Abstract
Prolyl 4-hydroxylase (P4H) catalyzes the post-translational hydroxylation of (2S)-proline (Pro) residues in procollagen strands. The resulting (2S,4R)-4-hydroxyproline (Hyp) residues are essential for the folding, secretion, and stability of the collagen triple helix. Even though its product (Hyp) differs from its substrate (Pro) by only a single oxygen atom, no product inhibition has been observed for P4H. Here, we examine the basis for the binding and turnover of substrates by human P4H. Synthetic peptides containing (2S,4R)-4-fluoroproline (Flp), (2S,4S)-4-fluoroproline (flp), (2S)-4-ketoproline (Kep), (2S)-4-thiaproline (Thp), and 3,5-methanoproline (Mtp) were evaluated as substrates for P4H. Peptides containing Pro, flp, and Thp were found to be excellent substrates for P4H, forming Hyp, Kep, and (2S,4R)-thiaoxoproline, respectively. Thus, P4H is tolerant to some substitutions on C-4 of the the pyrrolidine ring. In contrast, peptides containing Flp, Kep, or Mtp did not even bind to the active site of P4H. Each proline analog that does bind to P4H is also a substrate, indicating that discrimination occurs at the level of binding rather than turnover. As the iron(IV)-oxo species that forms in the active site of P4H is highly reactive, P4H has an imperative for forming a snug complex with its substrate, and appears to do so. Most notably, those proline analogs with a greater preference for a Cγ-endo pucker and cis peptide bond were the ones recognized by P4H. As Hyp has a strong preference for Cγ-exo pucker and trans peptide bond, P4H appears to discriminate against the conformation of proline residues in a manner that diminishes product inhibition during collagen biosynthesis.
Collagens are the major structural proteins in the extracellular matrix. All collagens are comprised of three polypeptide strands that coil together into a right-handed triple helix. Each strand contains a repeating three amino-acid sequence in which every third residue is a glycine (Gly1): Xaa–Yaa–Gly. The Xaa position is often (2S)-proline (Pro), and the Yaa position is often (2S,4R)-4-hydroxyproline (Hyp) (1). Hyp is produced by the action of prolyl 4-hydroxylase (P4H; EC 1.14.11.2). This post-translational modification is the most prevalent in the kingdom Animalia, where Hyp is more common than seven of the “common” amino-acid residues: Cys, Gln, His, Met, Phe, Trp, and Tyr.2 The presence of Hyp is crucial for the folding, secretion, and stability of the collagen triple helix under physiological conditions (4–7), and P4H activity is necessary for the viability of the nematode Caenorhabditis elegans (8–10) and the mouse Mus musculus (11). The enzyme-catalyzed hydroxylation of proline residues also plays a role in the sensing of molecular oxygen, though those enzymes are distinct from the P4H that acts on collagen (12).
P4H is a tetrameric enzyme comprised of two α subunits and two β subunits. Each α subunit (59 kDa) contains a catalytic domain and a peptide-binding domain (13). The β subunit (55 kDa) is protein disulfide isomerase (14), which is necessary to keep the α subunit from aggregating (15, 16) and to retain the enyzme in the endoplasmic reticulum (17). The study of P4H has been facilitated by the recent development of recombinant DNA systems for the high-level production of active P4H tetramers in Escherichia coli (18, 19). Still, the three-dimensional structure of P4H remains unknown.
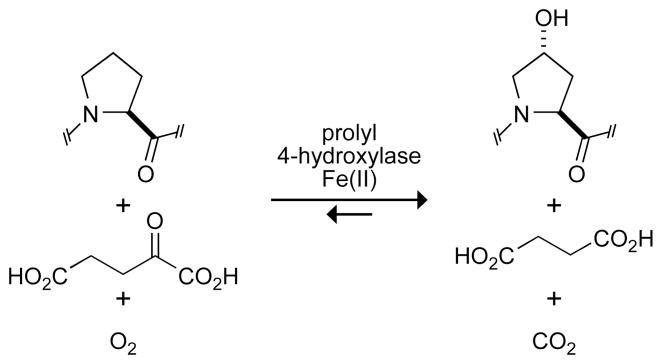
P4H is a non-heme iron(II) dioxygenase (20–22). This class of enzymes uses α-ketoglutarate and O2 as co-substrates (Figure 1). Hydroxylation of proline is accompanied by the oxidative decarboxylation of α-ketoglutarate to form succinate (23). During the reaction, one atom of molecular oxygen is incorporated into Hyp, and the other into succinate (24). Uncoupling of the decarboxylation of α-ketoglutarate from substrate hydroxylation results in inactivated P4H, which can be rescued by ascorbate (25–27). Details of the mechanism by which P4H catalyzes hydroxylation have been proposed based on related α-ketoglutarate-dependent dioxygenases (28–30). In the initial steps of catalysis, decarboxylation of α-ketoglutarate produces an iron(IV)-oxo species. This highly reactive species abstracts the proR hydrogen from C-4 of a proline residue in the substrate (31). A hydroxyl group is transferred to the radical intermediate to form the Hyp product.
Figure 1.
Reaction catalyzed by prolyl 4-hydroxylase.
P4H does not hydroxylate all proline residues. Free proline amino acids are not hydroxylated by P4H (32). Nor is polyproline hydroxylated, though it does bind to the enzyme and is a competitive inhibitor of enzymatic activity (33). Hydroxylation occurs only on proline residues in an Xaa–Pro–Gly sequence within a polypeptide. The strength of substrate binding increases with the length of the polypeptide but is obviated by triple-helical structure (34).
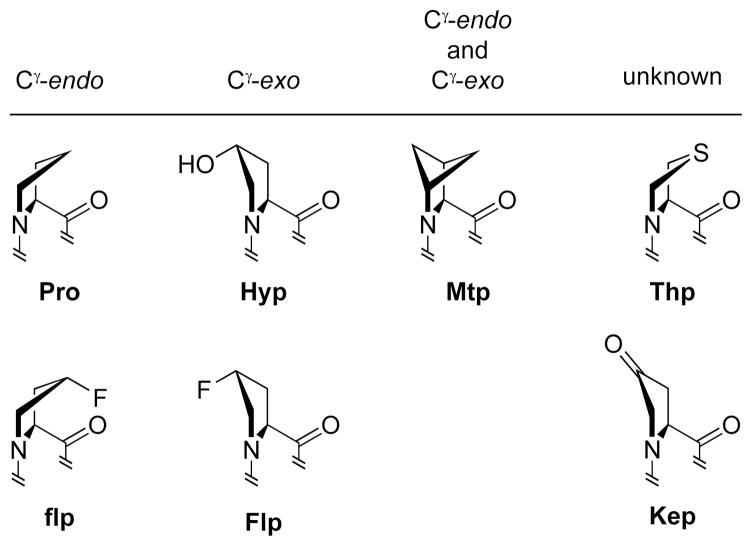
Computational, structural, and spectral analyses of Pro–Gly sequences in peptides and proteins have shown that adoption of a β-turn conformation is energetically favorable (5, 35–37). Other conformation preferences for the substrates of prolyl 4-hydroxylase are less clear. Because proline is a secondary amine and prolyl peptide bonds are tertiary amides, the cis isomer of prolyl peptide bonds occurs much more frequently than in peptide bonds between non-prolyl residues (38). The conformation of the prolyl peptide bond is correlated with another conformational feature of proline—its ring pucker (39, 40). The pyrrolidine ring of proline primarily adopts two puckered conformations: Cγ-endo and Cγ-exo (Figure 2).3 The trans/cis ratio and ring pucker are both influenced by substituents on the pyrrolidine ring that evoke stereoelectronic effects (42).
Figure 2.
Structures of prolines and its analogs studied herein, along with their predominant ring pucker.3 Each amino acid was incorporated into a tetrapeptide and examined as a substrate or inhibitor of P4H.
In this work, we create variations in trans/cis ratio and preferred ring pucker by using proline analogs with subtle substitutions at the 4-position of the pyrrolidine ring (Figure 2). We then demonstrate that the ability of P4H to bind to a proline analog and catalyze its oxidation correlates with the trans/cis ratio and preferred ring pucker. The results provide new insight into how this essential enzyme recognizes its substrate.
Experimental Procedures
Materials
Boc-FlpOH and Boc-flpOH were from OmegaChem (Lévis, Québec). Boc-protected amino acids were converted to their Fmoc derivatives via standard methods and used without further purification (43). Fluorenylmethoxycarbonyl (Fmoc)-mini-PEG-3™ (Fmoc-11-amino-3,6,9-trioxaundecanoic acid) was from Peptides International (Louisville, KY). Fmoc-ThpOH was from Advanced ChemTech (Louisville, KY). All other Fmoc-protected amino acids were from Novabiochem (La Jolla, CA). [1-14C]α-Ketoglutarate was from American Radiolabeled Chemicals (St. Louis, MO). All other chemicals were of reagent grade or better, and were used without further purification.
