10.1158/1078-0432.CCR-07-0266. [DOI] [PubMed] [Google Scholar]
- 7.Ikediobi ON, Davies H, Bignell G, et al. Mutation analysis of 24 known cancer genes in the NCI-60 cell line set. Mol Cancer Ther. 2006;5(11):2606–12. doi: 10.1158/1535-7163.MCT-06-0433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crowder RJ, Phommaly C, Tao Y, et al. PIK3CA and PIK3CB inhibition produce synthetic lethality when combined with estrogen deprivation in estrogen receptor-positive breast cancer. Cancer Res. 2009;69(9):3955–62. doi: 10.1158/0008-5472.CAN-08-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis MJ, Coop A, Singh B, et al. Letrozole is more effective neoadjuvant endocrine therapy than tamoxifen for ErbB-1- and/or ErbB-2-positive, estrogen receptor-positive primary breast cancer: evidence from a phase III randomized trial. J Clin Oncol. 2001;19(18):3808–16. doi: 10.1200/JCO.2001.19.18.3808. [DOI] [PubMed] [Google Scholar]
- 10.Baselga J, Semiglazov V, van Dam P, et al. Phase II Randomized Study of Neoadjuvant Everolimus Plus Letrozole Compared With Placebo Plus Letrozole in Patients With Estrogen Receptor-Positive Breast Cancer. J Clin Oncol. 2009 doi: 10.1200/JCO.2008.18.8391. [DOI] [PubMed] [Google Scholar]
- 11.Olson JA, Jr., Budd GT, Carey LA, et al. Improved surgical outcomes for breast cancer patients receiving neoadjuvant aromatase inhibitor therapy: results from a multicenter phase II trial. J Am Coll Surg. 2009;208(5):906–14. doi: 10.1016/j.jamcollsurg.2009.01.035. discussion 15–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellis MJ, Coop A, Singh B, et al. Letrozole inhibits tumor proliferation more effectively than tamoxifen independent of HER1/2 expression status. Cancer Res. 2003;63(19):6523–31. [PubMed] [Google Scholar]
- 13.Ellis MJ, Tao Y, Young O, et al. Estrogen-independent proliferation is present in estrogen-receptor HER2-positive primary breast cancer after neoadjuvant letrozole. J Clin Oncol. 2006;24(19):3019–25. doi: 10.1200/JCO.2005.04.3034. [DOI] [PubMed] [Google Scholar]
- 14.Serra V, Markman B, Scaltriti M, et al. NVP-BEZ235, a dual PI3K/mTOR inhibitor, prevents PI3K signaling and inhibits the growth of cancer cells with activating PI3K mutations. Cancer Res. 2008;68(19):8022–30. doi: 10.1158/0008-5472.CAN-08-1385. [DOI] [PubMed] [Google Scholar]
- 15.Ellis MJ, Tao Y, Luo J, et al. Outcome prediction for estrogen receptor-positive breast cancer based on postneoadjuvant endocrine therapy tumor characteristics. J Natl Cancer Inst. 2008;100(19):1380–8. doi: 10.1093/jnci/djn309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marone R, Cmiljanovic V, Giese B, Wymann MP. Targeting phosphoinositide 3-kinase: moving towards therapy. Biochim Biophys Acta. 2008;1784(1):159–85. doi: 10.1016/j.bbapap.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 17.Di Cosimo S, Baselga J. Phosphoinositide 3-kinase mutations in breast cancer: a “good” activating mutation? Clin Cancer Res. 2009;15(16):5017–9. doi: 10.1158/1078-0432.CCR-09-1173. [DOI] [PubMed] [Google Scholar]