Abstract
Leinamycin is a structurally novel Streptomyces-derived natural product that displays very potent activity against various human cancer cell lines (IC50 values in the low nanomolar range). Previous in vitro biochemical studies have revealed that leinamycin alkylates DNA, generates apurinic (AP) sites and reactive oxygen species (ROS), and causes DNA strand breaks. However, it is not clear whether these events occur inside cells. In the present study, we have determined the endogenous amount of AP sites and DNA strand breaks in genomic DNA and the amount of oxidative stress in a human pancreatic carcinoma cell line, MiaPaCa, treated with leinamycin by utilizing the aldehyde-reactive probe (ARP) assay, the comet assay, and fluorescent probes, respectively. We demonstrated that AP sites are formed rapidly following exposure to leinamycin, and the number of AP sites was increased up to seven- fold in a dose dependent manner. However, only 25–50% of these sites remain 2 h after media containing drug molecules was aspirated and replaced with fresh media. We also observed leinamycin induced ROS generation and a concomitant increase in apoptosis of MiaPaCa cells. Because both AP sites and ROS have the potential to generate strand breaks in cellular DNA, the comet assay was utilized to detect damage to nuclear DNA in leinamycin-treated MiaPaCa cell cultures. Both alkaline and neutral electrophoretic analysis revealed that leinamycin produces both single- and double-stranded DNA damage in drug-treated cells in a dose-dependent manner. Taken together, the results suggest that rapid conversion of leinamycin-guanine (N7) adducts into AP sites to produce DNA strand breaks, in synergy with leinamycin-derived ROS, account for the exceedingly potent biological activity of this natural product.
Keywords: Leinamycin, Apurinic site, Comet assay, DNA strand breaks
INTRODUCTION
Leinamycin is an antitumor antibiotic, which was first isolated from Streptomyces sp. by researchers at Kyowa Hakko Kogyo Co., Ltd. in Tokyo, Japan in 1989 (1–2). Over the past two decades, much has been learned about the structure and function of leinamycin. This natural product antibiotic possesses several novel structural elements, including a 1,2-dithiolan-3-one 1-oxide moiety and an 18-membered lactam ring containing an embedded Z,E-thiazol-5-yl-penta-2,4-dienone fragment (1–4). Leinamycin displays potent activity against cancer cell lines in both in vitro and in vivo tumor models and is currently in development as a potential antitumor agent (5–9).
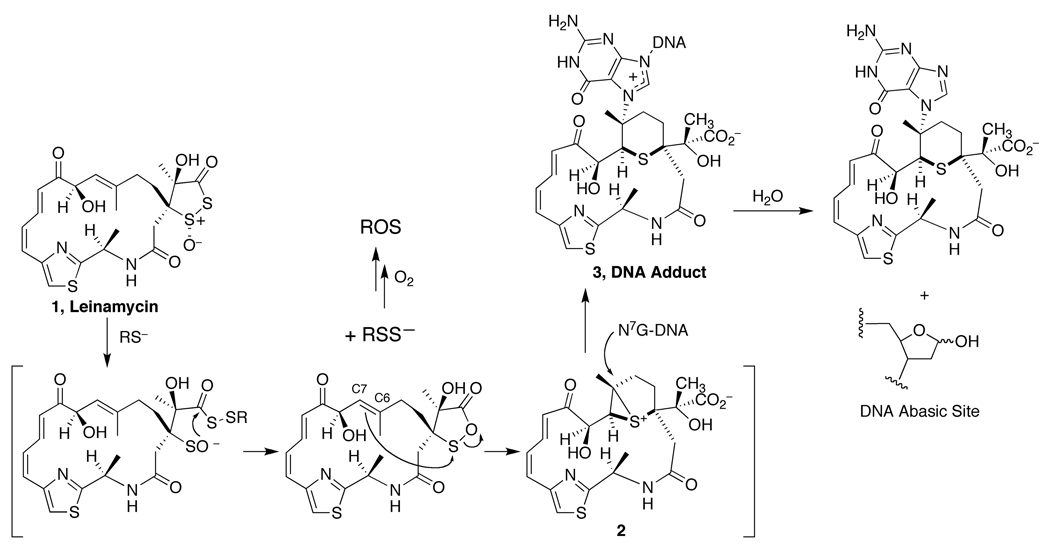
Early studies by the discoverers of leinamycin indicated that the natural product causes thiol-triggered DNA strand breaks (5). Subsequent studies showed that attack of a thiol on the 1,2-dithiolan-3-one 1-oxide in leinamycin initiates conversion of this heterocycle to a 1,2-oxathiolan-5-one (Figure 1, step 2) that undergoes further rearrangement to an episulfonium ion (Figure 1, step 3) (10–16). This episulfonium ion associates non-covalently with duplex DNA and alkylates the N7-position of guanine residues in double-stranded DNA with very high efficiency (Figure 1, step 4) (14, 17, 18). The guanine adduct is the only covalent DNA lesion resulting from the reaction of thiol-activated leinamycin with duplex DNA (17).
Figure 1.
Chemistry leading to DNA-alkylation by leinamycin followed by a rapid decomposition of a leinamycin-guanine adduct to yield an AP site in duplex DNA.
Recent in vitro studies by the Gates group revealed that leinamycin-guanine adducts in double-stranded DNA undergo unusually rapid depurination (t1/2 = 3 h) to generate AP sites (Figure 1) (19–21). In general, the depurination of N7-alkylguanine residues in double-stranded DNA occurs with a half-life in the range of 50–100 h under physiological conditions (20). While the chemical basis for the rapid loss of the leinamycin-guanine adduct from duplex DNA remains uncertain, this rapid depurination reaction might be biologically important because AP sites generated in cellular DNA are known to be cytotoxic lesions (22–26). In addition, AP sites are readily converted into DNA strand breaks, which are also potentially toxic lesions (27–28). Finally, recent evidence shows that AP sites can generate interstrand crosslinks in DNA under physiologically relevant conditions (29). Although both AP sites and single-strand breaks are subject to repair by various cellular systems (30–32), rapid generation of large numbers of AP sites by leinamycin has the potential to overwhelm the capacity of these repair enzymes (33–36).