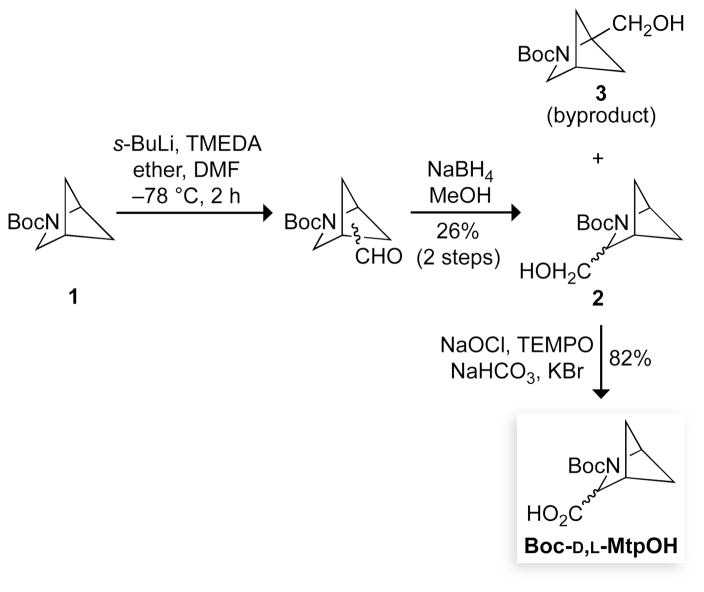
Synthesis of Boc-D,L-MtpOH
Boc-D,L-MtpOH was prepared by the route shown in Scheme 1, which is less costly and more convenient than those described previously (44, 45). Briefly, carbamate 1 was prepared as in ref. 46, and then used in a manner similar to that in ref. 47. Specifically, a solution of carbamate 1 (2.0 g, 10.91 mmol) and TMEDA (2.1 mL, 14.18 mmol, 1.3 equiv) in dry ether (50 mL) was cooled to −78 °C, and s-BuLi (10.1 mL, 14.18 mmol, 1.3 equiv, 1.4 M solution in cyclohexane) was added dropwise. The resulting solution was stirred for 2 h at −78 °C and then transferred via a canula to a solution of distilled DMF (6.3 mL, 81.83 mmol, 7.5 equiv) in dry diethyl ether (30 mL) that had been cooled to −78 °C. The solution was warmed slowly to room temperature, and then quenched with 10 mL saturated NH4Cl(aq). The ether layer was washed with distilled water (2 × 20 mL) and dried over Na2SO4(s). After filtration and concentration, the oil was taken up in 10 mL of dry MeOH, and this solution was cooled to 0 °C. NaBH4 (2.1 g, 54.56 mmol, 5.0 equiv) was added slowly to the solution, which was then stirred at 0 °C for 15 min. The reaction mixture was warmed to room temperature, and 10 mL of saturated NH4Cl(aq) was added slowly, followed by 30 mL of CH2Cl2. The aqueous layer was extracted with CH2Cl2 (3 × 30 mL). The organic extracts were combined, dried over Na2SO4(s), and filtered. The solvent was removed under reduced pressure, and the residue was purified by chromatography on silica gel (3:2 hexanes/diethyl ether) to furnish 3-CH2OH (2) (609 mg, 26%, Rf = 0.36 with 1:1 hexanes/diethyl ether), 1-CH2OH (3) (638 mg, 27%, Rf = 0.51), and starting material 1 (190 mg, 10%, Rf = 0.71).
Scheme 1.
Following the procedure in refs. 44 and 47, 3-CH2OH (2) (600 mg, 2.813 mmol) was dissolved in a solution of CH2Cl2 (14 mL), TEMPO (27 mg), saturated NaHCO3(aq) (11 mL), KBr (54 mg), and Bu4NCl (67 mg). This solution was cooled to 0 °C, and a solution of NaOCl (14 mmol), saturated NaHCO3(aq) (6 mL), and brine (12 mL) was added dropwise over 45 min. The reaction mixture was stirred for an additional 1 h, and then warmed to room temperature. The aqueous layer was separated, and the organic layer was washed with 50% w/v NaHCO3(aq) (3 × 65 mL). The aqueous layers were combined, and washed with CH2Cl2 (2 × 25 mL), acidified with dilute HCl solution until the pH was <3, and then extracted with ethyl acetate (5 × 200 mL). The combined organic extracts were dried over Na2SO4(s), and the solvent was removed to furnish Boc-D,L-MtpOH (524 mg, 82%) as off-white solid of sufficient purity (44, 45, 47).
Instrumentation
Measurements of UV and visible absorbance were made with a Cary model 3 spectrophotometer (Varian, Palo Alto, CA). Peptide synthesis was conducted with a Pioneer (PerCeptive Biosystems) or Symphony (Protein Technologies) automated synthesizer at the University of Wisconsin Biotechnology Center. Preparative high-performance liquid chromatography (HPLC) was performed with a system from Waters (Milford, MA) equipped with two 510 pumps and a 486 tunable absorbance detector. Analytical HPLC was performed with a Waters system equipped with two 515 pumps, a 717 plus autosampler, and a 996 photodiode array detector. Fast protein liquid chromatography (FPLC) was performed with an ÄKTA system from Amersham Pharmacia (Piscataway, NJ). Matrix-assisted laser desorption/ionization time-of-flight (MALDI–TOF) mass spectrometry was performed with a Perkin–Elmer (Wellesley, MA) Voyager MALDI–TOF mass spectrometer at the University of Wisconsin–Madison Biophysics Instrumentation Facility. Scintillation counting was performed with a Wallac 1450 MicroBeta TriLux liquid scintillation counter from Perkin–Elmer (Wellesley, MA).
Production and Purification of P4H
P4H was produced and purified by using procedures reported previously (18).
Synthesis of Peptides
Peptides were synthesized on a solid support (PEG-PS, Applied Biosystems) by standard Fmoc-protection methods using HATU activation. To increase the water-solubility of the Pro-, Flp-, flp-, Kep-, and Thp-containing peptides, a short PEG chain was added to their N-terminus. The PEG was added by coupling Fmoc-mini-PEG-3™ (Peptides International, Louisville, KY) to the N-terminus of the peptide while on solid support. The N-terminus remained protected with the Fmoc while the peptides were cleaved from the solid support with 8 mL of 95:2.5:2.5 trifluoroacetic acid (TFA)/triisopropylsilane/water, and then washed with CH2Cl2 and dried under vacuum. The dried peptide was heated at reflux with SOCl2 (15-fold molar excess) in ethanol for 2 h. The solvent was removed by rotary evaporation under reduced pressure. The N-terminal Fmoc was then removed by stirring the peptide in 20% v/v piperidine in DMF for 20 min. The peptides were purified by preparatory HPLC with a gradient (10–30% v/v) of acetonitrile in water containing TFA (0.1% v/v) to yield PEG-Gly–Tyr–Yaa–GlyOEt. MALDI MS m/z: [M + Na]+ Yaa = Pro (14% yield; calcd 632.3, found 632.6), Flp (6% yield; calcd 650.3; found 650.5), flp (72% yield; calcd 650.3; found 650.6), Kep (1.8% yield; calcd 646.3, found 646.5), Thp (8% yield; calcd 650.3; found 650.7).
Cbz-Gly–Tyr–Mtp–GlyOEt was synthesized by standard Fmoc-protection methods on solid support. The cleavage from solid support and esterification were performed as described for the PEGylated peptides. Purification was done by preparatory HPLC with a gradient (20–40% v/v) of acetonitrile in water. The synthesis was done with racemic Fmoc-D,L-Mtp, and the diastereomeric tetrapepetides were not separated. Cbz-Gly–Tyr–Mtp–GlyOEt was analyzed by MALDI–MS m/z: [M + Na]+ (calcd 589.2, found 589.5).
Peptide concentrations were determined by absorbance measurement in 6 M guanidine hydrochloride at pH 6.5 using ε = 1450 M−1cm−1 at 276 nm (48).
HPLC-based Assay of Enzymatic Activity
An HPLC-based assay described previously (18) was used to monitor product formation by P4H. Assays were performed for 5 min at 30 °C in 100 μL of 50 mM Tris–HCl buffer, pH 7.8, containing bovine serum albumin (1 mg/mL), catalase (100 μg/mL), dithiothreitol (100 μM), ascorbate (2 mM), FeSO4 (50 μM), P4H (90 nM), and α-ketoglutarate (500 μM). The tetrapeptide substrate (stock solution in ethanol) was added to initiate the reaction. The reactions were quenched by boiling for 60 s. All assays were performed in triplicate. A reversed-phase analytical Alltima HP C18 AQ column (4.6 × 250 mm) from Alltech (Deerfield, IL) was used to separate peptides by elution with aqueous acetonitrile (12–30 % v/v in 20 min) containing TFA (0.1% v/v) at 1.0 mL/min. Product formation was quantified by the substrate: product ratio, as determined by integration of the A214 nm with the Millennium32 software from Waters (Millford, MA).
[14C]CO2-Release Assay for Enzymatic Activity
An alternative means to assess P4H activity is to monitor the release of CO2, which is a product of catalysis (Figure 1). Procedures for monitoring the release of [14C]CO2 from [1-14C]α-ketoglutarate were as described elsewhere (23, 49). Concentrations were the same as above, except for that of P4H (395 nM). All reactions were performed in duplicate and corrected for the rate of decarboxylation in the absence of the peptide substrate.
Reduction and Conversion of Kep
For analysis, the ketone in PEG-Gly–Tyr–Kep–GlyOEt was either reduced to the alcohol with sodium borohydride (10 equiv) for 30 min at room temperature or converted to the oxime via reaction with hydroxylamine (10 equiv) in 250 mM sodium phosphate buffer, pH 5.0, for 1 h at 100 °C.
Chemical Oxidation of Thp
A mixture of PEG-Gly–Tyr–Thp(O)–GlyOEt and PEG-Gly–Tyr–thp(O)–GlyOEt was produced by chemical oxidation of the Thp-containing peptide with either 3-chloroperoxybenzoic acid (MCPBA) (1 equiv) in chloroform for 2.5 h at room temperature or sodium periodate (1.1 equiv) in aqueous methanol (75%) for 30 min at 50 °C (50). A peptide containing Thp(O,O) was produced by reaction of the Thp-containing peptide with MCPBA (10 equiv) in chloroform for 5 h at room temperature.
Results
Design of Peptide Substrates for P4H
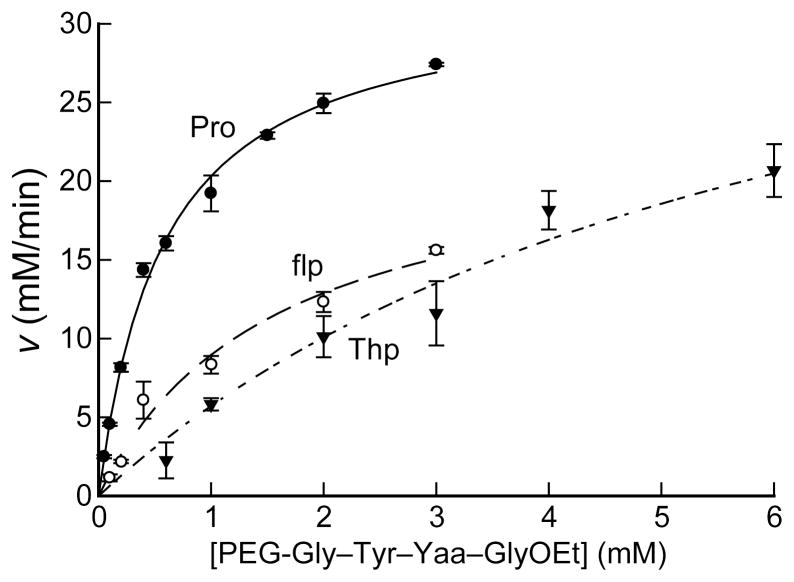
Previously, we reported on an HPLC-based assay for P4H activity using the peptide substrate dansyl-Gly–Phe–Pro–GlyOEt (18). This peptide is not especially soluble, and is thus not useful for assays at high concentration. To increase its solubility, the dansyl moiety was replaced with a short PEG segment. In addition, Phe was replaced by Tyr to aid in the determination of peptide concentration. PEG-Gly–Tyr–Pro–GlyOEt is a substrate for P4H with kcat = 360 min−1, KM = 0.58 mM, and kcat/KM = 1.0 × 104 M−1s−1 (Table 1, kFigure 3). This cat/KM value is 3-fold greater than that for dansyl-Gly–Phe–Pro–GlyOEt (18).
Table 1.
Comparison of PEG-Gly–Tyr–Yaa–GlyOEt as substrates or inhibitors for P4Ha
| Yaa | Substrate or Inhibitor | kcat (min−1) | KM (mM) | kcat/KM (103 M−1s−1) |
|---|---|---|---|---|
| Pro | Substrate | 360 ± 7 | 0.58 ± 0.03 | 10 ± 0.6 |
| flp | Substrate | 250 ± 21 | 1.6 ± 0.3 | 2.7 ± 0.5 |
| Thp | Substrate | 470 ± 59 | 6.4 ± 1.4 | 1.2 ± 0.3 |
| Flp | Neither | – | – | – |
| Hyp | Neitherb | – | – | – |
| Kep | Neither | – | – | – |
| Mtp | Neither | – | – | – |
Reaction components and conditions were as described in the text. Values represent the mean (± SE) of three replicates.
Data from ref. 37.
Figure 3.
Comparison of PEG-Gly–Tyr–Yaa–GlyOEt peptides, where Yaa = Pro (●), flp (○), or Thp (▼;), as substrates of P4H. Plot shows the rate of product formation at varying substrate concentrations. Reactions were performed as described in the Experimental Methods section. Reactions were initiated by the addition of tetrapeptide substrate (0.05–3 mM), and run at 30 °C for 5 min. Product formation was analyzed by HPLC. Individual points are the average (± SE) of three independent reactions. Data were fitted to the Michaelis–Menten equation to obtain kinetic parameters.
Flp does not Inhibit P4H
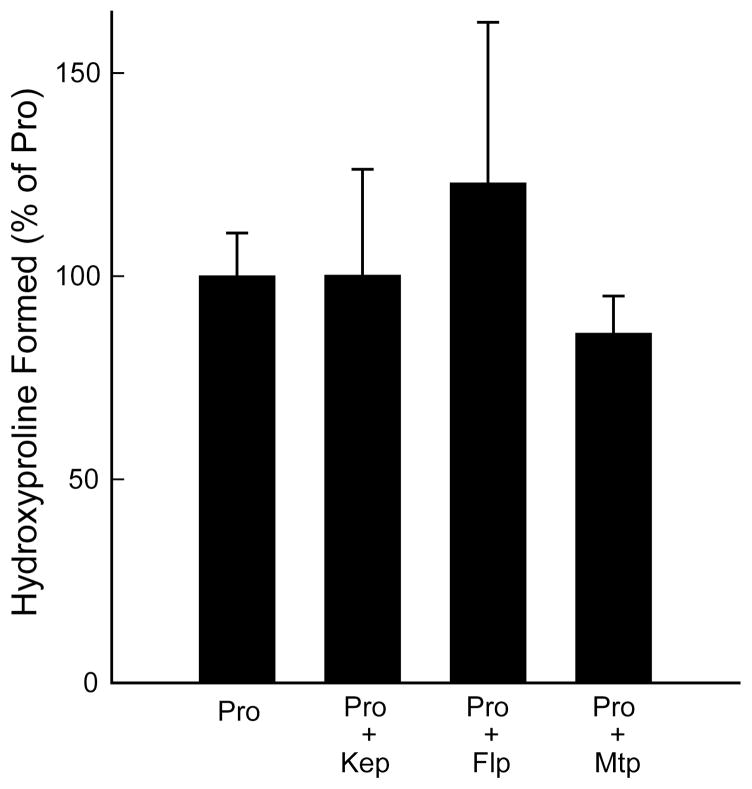
No product formation was detected when Flp was used as a substrate for P4H (data not shown). It is not surprising that P4H cannot turn over Flp because P4H abstracts the proR hydrogen from C-4 of a proline residue, and that hydrogen is replaced with fluorine in Flp. Likewise, Flp does not inhibit the P4H-catalyzed conversion of Pro to Hyp. Under standard P4H reaction conditions as described in the experimental procedures with PEG-Gly–Tyr–Pro–GlyOEt at 0.06 mM, which is an order of magnitude below its KM value, PEG-Gly–Tyr–Flp–GlyOEt had no measurable effect on hydroxylation of the Pro-containing peptide (Figure 4).
Figure 4.