In addition to the DNA-alkylation chemistry described above, attack of a thiol on leinamycin releases a persulfide intermediate (RSSH, Figure 1) that can mediate the generation of reactive oxygen species (ROS) in vitro (37–43). Persulfides generate ROS via an initial reduction of molecular oxygen to superoxide radical, followed by disproportionation to hydrogen peroxide, and finally production of the hydroxyl radical via the Fenton reaction (37–43). Such chemistry could contribute to the biological activity of leinamycin because intracellular generation of ROS can cause DNA strand cleavage and lead to cell death via general oxidative stress (44–48).
Our recent study demonstrated that exposure to leinamycin caused DNA strand breaks in both time- and dose-dependent manners, with strand cleavage occurring as early as 3 h after MiaPaCa cells were exposed to the drug (49). Most DNA-damaging agents are known to activate DNA checkpoint mechanisms irrespective of their mode of actions (50). The checkpoint kinase Chk2 is known to be activated in response to direct DNA cleaving agents such as ionizing radiation (IR), while replication arresting agents, such as the alkylating agents, aphidicolin and hydroxyurea, activate the checkpoint kinase Chk1 (50). Thus, the observed activation of Chk2, but not Chk1, in MiaPaCa cells treated with leinamycin is consistent with a cellular response to DNA strand break formation rather than DNA adduct formation (50). In the present study, we set out to determine whether leinamycin; increases the number of AP sites, forms DNA strand breaks, and produces ROS inside human cancer cell lines as previous chemical and biochemical studies have suggested. Our results showed that the ability of leinamycin to produce unique types of DNA damage and ROS might represent a new biochemical route to very potent cytotoxic activity against human cancer cells. Overall, this work makes important connections between the chemical reactivity of leinamycin (51–52) and its potent biological activity against human cancer cell lines (5, 9).
MATERIALS and METHODS
Materials
Oligonucleotides were constructed by Sigma Genosys (Woodlands, Texas). T4 polynucleotide kinase was purchased from New England Biolabs, and all radioactive nucleotides were from Amersham Life Sciences.
Cell lines
The MiaPaCa cell line was obtained from American Type Culture Collection (Rockville, MD) and cultured according to their instructions in RPMI 1640 supplemented with 10% FCS. All chemical reagents were purchased from Sigma Aldrich, and leinamycin was a kind gift from Dr. Yutaka Kanda of Kyowa Hakko Kogyo Ltd. All chemicals were dissolved in DMSO (5 mg/mL stock solution) and stored at −80°C.
Assay for AP sites in cultured cancer cells
The cultured MiaPaCa cells were treated with various concentrations of leinamycin, harvested, washed, and incubated in 1 mL of PBS/5 mM glucose with 3 mM Aldehyde Reactive Probe (ARP), which is N’-aminooxymethylcarbonylhydrazino-D-biotin (Dojindo Molecular Technologies Inc, Gaithersburg, MD) for 60 min at 37°C as previously described (53). The cells were then collected and washed twice with 1 mL of PBS to remove residual ARP. The DNA was then isolated and immobilized on a 96-well plate with DNA binding solution. The 96-well plate was then incubated with streptavidin-conjugated horseradish peroxidase (HRP) (Amersham, Piscataway, NJ) and rinsed with a washing buffer. After adding 100 µL of substrate solution (Dojindo Molecular Technologies Inc, Gaithersburg, MD) to each well and incubating the microplate at 37 °C for 1 h as the manufacturer suggested, the enzymatic activity of HRP was detected colorimetrically by measuring the absorbance at 650 nm. The number of AP sites was calculated based upon a standard curve generated using ARP standard DNA solutions (Dojindo Molecular Technologies Inc, Gaithersburg, MD) as described previously (54). In addition to colorimetric detection, the relative amount of AP sites was determined by flow cytometry. Harvested cells were fixed and incubated with FITC-conjugated streptavidin (Sigma Aldrich). Cellular fluorescence was determined with the Coulter ELITE ESP flow cytometer, and fluorescent signals were collected using the standard configuration of the flow cytometer (green fluorescence for FITC).
Comet assay
The neutral comet assay was performed as previously described (55). Briefly, following treatment of MiaPaCa cells with leinamycin, cells were trypsinized, pelleted, and suspended in 100 µL of PBS. The cell suspension (10 µL) was mixed with 100 µl of LMAgarose (Trevigen) at a ratio of 1: 10 (v/v), and 75 µL of the mixture was pipetted onto a CometSlide (Trevigen) and allowed to gel at 4°C until a clearing appeared around the agarose. Slides were immersed in cold lysis solution (Trevigen) for 1 h at 4°C and then rinsed several times in TBE buffer. Slides were then electrophoresed in either TBE buffer or alkaline electrophoresis solution (300 mM NaOH, 1 mM EDTA, pH> 13) at 1 volt per cm, electrode to electrode. After electrophoresis, slides were rinsed 3 times with distilled water and allowed to dry overnight. The slides were then stained with 50 µl of SYBR Green I (Trevigen) and scored with a Zeiss Axioplan fluorescent microscope with 516–560 nm emission from a 70 W mercury lamp connected to a camera. For each dose, about 30–60 cells per slide were scored and recorded for analysis. Images were acquired and analyzed using a self-developed image system. Comet length, percentage of DNA in the tail, and comet moment were calculated as previously described (56).
Purification and radiolabeling of oligonucleotides
5'-end labeled duplex oligonucleotides shown in Figure 5 were prepared as described below. Each strand was 32P-5′-end labeled with T4 polynucleotide kinase and γ-32P-ATP, heat-treated to inactivate the enzyme, and annealed with the unlabeled complementary strand. The resulting duplex oligonucleotides were run through a Sephadex G-10 column to remove unincorporated γ-32P-ATP. The annealed duplex oligonucleotides were further purified with a 16% nondenaturing PAGE gel. The DNA was visualized by exposure to X-ray film and purified from the gel by using the crush and soak method.
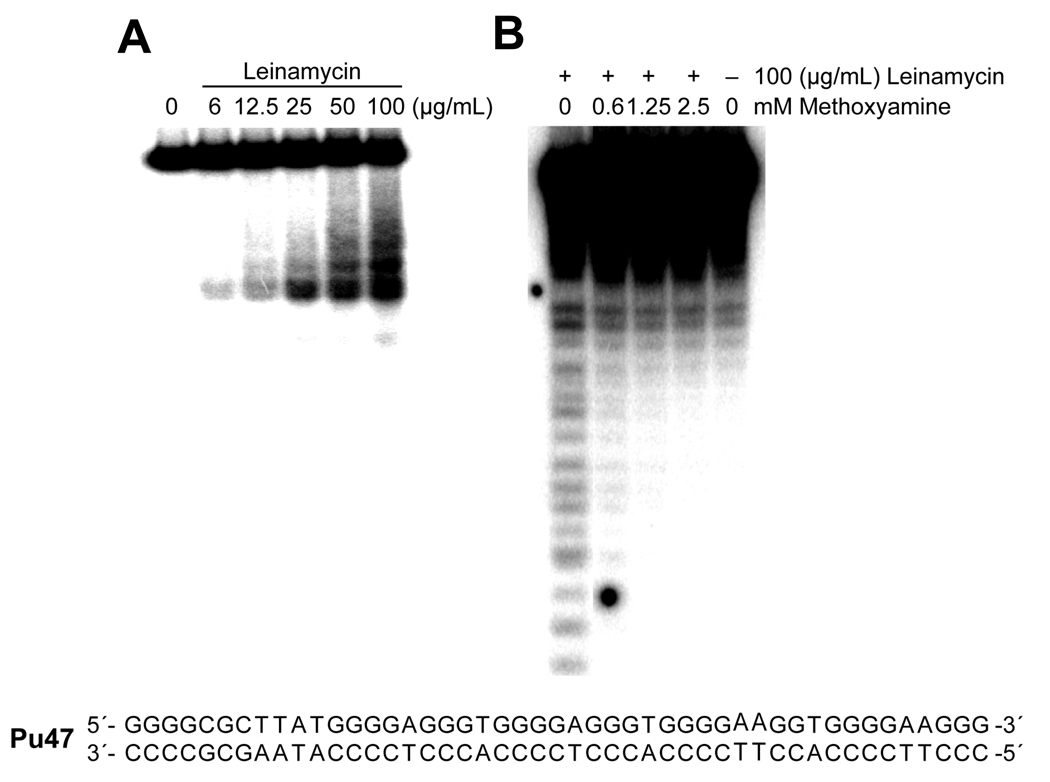
Figure 5.
A) A native PAGE gel analysis showing double stranded cleavage of a 5'-end-labeled 47-bp oligomer duplex (Pu47) by treatment with leinamycin. B) Protective effect of methoxyamine on leinamycin-induced double-stranded DNA breaks. A 5'-end-labeled Pu47 was treated with leinamycin in the presence of increasing concentrations of methoxyamine. To determine true DNA strand breakage sites, the DNA samples were dissolved in 20 µL of a neutral formamide dye and were electrophoresed in a 16% denaturing PAGE gel without heat denaturation.
Leinamycin cleavage reaction
End-labeled duplex oligonucleotides (100 fmol) were incubated with 100 µg/mL leinamycin in a 20 µl reaction mix containing 40 mM Tris–HCl, pH 8.0, 1 mM DTT, and 20 mM NaCl. The reaction was terminated by adding 80 µl of a drug stop buffer containing 5 µg of calf thymus DNA in 0.4 M NaOAc (pH 6.0) solution, and DNA was precipitated with 3 volumes of ethanol. For nondenaturing gel electrophoresis, DNA pellets were resuspended in a gel loading dye and electrophoresed in a 16% native PAGE gel. To determine true DNA strand breakage sites, the DNA pellet was suspended in 20 µL of a neutral formamide dye and the products were resolved by electrophoresis in a 16% denaturing PAGE gel without heat denaturation. The dried gel was exposed to a PhosphorImager screen and visualized using ImageQuant software.
ROS assay
The cellular production of ROS by leinamycin in the human pancreatic cancer cell line MiaPaCa was measured using dichlorofluorescin diacetate (DCFDA) and flow cytometry as previously described (57). In brief, following treatment of MiaPaCa cells with various concentrations of leinamycin for 2h, cells were trypsinized, pelleted, washed, and incubated in 1 mL of PBS/5 mM glucose with 50 µM DCFDA for 60 min at 37°C. The cells were then collected and washed twice with 1 ml PBS to remove residual DCFDA. DCF fluorescence was quantified at 525 nm, when excited with 488 nm light with the Coulter ELITE ESP flow cytometer.
Caspase activity assay
Caspase 3-like activity was determined using the ApoAlert Caspase Fluorescent Assay Kit (Clontech Laboratory, Inc.) according to the manufacturer's protocol. Briefly, after treatment with leinamycin for 2 hours at 0, 12.5, 25, or 100 ng/mL, cells were washed with ice-cold PBS and lysed in cell lysis buffer. Cell lysates were mixed with caspase assay buffer containing 20 µmol/L Ac-DEVD-AFC as a caspase-3 substrate and incubated for 1 h at 37 °C. Enzyme catalyzed release of the fluorophore 7-amino-4-trifluoromethyl coumarin (AFC) from the labeled substrate, DEVD-AFC, was monitored using a Synergy HT Multi-Detection Microplate Reader (BioTek) with an excitation wavelength of 395 nm and an emission wavelength of 509 nm.