Peptides containing Flp, Kep, or Mtp do not inhibit Hyp formation from Pro by P4H. Graph shows the hydroxyproline formed from proline in the presence of no proline analog, Kep, Flp, or Mtp. The reaction with no proline analog was designated as 100%. Reactions contained 0.6 mM PEG-Gly–Tyr–Yaa–GlyOEt, where Yaa = Kep, Flp or Mtp, and 0.06 mM PEG-Gly–Tyr–Pro–GlyOEt. Individual points are the average (± SE) of three independent reactions.
flp is a Substrate for P4H
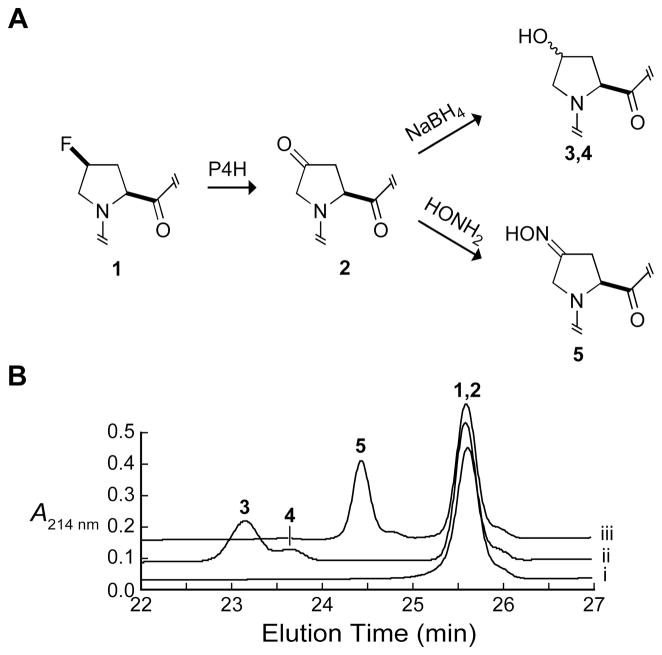
Unlike Flp, flp has a hydrogen in the 4R position of the pyrrolidine ring, and PEG-Gly–Tyr–flp–GlyOEt was determined to be a substrate for P4H (Figures 3 and 5). The product was identified as 4-ketoproline (Kep) by MALDI–MS (m/z 624.4, calc 624.3), and coeluted with a standard during HPLC (data not shown). Kep was characterized further by reduction of its ketone to a hydroxyl group with sodium borohydride (MALDI–MS m/z 626.6, calc 626.3) and reaction of its ketone with hydroxylamine to form an oxime (MALDI–MS m/z 639.5, calc 639.3) (Figure 5). Kep can form by hydroxylation of flp to produce a fluorohydrin, followed by the elimination of a fluoride ion. From triplicate P4H reactions under standard conditions with varying concentrations of PEG-Gly–Tyr–flp–GlyOEt, the value of kcat/KM was determined to be (2.7 ± 0.5) × 103 M−1s−1. This value is 27% that of the Pro-containing peptide, which has a kcat/KM value of (10 ± 0.6) × 103 M−1s−1 (Table 1, Figure 3).
Figure 5.
Characterization of the products for the turnover of PEG-Gly–Tyr–flp–GlyOEt by P4H. (A) Kep (2) is formed from flp (1) by P4H. Kep is reduced with sodium borohydride (10 equiv) for 30 min to form Hyp and hyp (3, 4), or Kep was converted to an oxime (5) by reaction with hydroxylamine (10 equiv) in 250 mM sodium phosphate buffer, pH 5.0, for 1 h at 100 °C. (B) HPLC analysis of PEG-Gly–Tyr–flp–GlyOEt reactions. (i) Turnover by P4H; (ii) turnover by P4H and treatment with sodium borohydride; (iii) turnover by P4H and treatment with hydroxylamine. Peak 1,2 is from the coeluting peptides containing flp or Kep; peaks 3 and 4 are from the peptides containing Hyp or hyp; peak 5 is from the peptide of containing the oxime of Kep. HPLC conditions are described in the Experimental Procedures section.
Kep does not Inhibit P4H
Kep, the product of the P4H-catalyzed reaction of flp, was not found to be a substrate for P4H (data not shown). Like Flp, Kep did not even inhibit the P4H-catalyzed conversion of Pro to Hyp (Figure 4).
Thp is a Substrate for P4H
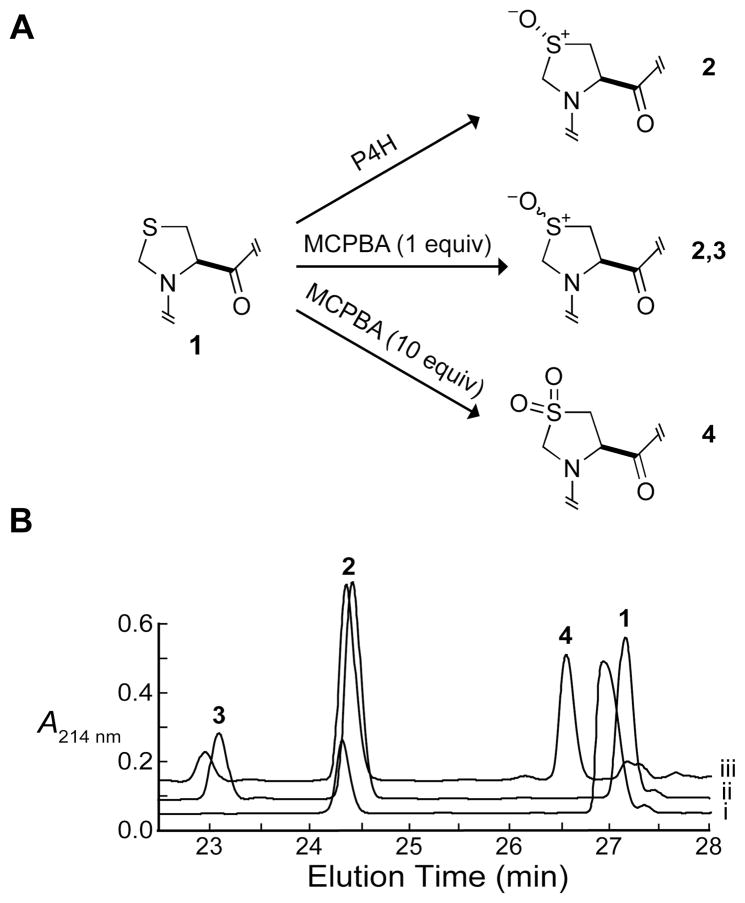
Thp has a sulfur atom bearing no hydrogen atoms at the 4-position of the thiazolidine ring. Nonetheless, PEG-Gly–Tyr–Thp–GlyOEt was found to be a substrate for P4H (Figure 6). P4H added a single oxygen atom to Thp to form a sulfoxide (Thp(O)) in the peptide product (MALDI–MS m/z 666.6, calc 666.3 [M + Na]+). Thp(O) was also formed, along with its thp(O), in the Thp-containing peptide by chemical oxidation using MCPBA or sodium periodate. Oxidation by sodium periodate yielded two products with the same mass according to HPLC and MALDI–MS. Kanai and coworkers reported that this nonenzymatic reaction with a thioprolylpyrrolidine derivative substrate produces an 85:15 mixture of the 4R and 4S diastereomers (50). HPLC analysis of the sodium periodate reaction compared to the P4H-catalyzed formation of the sulfoxide showed that P4H produces the 4R diastereomer exclusively. Chemical oxidation by MCPBA yielded the sulfoxide diastereomers, as well as the sulfone (Thp(O,O); MALDI–MS m/z 682.4, calc 682.3 [M + Na]+), which was not formed by P4H. From triplicate P4H reactions under standard conditions with varying concentrations of PEG-Gly–Tyr–Thp–GlyOEt, the kcat/KM value was determined to be (1.2 ± 0.3) × 103 M−1s−1, which is 12% that of the Pro-containing peptide (Table 1, Figure 3).
Figure 6.
Characterization of the products for the turnover of PEG-Gly–Tyr–Thp–GlyOEt by P4H. (A) Thp(O) (2) is formed from Thp (1) upon catalysis by P4H. Thp(O) and thp(O) (2,3) are formed from Thp upon reaction with MCPBA (1 equiv) in chloroform for 2.5 h. Thp(O,O) (4) is formed from Thp upon reaction with MCPBA (10 equiv) in chloroform for 5 h. (B) HPLC analysis of reactions of PEG-Gly–Tyr–Thp–GlyOEt. (i) Turnover by P4H; (ii) treatment with MCPBA (1 equiv); (iii) treatment with MCPBA (10 equiv). Peak 1 is from the peptide containing Thp peptide; peaks 2 and 3 are for the peptides containing Thp(O) or thp(O); peak 4 is for the peptide containing Thp(O,O). HPLC conditions are described in the Experimental Procedures section.