RESULTS
Determination of AP sites in genomic DNA isolated from leinamycin-treated cells using the aldehyde-reactive probe (ARP)
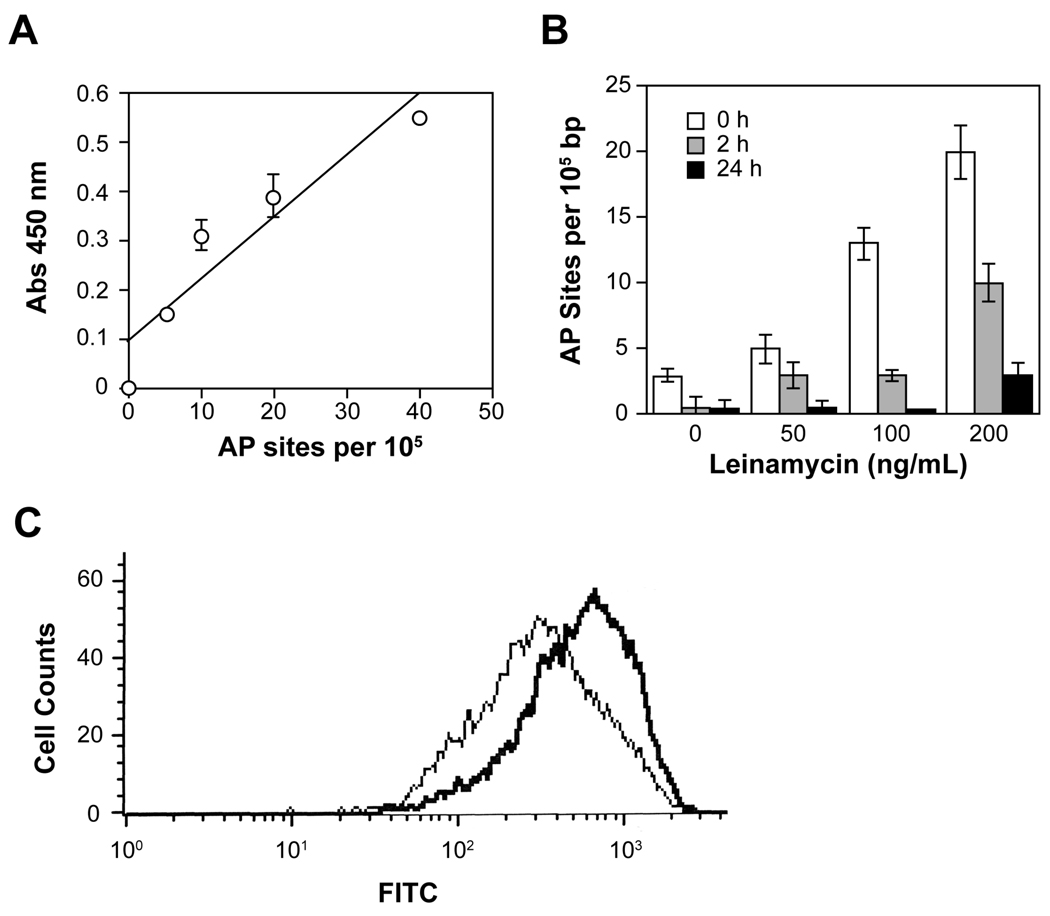
To determine whether leinamycin increases the number of AP sites in living cells, the number of AP sites in genomic DNA was determined following treatment of MiaPaCa cells with leinamycin as previously described (53). Previous studies revealed that the ARP reagent can penetrate viable cells and react directly with AP sites (54). Thus, by directly labeling AP sites in cultured cells with ARP reagent, we tried to minimize potential experimental errors resulting from the accidental generation of the AP sites or loss of the AP sites occurring during the DNA extraction process at high temperature (54). The number of AP sites in isolated DNA from cells was then determined by measuring the enzymatic activity of horseradish peroxidase (HRP) conjugated to streptavidin and calculated based upon a standard curve generated using ARP standard DNA solutions (see Fig. 2A). As shown in Figure 2B, there is a concentration-dependent increase in the number of AP sites in MiaPaCa cells after a transient exposure (2 h) to leinamycin. We observed that the levels of AP sites increased up to seven-fold in a dose dependent manner. Specifically, control cells contained expected (53) background levels of 3 AP sites per 105 bp, whereas leinamycin-treated cells contained between 5–20 AP sites per 105 bp after a transient exposure (2 h) to a leinamycin concentration of 200 ng/mL. However, the burst of AP sites generated by leinamycin was very brief; only 25 to 50% of these sites remain within 2 h after media containing the drug was aspirated and replaced with fresh media. A flow cytometric method was also independently used to directly measure the AP sites in MiaPaCa cells after labeling the AP sites in chromosomal DNA of living cells with biotin-containing ARP followed by treatment with streptavidin-FITC. As shown in Figure 2C, about a three-fold increase in the FITC fluorescence signal was observed in MiaPaCa cells exposed to 100 ng/mL leinamycin for 2 h compared to the untreated cells. Overall, these data are consistent with the recent in vitro studies showing that the leinamycin-guanine adduct in double-stranded DNA undergoes unusually rapid depurination to generate AP sites in duplex DNA (19–21).
Figure 2.