P4H Decarboxylates [1-14C]α-Ketoglutarate when Pro, flp or Thp is a Substrate
The HPLC-based assay described above identified flp- and Thp-containing peptides as novel substrates of P4H. These proline analogs, along with the Pro-containing peptide, were also tested in an assay for P4H activity that measures the release of [14C]CO2 from [1-14C]α-ketoglutarate. The rate of decarboxylation in the absence of the peptide substrate was subtracted from each measurement. Substrate concentrations for assays with the Pro-, flp-, and Thp-containing peptides were 58, 160, and 640 μM, which were 10% of the KM values determined in the HPLC-based assay. Under these conditions the rate of CO2 released in the presence of the Pro-containing peptide was 1.9 ± 0.7 μM/min (Table 2). The rate with flp in the peptide substrate was 0.5 ± 0.1 μM/min, which is 26% that of Pro; the rate with Thp was 0.1 ± 0.1 μM/min, 6% that of Pro. These rates follow closely the trend observed with the HPLC-based assay.
Table 2.
Comparison of [14C]CO2 release from [1-14C] α-ketoglutarate in P4H assays with PEG-Gly–Tyr–Yaa–GlyOEt as the substratea
| Yaa | v (μM/min) | % of Pro |
|---|---|---|
| Pro | 1.9 ± 0.7 | 100 |
| flp | 0.5 ± 0.1 | 26 |
| Thp | 0.1 ± 0.1 | 6 |
Reactions were run in duplicate. The rate of decarboxylation in the absence of the peptide substrate was subtracted from each measurement. Components and conditions were as described in the text. Substrate concentrations were 58, 160, and 640μM, for Pro, flp, and Thp, respectively, which are 10% of each KM value determined in the HPLC-based assay.
Mtp does not Inhibit P4H
Mtp, the bicyclic proline analog, was not found to be a substrate for P4H. No product was detected by either HPLC or mass spectrometry (data not shown). Like Flp and Kep, Mtp did not inhibit the P4H-catalyzed conversion of Pro to Hyp (Figure 4).
Discussion
P4H catalyzes an extremely difficult chemical reaction—the hydroxylation of an unactivated methylene group to form a secondary alcohol. The putative mechanism involves the oxidative decarboxylation of α-ketoglutarate, which promotes the formation of a highly reactive iron(IV)-oxo species from molecular oxygen (28–30). This iron(IV)-oxo species then abstracts the proR hydrogen from C-4 of a proline residue, replacing it with a hydroxyl group to form Hyp (31).
In previous work, proline analogs with substituents at C-3 and C-5 have been used to probe catalysis by P4H. Peptides containing a racemic mixture of 3-fluoroprolines in the Yaa position were found to be substrates for the enzyme (51). Peptides containing the proline analogs 3-exomethyleneproline (51), 5-oxaproline (52), and 3,4-dehydroproline (53–55) inhibited P4H activity. In our work, we have focused instead on C-4, which is the carbon that undergoes a change in covalency during the reaction catalyzed by P4H.4
Requirements of Nonnatural Proline Analogs for Turnover by P4H
P4H can elicit the homolytic cleavage of a C–H bond but not a C–F bond. The change from Pro to Flp is conservative from the perspective of sterics, as hydrogen and fluorine have comparable van der Waals radii (rH = 1.20 Å; rF = 1.35 Å (56)). A C–F bond (ΔH° = 116 kcal/mol) is, however, much stronger than a C–H bond (ΔH° = 98 kcal/mol). Accordingly, we were not surprised to learn that P4H cannot turn over Flp. This finding contrasts with reports by others in the 1960’s that relied on less direct assays (57, 58). We did, however, expect P4H to bind to Flp as it does to Pro. Yet, no inhibition of P4H activity by Flp was detectable (Table 1).
Unlike Flp, flp is a substrate for P4H. The stability of a carbon radical with an α-fluoro substituent is ΔH = 0.7 kcal/mol greater than that of an unsubstituted carbon radical, according to homolytic bond dissociation enthalpies of 2-fluoropropane and propane calculated at 25 °C (59). Hence, the hydroxylation of flp should be somewhat faster than that of Pro. Of course, other factors contribute to the rate of an enzymatic reaction, and the actual values of kcat are 360 and 250 min−1 for Pro and flp, respectively (Table 1).
The product of the turnover of flp by P4H is Kep (Figure 5). This product has notable utility for future studies of P4H in biological systems. The turnover of flp introduces a functional group—a ketone—with orthogonal reactivity into a protein. This group could serve as a handle for a proteomic analyses of P4H substrates (60), as flp can be incorporated into proteins by biosynthesis (61–65). Furthermore, the turnover of flp not only produces Kep, but also releases fluoride ion, whose detection could provide the basis for a direct, continuous assay of P4H activity.
The sulfide in the Thp-containing peptide is oxidized to a sulfoxide by P4H (Table 1; Figure 6). Sulfides can be oxidized by other dioxygenases, such as thymine hydroxylase (66), 4-hydroxyphenylpyruvate dioxygenase (67), and cysteine dioxygenase (68). Although the chemical oxidation of Thp produces both sulfoxide diastereomers (50), turnover by P4H produces only the 4R sulfoxide, which has the same relative stereochemistry as the natural product, Hyp. α-Ketoglutarate is decarboxylated when Thp, flp, or Pro is the substrate in a reaction with P4H (Table 2). The relative rates of the decarboxylation reactions (Thp < flp < Pro) is similar to that for Hyp production (Table 1). These data are consistent with Thp, flp, and Pro being turned over by the same mechanism.
Role of Proline Conformation in Substrate Binding by P4H
Proline is the only proteinogenic amino acid whose trans peptide bond isomer is favored only slightly over the cis isomer (38). In other peptide bonds, the trans conformation is favored greatly. Proline is also the only proteinogenic amino acid to contain a saturated ring. The five-membered pyrrolidine ring of proline primarily adopts two conformations, Cγ-endo or Cγ-exo (Figure 2).3 The pucker of its pyrrolidine ring and the isomerization of its peptide bond are correlated attributes of proline (39, 40). Electronegative atoms such as fluorine (χF = 4.0 (56); cf.: χH = 2.1) alter the ring pucker via the gauche effect. A Cγ-exo pucker allows for an n→π* interaction between the oxygen of the Xaa–Pro peptide bond and the carbon of the Pro–Gly peptide bond. In turn, this n→π* interaction stabilizes a trans peptide bond. The absence of a significant n→π* interaction in the Cγ-endo pucker favors a cis peptide bond (39, 40).
The proline analogs used in this study differ in their predominant ring pucker (Figure 2). Hyp and Flp adopt a Cγ-exo pucker and have a high trans/cis ratio, whereas flp and Pro adopt a Cγ-endo pucker and have a low trans/cis ratio (40, 69). The ring puckers of Kep and Thp have not been examined directly. Nonetheless, the trans/cis ratio of Kep is higher than that of Pro (70), and the trans/cis ratio of Thp is lower than that of Pro (71). We find that Pro, flp and Thp are substrates for P4H, and that Hyp, Flp, and Kep are neither substrates nor inhibitors of the enzyme. We therefore propose that proline analogs that favor the Cγ-endo pucker and have a low trans/cis ratio bind to the active site of P4H, and those that favor a Cγ-exo or other pucker and have a high trans/cis ratio do not.
To test our proposal, we determined the ability of P4H to recognize Mtp (Figure 2). In essence, this analog has two C-4 carbons and thus always displays both ring puckers (72). Mtp has a trans/cis ratio that is between that of Pro and flp (44). We find that P4H does not hydroxylate either C-4 carbon of Mtp, and that Mtp does not inhibit P4H activity. These results refine our proposal by suggesting a proline residue with Cγ-exo pucker does not bind to the active site of the enzyme.
Our data suggest a means by which P4H diminishes product inhibition, which is often detrimental to enzyme function (73, 74). The stereoelectronic consequences of catalysis by P4H convert the Pro substrate into a Hyp product, which has a greater preference for a Cγ-exo ring pucker and trans peptide bond. These changes discourage P4H from binding previously hydroxylated collagen strands. A similar mechanism to avert product inhibition is employed by oligosaccharyl transferase, which catalyzes the formation of a cis amide bond during the N-glycosylation of asparagine residues. Isomerization to the trans conformations takes place after product release and prevents the product from binding again to the enzyme (75).
A summarial model for substrate recognition by P4H is shown in Figure 7. This model is based on the data herein, as well as known structures of Pro and Hyp. Previously, we used X-ray diffraction analysis to determine the three-dimensional structures of crystalline AcProOMe and AcHypOMe (76). In these structures, AcProOMe has a cis peptide bond and Cγ-endo ring pucker, and AcHypOMe has a trans peptide bond and Cγ-exo ring pucker. In the model, the iron(IV)-oxo species is proximal to the proR hydrogen on C-4 of Pro, which is the single atom on the pyrrolidine ring that is farthest from the main chain of the peptide substrate. Thus, the hydrogen atom to be abstracted reaches most deeply into the enzymic active site. Hydroxylation then promotes a conformational change in the product that the P4H active site is unable to accommodate.
Figure 7.
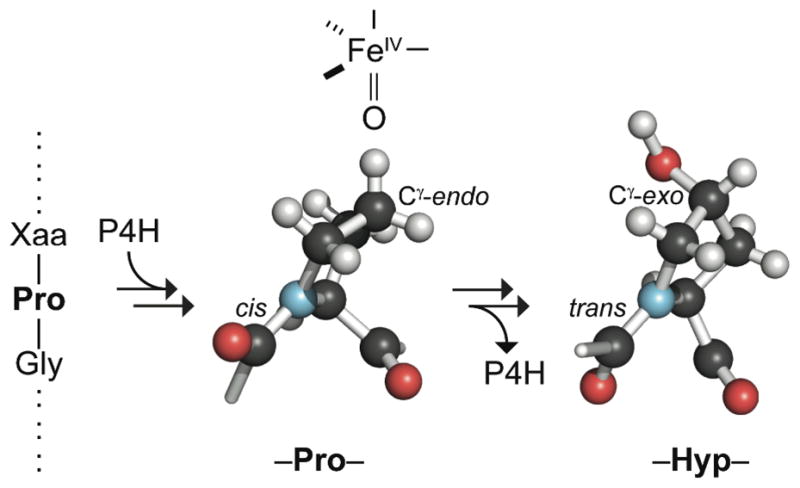
Putative model for substrate recognition by P4H. The Pro residue of the substrate has a cis peptide bond and Cγ-endo ring pucker; the iron(IV)-oxo speices is in a position to remove the proR hydrogen on C-4 (31). Hydroxylation changes the preferred conformation, and the Hyp residue of the product has a trans peptide bond and Cγ-exo ring pucker. The structures of Pro and Hyp are actual fragments of the crystal structures of AcProOMe and AcHypOMe (76), and are aligned such that the nitrogen and its three pendant carbons are in spatial alignment.
Conclusions
We have discovered two novel substrates for P4H: flp and Thp. These analogs demonstrate for the first time that perturbations can be made at the 4-position of a proline substrate with retention of high P4H activity. Other proline analogs investigated, Flp, Kep and Mtp, are not recognized by P4H. A comparison of the conformational preferences of these five proline analogs, along with Pro and Hyp, shows that proline analogs that prefer conformations similar to Pro—the substrate—are accepted by P4H, whereas analogs that are similar to Hyp—the product—are not. Therefore, the P4H-catalyzed formation of Hyp promotes conformational changes that limit product inhibition. Moreover, each analog that binds to the active site of P4H is also a substrate, indicating that the enzyme discriminates at the level of substrate binding rather than substrate turnover. The highly reactive iron(IV)-oxo species that forms in the active site of P4H is perhaps the most powerful oxidizing agent in biology (30), and must therefore be sequestered deeply within its enzymic active site. Hence, P4H has an imperative for forming a snug complex with its substrate, and appears to do so.
Acknowledgments
We are grateful to M. D. Shoulders and L. D. Lavis for contributive discussions, and G. L. Case of the Peptide Synthesis Facility at the University of Wisconsin Biotechnology Center for technical assistance.
Footnotes
Abbreviations: Boc, tert-butyloxycarbonyl; DMF, dimethylformamide; ESI–MS, electrospray ionization mass spectrometry; Flp, (2S,4R)-4-fluoroproline; flp, (2S,4S)-4-fluoroproline; Fmoc, fluorenylmethoxycarbonyl; FPLC, fast protein liquid chromatography; Gly, glycine; HPLC, high-performance liquid chromatography; Hyp, (2S,4R)-4-hydroxyproline; hyp, (2S,4S)-4-hydroxyproline; Kep, (2S)-4-ketoproline; MALDI–TOF, matrix-assisted laser desorption/ionization time-of-flight; MCPBA, 3-chloroperoxybenzoic acid; Mtp, 3,5-methanoproline or 2-azabicyclo[2.1.1]hexane-3-carboxylic acid; PEG, poly(ethylene glycol); P4H, prolyl 4-hydroxylase; Pro, (2S)-proline; TEMPO, 2,2,6,6-tetramethylpiperidine-1-oxyl; TFA, trifluoroacetic acid; Thp, (2S)-4-thiaproline; Thp(O), (2S,4R)-4-thiaoxoproline; thp(O), (2S,4S)-4-thiaoxoproline; Thp(O,O), (2S)-4-thiadioxoproline.
The abundance of Hyp in animal proteins is <4%, a value calculated from the abundance of collagen amongst animal proteins (⅓) and the prevalence of Hyp within the Xaa–Yaa–Gly sequence of collagen (<38% × ⅓) (2). The abundance of the “common” amino acids is given in ref. 3.
The pyrrolidine ring of proline actually prefers two distinct twist, rather than envelope, conformations (41). As Cγ experiences a large out-of-plane displacement in these twisted rings, we refer to pyrrolidine ring conformations simply as “Cγ-exo” and “Cγ-endo”.
The diastereomers of (2S)-4-methylproline have been reported to be neither substrates nor inhibitors of human P4H (51).
This work was supported by Grants AR044276 (NIH to R.T.R) and CHE 0515635 (NSF to G.R.K.). K.L.G. was supported by Chemistry–Biology Interface Training Grant T32 BM008505 (NIH). MALDI–MS experiments were performed at the University of Wisconsin–Madison Biophysics Instrumentation Facility, which was established with Grants BIR-9512577 (NSF) and RR13790 (NIH).
References
- 1.Prockop DJ, Kivirikko KI. Collagens: Molecular biology, diseases, and potentials for therapy. Annu Rev Biochem. 1995;64:403–434. doi: 10.1146/annurev.bi.64.070195.002155. [DOI] [PubMed] [Google Scholar]
- 2.Ramshaw JAM, Shah NK, Brodsky B. Gly–X–Y tripeptide frequencies in collagen: A context for host–guest triple-helical peptides. J Struct Biol. 1998;122:86–91. doi: 10.1006/jsbi.1998.3977. [DOI] [PubMed] [Google Scholar]
- 3.McCaldon P, Argos P. Oligopeptide biases in protein sequences and their use in predicting protein coding regions in nucleotide sequences. Proteins: Struct Funct Genet. 1988;4:99–122. doi: 10.1002/prot.340040204. [DOI] [PubMed] [Google Scholar]
- 4.Berg RA, Prockop DJ. The thermal transition of a non-hydroxylated form of collagen. Evidence for a role for hydroxyproline in stabilizing the triple-helix of collagen. Biochem Biophys Res Commun. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 5.Chopra RK, Ananthanarayanan VS. Conformational implications of enzymatic proline hydroxylation in collagen. Proc Natl Acad Sci USA. 1982;79:7180–7184. doi: 10.1073/pnas.79.23.7180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bulleid NJ, Wilson R, Lees JF. Type-III procollagen assembly in semi-intact cells: Chain association, nucleation and triple-helix folding do not require formation of inter-chain disulphide bonds but triple-helix nucleation does require hydroxylation. Biochem J. 1996;317:195–202. doi: 10.1042/bj3170195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins CL, Raines RT. Insights on the conformational stability of collagen. Nat Prod Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 8.Winter AD, Page AP. Prolyl 4-hydroxylase is an essential procollagen-modifying enzyme required for exoskeleton formation and the maintenance of body shape in the nematode Caenorhabditis elegans. Mol Cell Biol. 2000;20:4084–4093. doi: 10.1128/mcb.20.11.4084-4093.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Friedman L, Higgin JJ, Moulder G, Barstead R, Raines RT, Kimble J. Prolyl 4-hydroxylase is required for viability and morphogenesis in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:4736–4741. doi: 10.1073/pnas.97.9.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Myllyharju J, Kukkola L, Winter AD, Page AP. The exoskeleton collagens in Caenorhabditis elegans are modified by prolyl 4-hydroxylases with unique combinations of subunits. J Biol Chem. 2002;277:29187–29196. doi: 10.1074/jbc.M203824200. [DOI] [PubMed] [Google Scholar]
- 11.Holster T, Pakkanen O, Soininen R, Sormunen R, Nokelainen M, Kivirikko KI, Myllyharju J. Loss of assembly of the main basement membrane collagen, Type IV, but not fibril-forming collagens and embryonic death in collagen prolyl 4-hydroxylase I null mice. J Biol Chem. 2007;282:2512–2519. doi: 10.1074/jbc.M606608200. [DOI] [PubMed] [Google Scholar]
- 12.Kaelin WG. Proline hydroxylation and gene expression. Annu Rev Biochem. 2005;74:115–128. doi: 10.1146/annurev.biochem.74.082803.133142. [DOI] [PubMed] [Google Scholar]
- 13.Lamberg A, Pihlajaniemi T, Kivirikko KI. Site-directed mutagenesis of the alpha subunit of human prolyl 4-hydroxylase. Identification of three histidine residues critical for catalytic activity. J Biol Chem. 1995;270:9926–9931. doi: 10.1074/jbc.270.17.9926. [DOI] [PubMed] [Google Scholar]