Increased production of AP sites in MiaPaCa cells treated with leinamycin. A, standard curve for the detection of AP sites in DNA containing known numbers of AP sites using an ARP assay kit (Dojindo Molecular Technologies) (32). B, colorimetric measurement of AP sites in DNA isolated from MiaPaCa cells treated with leinamycin. MiaPaCa cells were treated for 2 hours with 0, 50, 100, or 200 ng/mL leinamycin, and media containing drug molecules was aspirated, washed with PBS twice, and replaced with fresh media. At 0, 2, and 24 hours after incubation with fresh media, the cells were then incubated with 3 mM ARP for 60 min at 37°C in 1 ml PBS containing 5 mM glucose to directly label the AP sites in chromosomal DNA of living cells as previously described [31]. The number of AP sites was calculated based upon a standard curve generated using ARP standard DNA solutions as described in Fig. 2A. C, flow cytometric measurement of the relative amount of AP sites in MiaPaCa cells treated with 0 or 100 ng/mL leinamycin. The drug-treated cells were harvested, fixed and incubated with FITC-conjugated streptavidin (Sigma Aldrich). Cellular fluorescence was determined with the Coulter ELITE ESP flow cytometer, and fluorescence signals were collected using the standard configuration of the flow cytometer (green fluorescence for FITC).
Leinamycin causes oxidative stress in human cancer cells
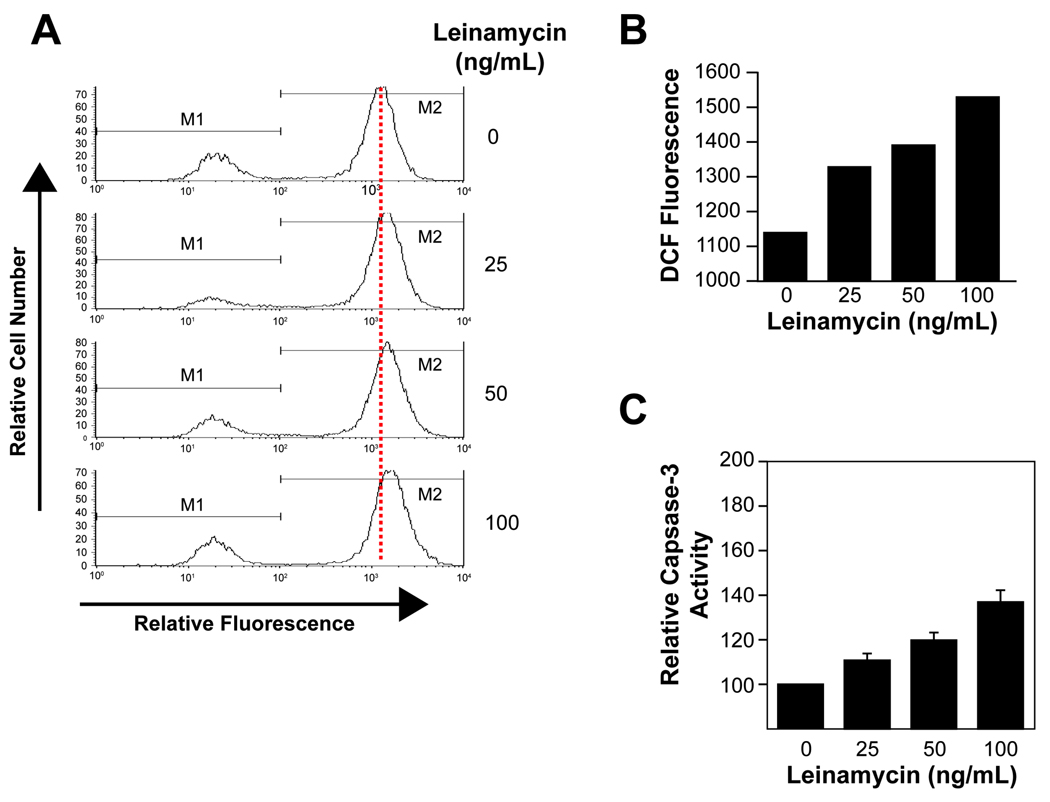
Previous chemical studies predicted that leinamycin could generate reactive oxygen species (ROS) in cultured cells from persulfide species generated in the reaction of thiols with leinamycin, in addition to its DNA-alkylating property (37–43). To determine whether leinamycin causes oxidative stress in a human cancer cell line, we used a fluorescent probe, DCFDA and flow cytometric analysis to determine the DCF fluorescence of leinamycin-treated versus untreated cells as previously described (57). As shown in Figure 3A and 3B, the fluorescence signal increased up to 50% in MiaPaCa cells treated with 100 ng/mL leinamycin for 2 hours as compared to untreated cells. The role of ROS in drug-induced apoptosis has been well established in various human cancer cells, although the exact mechanism by which ROS promotes apoptosis remains to be discovered. Based on an increased generation of ROS in leinamycin-treated MiaPaCa cells, we examined the effect of leinamycin on apoptosis in these cells. In order to quantify the proteolytic activation of caspase-3, lysates normalized for the protein from the MiaPaCa cells treated with leinamycin were assayed for their caspase-3 activity using Ac-DEVD-AFC as a substrate. As shown in Figure 3C, leinamycin treatment increases caspase-3 activity up to 30%, after 2-h exposure to 100 ng/mL leinamycin.
Figure 3.
A, flow cytometric analyses of the fluorescence of leinamycin-treated MiaPaCa cells at various concentrations for 2 h. MiaPaCa cells were treated with 0, 25, 50, or 100 ng/mL leinamycin for 2 hours. Drug-treated MiaPaCa cells were incubated with 0.4 mM DCFH-DA for 30 min, and then washed with PBS. DCF fluorescence was quantified by its fluorescence at 525 nm, when excited with 488 nm light with the Coulter ELITE ESP flow cytometer. B, a graphical representation of data presented in A. C, Activation of caspase-3 in leinamycin-treated MiaPaCa cells. MiaPaCa cells were incubated with 0, 25, 50, or 100 ng/mL leinamycin for 2 hours, and caspase-3-like activity was measured by the DEVD-AFC-cleavage assay. Data are mean ± SEM of three separate experiments.