- 14.Koivu J, Myllyla R, Helaakoski T, Pihlajaniemi T, Tasanen K, Kivirikko KI. A single polypeptide acts both as the β subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J Biol Chem. 1987;262:6447–6449. [PubMed] [Google Scholar]
- 15.Vuori K, Pihlajaniemi T, Myllyla R, Kivirikko KI. Site-directed mutagenesis of human protein disulphide isomerase: Effect on the assembly, activity and endoplasmic reticulum retention of human prolyl 4-hydroxylase in Spodoptera frugiperda insect cells. EMBO J. 1992;11:4213–4217. doi: 10.1002/j.1460-2075.1992.tb05515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vuori K, Pihlajaniemi T, Marttila M, Kivirikko KI. Characterization of the human prolyl 4-hydroxylase tetramer and its multifunctional protein disulfide-isomerase subunit synthesized in a baculovirus expression system. Proc Natl Acad Sci USA. 1992;89:7467–7470. doi: 10.1073/pnas.89.16.7467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kivirikko KI, Myllyla R, Pihlajaniemi T. Protein hydroxylation: Prolyl 4-hydroxylase, an enzyme with four cosubstrates and a multifunctional subunit. FASEB J. 1989;3:1609–1617. [PubMed] [Google Scholar]
- 18.Kersteen EA, Higgin JJ, Raines RT. Production of human prolyl 4-hydroxylase in Escherichia coli. Protein Expr Purif. 2004;38:279–291. doi: 10.1016/j.pep.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 19.Neubauer A, Neubauer P, Myllyharju J. High-level production of human collagen prolyl 4-hydroxylase in Escherichia coli. Matrix Biol. 2005;24:59–68. doi: 10.1016/j.matbio.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Guzman NA. Prolyl 4-Hydroxylase, Protein Disulfide Isomerase, and Other Structurally Related Proteins. Marcel Dekker; New York: 1998. [Google Scholar]
- 21.Fox BG. Catalysis by non-heme iron. In: Sinnott M, editor. Comprehensive Biological Catalysis: A Mechanistic Reference. Academic Press; San Diego: 1998. pp. 261–348. [Google Scholar]
- 22.Kivirikko KI, Pihlajaniemi T. Collagen hydroxylases and the protein disulfide isomerase subunit of prolyl 4-hydroxylases. Adv Enzymol Relat Areas Mol Biol. 1998;72:325–398. doi: 10.1002/9780470123188.ch9. [DOI] [PubMed] [Google Scholar]
- 23.Rhoads RE, Udenfriend S. Decarboxylation of α-ketoglutarate coupled to collagen proline hydroxylase. Proc Natl Acad Sci USA. 1968;60:1473–1478. doi: 10.1073/pnas.60.4.1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cardinale GJ, Rhoads RE, Udenfriend S. Simultaneous incorporation of 18O into succinate and hydroxyproline catalyzed by collagen proline hydroxylase. Biochem Biophys Res Commun. 1971;43:537–543. doi: 10.1016/0006-291x(71)90647-4. [DOI] [PubMed] [Google Scholar]
- 25.Myllyla R, Kuutti-Savolainen ER, Kivirikko KI. The role of ascorbate in the prolyl hydroxylase reaction. Biochem Biophys Res Commun. 1978;83:441–448. doi: 10.1016/0006-291x(78)91010-0. [DOI] [PubMed] [Google Scholar]
- 26.de Jong L, Albracht SP, Kemp A. Prolyl 4-hydroxylase activity in relation to the oxidation state of enzyme-bound iron. The role of ascorbate in peptidyl proline hydroxylation. Biochim Biophys Acta. 1982;704:326–332. doi: 10.1016/0167-4838(82)90162-5. [DOI] [PubMed] [Google Scholar]
- 27.Myllyla R, Majamaa K, Gunzler V, Hanauske-Abel HM, Kivirikko KI. Ascorbate is consumed stoichiometrically in the uncoupled reactions catalyzed by prolyl 4-hydroxylase and lysyl hydroxylase. J Biol Chem. 1984;259:5403–5405. [PubMed] [Google Scholar]
- 28.Costas M, Mehn MP, Jensen MP, Que L., Jr Dioxygen activation at mononuclear nonheme iron active sites: Enzymes, models, and intermediates. Chem Rev. 2004;104:939–986. doi: 10.1021/cr020628n. [DOI] [PubMed] [Google Scholar]
- 29.Hoffart LM, Barr EW, Guyer RB, Bollinger JM, Jr, Krebs C. Direct spectroscopic detection of a C-H-cleaving high-spin Fe(IV) complex in a prolyl-4-hydroxylase. Proc Natl Acad Sci USA. 2006;103:14738–14743. doi: 10.1073/pnas.0604005103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krebs C, Galonić Fujimori D, Walsh CT, Bollinger JM., Jr Non-heme Fe(IV)-oxo intermediates. Acc Chem Res. 2007;40:484–492. doi: 10.1021/ar700066p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita Y, Gottlieb A, Peterkofsky B, Udenfriend S, Witkop B. The preparation of cis- and trans-4-H3-L-prolines and their use in studying the mechanism of enzymatic hydroxylation in chick embryos. J Am Chem Soc. 1964;86:4709–4716. [Google Scholar]
- 32.Cardinale GJ, Udenfriend S. Prolyl hydroxylase. Adv Enzymol Relat Areas Mol Biol. 1974;41:245–300. doi: 10.1002/9780470122860.ch6. [DOI] [PubMed] [Google Scholar]
- 33.Prockop DJ, Kivirikko KI. Effect of polymer size on inhibition of protocollagen proline hydroxylase by polyproline II. J Biol Chem. 1969;244:4838–4842. [PubMed] [Google Scholar]
- 34.Kivirikko KI, Kishida Y, Sakakibara S, Prockop DJ. Hydroxylation of (X-Pro-Gly)n by protocollagen proline hydroxylase. Effect of chain length, helical conformation and amino acid sequence in the substrate. Biochim Biophys Acta. 1972;271:347–356. doi: 10.1016/0005-2795(72)90209-7. [DOI] [PubMed] [Google Scholar]
- 35.Rapaka RS, Renugopalakrishnan V, Urry DW, Bhatnagar RS. Hydroxylation of proline in polytripeptide models of collagen: Stereochemistry of polytripeptide-prolyl hydroxylase interaction. Biochemistry. 1978;17:2892–2898. doi: 10.1021/bi00607a030. [DOI] [PubMed] [Google Scholar]
- 36.Brahmachari SK, Ananthanarayanan VS. β-Turns in nascent procollagen are sites of posttranslational enzymatic hydroxylation of proline. Proc Natl Acad Sci USA. 1979;76:5119–5123. doi: 10.1073/pnas.76.10.5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Atreya PL, Ananthanarayanan VS. Interaction of prolyl 4-hydroxylase with synthetic peptide substrates. A conformational model for collagen proline hydroxylation. J Biol Chem. 1991;266:2852–2858. [PubMed] [Google Scholar]
- 38.Fischer G. Chemical aspects of peptide bond isomerisation. Chem Soc Rev. 2000;29:119–127. [Google Scholar]
- 39.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. Conformational stability of collagen relies on a stereoelectronic effect. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 40.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. Collagen stability: Insights from NMR spectroscopic and hybrid density functional computational investigations of the effect of electronegative substituents on prolyl ring conformations. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
- 41.Giacovazzo C, Monaco HL, Artioli G, Viterbo D, Ferraris G, Gilli G, Zanotti G, Catti M. Fundamentals of Crystallography. 2. Oxford University Press; Oxford, UK: 2002. [Google Scholar]
- 42.Raines RT. 2005 Emil Thomas Kaiser Award. Protein Sci. 2006;15:1219–1225. doi: 10.1110/ps.062139406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hodges JA, Raines RT. Stereoelectronic and steric effects in the collagen triple helix: Toward a code for strand association. J Am Chem Soc. 2005;127:15923–15932. doi: 10.1021/ja054674r. [DOI] [PubMed] [Google Scholar]
- 44.Jenkins CL, Lin GL, Duo JQ, Rapolu D, Guzei IA, Raines RT, Krow GR. Substituted 2-azabicyclo[2.1.1]hexanes as constrained proline analogues: Implications for collagen stability. J Org Chem. 2004;69:8565–8573. doi: 10.1021/jo049242y. [DOI] [PubMed] [Google Scholar]
- 45.Krow GR, Lin G, Yu F. The rearrangement route to 3-carboxy- and 3-hydroxymethyl-2-azabicyclo[2.1.1]hexanes: 3,5-Methanoprolines. J Org Chem. 2005;70:590–595. doi: 10.1021/jo0484171. [DOI] [PubMed] [Google Scholar]
- 46.Krow GR, Huang Q, Lin G, Centafont RA, Thomas AM, Gandla D, DeBrosse C, Carroll PJ. 5-Carboxy-2-azabicyclo[2.1.1]hexanes as precursors of 5-halo, amino, phenyl, and 2-methoxycarbonylethyl methanopyrrolidines. J Org Chem. 2006;71:2090–2098. doi: 10.1021/jo052506b. [DOI] [PubMed] [Google Scholar]
- 47.Krow GR, Herzon SB, Lin G, Qiu F, Sonnet PE. Temperature-dependent regiochemical diversity in lithiation-electrophilic substitution reactions on N-BOC-2-azabicyclo[2.1.1]hexane. 2,4- and 3,5-Methanoprolines. Org Lett. 2002;4:3151–3154. doi: 10.1021/ol026509b. [DOI] [PubMed] [Google Scholar]
- 48.Gill SC, von Hippel PH. Calculation of protein extinction coefficients from amino acid sequence data. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 49.Kivirikko KI, Myllyla R. Posttranslational enzymes in the biosynthesis of collagen: Intracellular enzymes. Methods Enzymol. 1982;82:245–304. doi: 10.1016/0076-6879(82)82067-3. [DOI] [PubMed] [Google Scholar]
- 50.Kanai K, Podanyi B, Bokotey S, Hajdu F, Hermecz I. Stereoselective sulfoxide formation from a thioproline derivative. Tetrahedron Asymmetry. 2002;13:491–495. [Google Scholar]
- 51.Tandon M, Wu M, Begley TP, Myllyharju J, Pirskanen A, Kivirikko K. Substrate specificity of human prolyl-4-hydroxylase. Bioorg Med Chem Lett. 1998;8:1139–1144. doi: 10.1016/s0960-894x(98)00183-8. [DOI] [PubMed] [Google Scholar]
- 52.Gunzler V, Brocks D, Henke S, Myllyla R, Geiger R, Kivirikko KI. Syncatalytic inactivation of prolyl 4-hydroxylase by synthetic peptides containing the unphysiologic amino acid 5-oxaproline. J Biol Chem. 1988;263:19498–19504. [PubMed] [Google Scholar]
- 53.Salvador RA, Tsai I, Marcel RJ, Felix AM, Kerwar SS. The in vivo inhibition of collagen synthesis and reduction of prolyl hydroxylase activity by 3,4-dehydroproline. Arch Biochem Biophys. 1976;174:381–392. doi: 10.1016/0003-9861(76)90366-0. [DOI] [PubMed] [Google Scholar]
- 54.Kerwar SS, Felix AM, Marcel RJ, Tsai I, Salvador RA. Effect of L-3,4-dehydroproline on collagen synthesis and prolyl hydroxylase activity in mammalian cell cultures. J Biol Chem. 1976;251:503–509. [PubMed] [Google Scholar]
- 55.Nolan JC, Ridge S, Oronsky AL, Kerwar SS. Studies on mechanism of reduction of prolyl hydroxylase-activity by D,L-3,4-dehydroproline. Arch Biochem Biophys. 1978;189:448–453. doi: 10.1016/0003-9861(78)90233-3. [DOI] [PubMed] [Google Scholar]
- 56.Pauling L. The Nature of the Chemical Bond. 3. Cornell University Press; Ithaca, NY: 1960. [Google Scholar]
- 57.Gottlieb AA, Fujita Y, Udenfriend S, Witkop B. Incorporation of cis- and trans-4-fluoro-L-prolines into proteins and hydroxylation of trans isomer during collagen biosynthesis. Biochemistry. 1965;4:2507–2513. [Google Scholar]
- 58.Hutton JJ, Jr, Witkop B, Kurtz J, Berger A, Udenfriend S. Synthetic polypeptides as substrates and inhibitors of collagen proline hydroxylase. Arch Biochem Biophys. 1968;125:779–785. doi: 10.1016/0003-9861(68)90514-6. [DOI] [PubMed] [Google Scholar]
- 59.Zhang XM. Radical substituent effects of α-fluorine and α-trifluoromethyl groups. J Org Chem. 1998;63:3590–3594. [Google Scholar]
- 60.Chen I, Ting AY. Site-specific labeling of proteins with small molecules in live cells. Curr Opin Biotechnol. 2005;16:35–40. doi: 10.1016/j.copbio.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 61.Renner C, Alefelder S, Bae JH, Budisa N, Huber R, Moroder L. Fluoroprolines as tools for protein design and engineering. Angew Chem, Int Ed. 2001;40:923–925. doi: 10.1002/1521-3773(20010302)40:5<923::AID-ANIE923>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 62.Kim W, George A, Evans M, Conticello VP. Cotranslational incorporation of a structurally diverse series of proline analogues in an Escherichia coli expression system. ChemBioChem. 2004;5:928–936. doi: 10.1002/cbic.200400052. [DOI] [PubMed] [Google Scholar]
- 63.Kim W, McMillan RA, Snyder JP, Conticello VP. A stereoelectronic effect on turn formation due to proline substitution in elastin-mimetic polypeptides. J Am Chem Soc. 2005;127:18121–18132. doi: 10.1021/ja054105j. [DOI] [PubMed] [Google Scholar]
- 64.Kim W, Conticello VP. Protein engineering methods for investigation of structure–function relationsips in protein-based elastomeric materials. Polym Rev. 2007;47:93–119. [Google Scholar]
- 65.Steiner T, Hess P, Bae JH, Wiltschi B, Moroder L, Budisa N. Synthetic biology of proteins: Tuning GFPs folding and stability with fluoroproline. PLoS One. 2008;3:e1680. doi: 10.1371/journal.pone.0001680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Thornburg LD, Lai MT, Wishnok JS, Stubbe J. A non-heme iron protein with heme tendencies: An investigation of the substrate specificity of thymine hydroxylase. Biochemistry. 1993;32:14023–14033. doi: 10.1021/bi00213a036. [DOI] [PubMed] [Google Scholar]
- 67.Pascal RA, Oliver MA, Chen YCJ. Alternate substrates and inhibitors of bacterial 4-hydroxyphenylpyruvate dioxygenase. Biochemistry. 1985;24:3158–3165. doi: 10.1021/bi00334a013. [DOI] [PubMed] [Google Scholar]
- 68.McCoy JG, Bailey LJ, Bitto E, Bingman CA, Aceti DJ, Fox BG, Phillips GN. Structure and mechanism of mouse cysteine dioxygenase. Proc Natl Acad Sci USA. 2006;103:3084–3089. doi: 10.1073/pnas.0509262103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Improta R, Benzi C, Barone V. Understanding the role of stereoelectronic effects in determining collagen stability. 1 A quantum mechanical study of proline, hydroxyproline, and fluoroproline dipeptide analogues in aqueous solution. J Am Chem Soc. 2001;123:12568–12577. doi: 10.1021/ja010599i. [DOI] [PubMed] [Google Scholar]
- 70.Thomas KM, Naduthambi D, Tririya G, Zondlo NJ. Proline editing: A divergent strategy for the synthesis of conformationally diverse peptides. Org Lett. 2005;7:2397–2400. doi: 10.1021/ol0506720. [DOI] [PubMed] [Google Scholar]
- 71.Kern D, Schutkowski M, Drakenberg T. Rotational barriers of cis/trans isomerization of proline analogues and their catalysis by cyclophilin. J Am Chem Soc. 1997;119:8403–8408. [Google Scholar]
- 72.Krow GR, Cannon KC. Azabicyclo[2.1.1]hexanes. A review. Heterocycles. 2004;62:877–898. [Google Scholar]
- 73.Walter C, Frieden E. The prevalence and significance of the product inhibition of enzymes. Adv Enzymol Relat Areas Mol Biol. 1963;25:1963. doi: 10.1002/9780470122709.ch4. [DOI] [PubMed] [Google Scholar]
- 74.Cook P, Cleland WW. Enzymes Kinetics and Mechanism. Garland Science; New York, NY: 2007. pp. 128–150. [Google Scholar]
- 75.Peluso S, Ufret MD, O’Reilly MK, Imperiali B. Neoglycopeptides as inhibitors of oligosaccharyl transferase: Insight into negotiating product inhibition. Chem Biol. 2002;9:1323–1328. doi: 10.1016/s1074-5521(02)00281-8. [DOI] [PubMed] [Google Scholar]
- 76.Panasik N, Jr, Eberhardt ES, Edison AS, Powell DR, Raines RT. Inductive effects on the structure of proline residues. Int J Pept Protein Res. 1994;44:262–269. doi: 10.1111/j.1399-3011.1994.tb00169.x. [DOI] [PubMed] [Google Scholar]