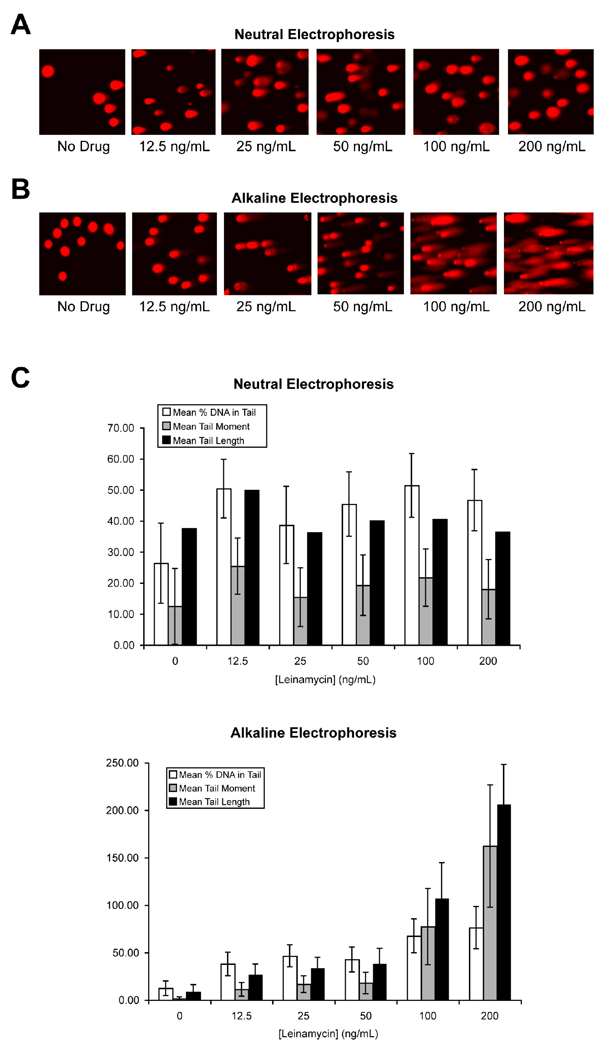
Measurement of single-strand and double-strand breaks in the DNA of leinamycin-treated cells
Leinamycin-derived AP sites have the potential to generate single strand breaks via a β-elimination reaction that cleaves the O–P bond on the 3'-side of the abasic site (22,28,58). Similarly, leinamycin-derived ROS may generate single-strand breaks in chromosomal DNA by reactions involving hydrogen atom abstraction from the DNA backbone (56–62). Therefore, we characterized the type of DNA strand breaks (single or double) produced by leinamycin in MiaPaCa cells by the neutral comet assay to identify double-strand breaks as previously described (33–34). As shown in Figure 4A, the formation of double strand DNA breaks was moderate in leinamycin-treated MiaPaCa cells in the comet assay in which lysis and electrophoresis were done under neutral conditions. However, the comets with a distinct head, consisting of intact DNA, and a tail, consisting of damaged or broken pieces of DNA, appeared in drug-treated cells in a dose- and time-dependent manner in the comet assay in which the electrophoresis was done under alkaline conditions (see Figure 4B). Results of the neutral comet assay were analyzed to determine the mean tail moment, mean tail length, and mean percentage of tail DNA, and are summarized in Fig. 4C. Taken together, these results suggest that leinamycin predominantly produces single-stranded DNA breaks in MiaPaCa cells in a dose- and time-dependent manner.
Figure 4.
Level of leinamycin-induced DNA strand breaks breaks in MiaPaCa cell as determined by a neutral Comet assay. MiaPaCa cells were treated with 0, 12.5, 25, 50, 100, or 200 ng/mL leinamycin for 2 hours, and the lysis of the harvested cells was done under neutral conditions. The electrophoresis was done under either neutral (A) or alkaline (B) condition. C, Mean tail moment, mean tail length, and mean percentage of tail DNA in MiaPaCa cells treated with 0, 12.5, 25, 50, 100, or 200 ng/mL leinamycin for 2 hours and analyzed by a neutral Comet assay using electrophoresis under either neutral or alkaline condition. The number of cells scored in each treatment was 30–60. Error bars denote SEM.
Leinamycin can produce both single-strand breaks (SSBs) and double-strand breaks (DSBs) in the DNA backbone
In order to further test whether leinamycin produces DNA strand breaks, the duplex DNA oligomer Pu47 was labeled at the 5’ end of the G-rich strand with 32P. As shown in Figure 5A, a native PAGE analysis of the sample revealed that leinamycin causes DNA fragmentation in a dose-dependent manner. The cleavage of double-stranded DNA by leinamycin could be initiated by single-strand breaks on one strand followed by formation of single-strand breaks on the opposite DNA strand close to the initial single strand break site, resulting in double-strand cuts in duplex DNA oligomers. Previous studies suggested that leinamycin induces a significant amount of DNA adducts that spontaneously depurinate to form AP sites in DNA (19). Although AP sites are known to be relatively stable under physiological conditions, spontaneous cleavage of AP sites can occur via β-elimination, resulting in single-strand breaks (SSBs) with 3’ and 5’-ends blocked with an unsaturated aldehydic (α,β-4-hydroxy-2-pentenal)-3’-phosphate and phosphate groups, respectively (28). Methoxyamine blocks strand cleavage at AP sites by reacting with the aldehyde group at the same sites in DNA and preventing β-elimination and the hydrolysis of the 3'-phosphodiester of an abasic site (29,30). To obtain clear evidence that leinamycin-induced DNA strand breaks may spontaneously occur via β-elimination reactions at aldehydic abasic sites, we tested whether methoxyamine decreases the yield of DNA strand breaks induced by the natural product. As shown in Figure 5B, treatment of oligomer duplexes with leinamycin in the presence of methoxyamine significantly decreased the number of true DNA strand breaks produced by leinamycin in the absence of thermal treatment. Collectively, these results confirm that DNA strand breaks induced by leimamycin are a result of spontaneous cleavage via β-elimination and the hydrolysis of the 3'-phosphodiester of an abasic site.
DISCUSSION
Leinamycin damages DNA by unique chemical mechanisms and displays potent anticancer activity against several human malignant cell lines (1,2,5–9). Recent in vitro studies revealed that the leinamycin-guanine adducts in double-stranded DNA undergo unusually rapid depurination to generate AP sites in the DNA duplex (19,21). The half life of the leinamycin-guanine adduct in converting to an AP site in duplex DNA was estimated to be about 3 h, while typical N7-guanine adducts undergo depurination slowly in duplex DNA, with a half-life of about 50–100 h at physiological conditions (20). Thus, the N7-alkylguanine residue generated by leinamycin decomposes to yield an AP site faster than any other guanine adduct ever reported (19,20,58). Therefore, it was of great importance to determine whether leinamycin generates AP sites in living cells to understand the mechanisms involved in its antitumor activity. In the current study, we utilized the ARP reagent to measure AP sites formed in genomic DNA of MiaPaCa cells treated with leinamycin, and both flow cytometry and colorimetric methods were independently used to measure the AP sites labeled with ARP reagent (43). We observed that the levels of AP sites increased approximately seven-fold after a transient exposure (2 h) to concentrations of leinamycin less than 200 ng/mL. This result is generally consistent with our earlier observations that the leinamycin-guanine adduct in double-stranded DNA undergoes unusually rapid depurination to generate an AP site in the DNA duplex.
In vitro biochemical experiments have shown that, in addition to its DNA-alkylating properties, leinamycin generates ROS under physiologically-relevant conditions (38–43). Thus, in this study, we measured oxidative stress (ROS generation) in MiaPaCa cells upon treatment with leinamycin using DCFDA and flow cytometry. We observed an approximately 50% increase in cellular production of ROS in the leinamycin-treated cells compared to the mock-treated cells in a dose- and time-dependent manner. In general, conversion of precursors like DCFDA to their fluorescent form is observed in cells that undergo an apoptotic-like cell death (44–48). Since leinamycin also induces apoptosis under similar conditions, as demonstrated by a 30% activation of caspase 3 (2h, 100 ng/mL), it is not yet clear whether the leinamycin-induced conversion of DCFDA to DCF is due directly to leinamycin-derived ROS or from the release of cytochrome c that occurs during the early phases of apoptosis, thus requiring further studies on the mechanism of ROS formation in leinamycin treated cells.
It is worthwhile to note that the AP sites in leinamycin treated cells have a short half life, since the levels of AP sites remain only 25–50% above background 2h after drugs were removed from the flasks and fresh medium was added. In a previous study, the kinetics of abasic site formation in methyl methanesulfonate (MMS)-treated HeLa cells were investigated using the ARP, revealing that the amount of AP sites in MMS-treated cells increases up to 3 h, and then disappears with a half-life of about 20 h, which is quite different from what we observed for leinamycin (63). The relatively rapid formation and disappearance of leinamycin-derived AP sites may be due to several factors, including rapid DNA alkylation by activated leinamycin (14,17,18), rapid spontaneous depurination of the leinamycin-guanine adduct (19–21) and, finally, rapid removal of the AP site by enzymatic repair processes (64, 65).
Our previous study suggested that the generation of apoptotic cells after treatment with leinamycin closely follows the rapid induction of DNA-strand breaks by leinamycin (49). Therefore, we evaluated the induction of DNA strand breaks after the exposure of MiaPaCa cells to leinamycin using the Comet assay, which is generally used for the detection of single- and double-strand breaks and alkaline-labile sites at the single cell level (55, 56). The results from both alkaline and neutral electrophoretic analysis confirmed that leinamycin predominantly produces single-stranded DNA breaks in drug-treated cells. Our results support the hypothesis that rapid conversion of N7-leinamycin-G adducts into AP sites is responsible for the increased formation of DNA strand breaks, since AP sites can be chemically converted into DNA strand breaks through β-elimination in vitro. The conversion of AP sites into DNA strand breaks can also be catalyzed by physiological concentrations of polyamines and histones inside cells (64). Furthermore, Swenberg has suggested that the rate-determining step in AP site repair is not the AP endonuclease-induced cleavage reaction but the trimming of the 5'-deoxyribose phosphate (dRP) group from the strand break (65). Thus, if the rapid depurination of the leinamycin-guanine adduct overwhelms the cell’s ability to repair AP sites it is likely that strand breaks (not AP sites) are the resulting intermediate that builds up in the cell. Other alkylating agents such as MMS that do not depurinate rapidly do not yield the build-up of strand breaks that is seen for leinamycin. In this study, DNA strand breaks (single or double) produced by leinamycin in DNA were further examined by using a native and denaturing polyacrylamide gel electrophoresis analysis of duplex DNA oligomers treated with leinamycin. This analysis revealed that this drug can produce both single and double strand breaks in a duplex DNA oligomer. Interestingly, methoxyamine effectively decreases the amount of DNA strand breaks produced by leinamycin in duplex DNA oligomers in a dose-dependent manner. Methoxyamine is known to block strand cleavage at abasic sites by reacting with the aldehyde group at AP sites in DNA and preventing β-elimination and the hydrolysis of the 3'-phosphodiester of an abasic site (29,30). Our study provides strong evidence that leinamycin-mediated strand breaks could arise primarily through the DNA alkylating properties of leinamycin and its ability to subsequently generate labile AP sites. DNA strand breaks induced by leinamycin may arise either via spontaneous cleavage via β-elimination or through processing of the abasic site by APE.
Taken together, our results are consistent with our hypothesis that the ability of leinamycin to efficiently alkylate DNA, generate a burst of AP sites along with DNA strand breaks, and generate oxidative stress represents an unrecognized biochemical route to potent anticancer activity against human cancer cells, establishing a strong link between the DNA-damaging properties of leinamycin and its potent activity against human cancer cell lines. Since DNA strand breaks are generally regarded as more lethal lesions to cells compared to base adducts or AP sites, our results suggest that rapid conversion of AP sites into DNA strand breaks may account for leinamycin’s very potent biological activity (32).
ACKNOWLEDGEMENT
This research was supported by the National Institutes of Health (CA119131). We are grateful to Dr. Kanda at Kyowa Hakko Kogyo Co., Ltd. in Tokyo, Japan for providing leinamycin for this study. We are grateful to Dr. Allison Hays for critical comments, discussion and editing the final version of the article.
REFERENCES
- 1.Hara M, Takahashi I, Yoshida M, Asano K, Kawamoto I, Morimoto M, Nakano H. DC 107, a novel antitumor antibiotic produced by a Streptomyces sp. J. Antibiot. 1989;42:333–335. doi: 10.7164/antibiotics.42.333. [DOI] [PubMed] [Google Scholar]
- 2.Hara M, Asano K, Kawamoto I, Takiguchi T, Katsumata S, Takahashi K, Nakano H. Leinamycin, a new antitumor antibiotic from Streptomyces: producing organism, fermentation and isolation. J. Antibiot. 1989;42:1768–1774. doi: 10.7164/antibiotics.42.1768. [DOI] [PubMed] [Google Scholar]
- 3.Hirayama N, Matsuzawa ES. Molecular Structure of a Novel Antitumor Antibiotic Leinamycin. Chem. Lett. 1993;11:1957–1958. [Google Scholar]
- 4.Kanda Y, Fukuyama T. Total Synthesis of (+)-Leinamycin. J. Am. Chem. Soc. 1993;115:8451–8452. [Google Scholar]
- 5.Hara M, Saitoh Y, Nakano H. DNA strand scission by the novel antitumor antibiotic leinamycin. Biochemistry. 1990;29:5676–5681. doi: 10.1021/bi00476a005. [DOI] [PubMed] [Google Scholar]
- 6.Kanda Y, Ashizawa T, Saitoh Y, Saito H, Gomi K, Okabe M. Synthesis and antitumor activity of leinamycin derivatives: modifications of C-8 hydroxy and C-9 keto groups. Bioorg. Med. Chem. Lett. 1998;8:909–912. doi: 10.1016/s0960-894x(98)00133-4. [DOI] [PubMed] [Google Scholar]
- 7.Kanda Y, Ashizawa T, Kakita S, Takahashi Y, Kono M, Yoshida M, Saitoh Y, Okabe M. Synthesis and antitumor activity of novel thioester derivatives of leinamycin. J. Med. Chem. 1999;42:1330–1332. doi: 10.1021/jm9900366. [DOI] [PubMed] [Google Scholar]
- 8.Ashizawa T, Kawashima K, Kanda Y, Gomi K, Okabe M, Ueda K, Tamaoki T. Antitumor activity of KF22678, a novel thioester derivative of leinamycin. Anticancer Drugs. 1999;10:829–836. doi: 10.1097/00001813-199910000-00006. [DOI] [PubMed] [Google Scholar]
- 9.Kanda Y, Ashizawa T, Kawashima K, Ikeda S, Tamaoki T. Synthesis and antitumor activity of novel C8-ester derivatives of leinamycin. Bioorg. Med. Chem. Lett. 2003;13:455–458. doi: 10.1016/s0960-894x(02)00949-6. [DOI] [PubMed] [Google Scholar]
- 10.Behroozi SB, Kim W, Gates KS. The Reaction of n-Propanethiol With 3H-1,2-Benzodithiol-3-one 1-oxide and 5,5-Dimethyl-1,2-dithiolan-3-one 1-oxide: Studies Related to the Reaction of Antitumor Antibiotic Leinamycin With DNA. J. Org. Chem. 1995;60:3964–3966. [Google Scholar]
- 11.Breydo L, Gates KS. Thiol-triggered activation of leinamycin: a theoretical study. J. Org. Chem. 2002;67:9054–9060. doi: 10.1021/jo020568l. [DOI] [PubMed] [Google Scholar]
- 12.Zang H, Breydo L, Mitra K, Dannaldson J, Gates KS. DNA alkylation by leinamycin can be triggered by cyanide and phosphines. Bioorganic Med. Chem. Lett. 2001;11:1511–1515. doi: 10.1016/s0960-894x(01)00196-2. [DOI] [PubMed] [Google Scholar]
- 13.Chatterji T, Kizil M, Keerthi K, Chowdhury G, Posposil T, Gates KS. Small molecules that mimic the thiol-triggered alkylating properties seen in the natural product leinamycin. J. Am. Chem. Soc. 2003;125:4996–4997. doi: 10.1021/ja029169y. [DOI] [PubMed] [Google Scholar]
- 14.Asai A, Hara M, Kakita S, Kanda Y, Yoshida M, Saito H, Saitoh Y. Thiol-mediated DNA alkylation by the novel antitumor antibiotic leinamycin. J. Am. Chem. Soc. 1996;118:6802–6803. [Google Scholar]
- 15.Asai A, Saito H, Saitoh Y. Thiol-independent DNA cleavage by a leinamycin degradation product. Bioorg. Med. Chem. 1997;5:723–729. doi: 10.1016/s0968-0896(97)00015-1. [DOI] [PubMed] [Google Scholar]
- 16.Keerthi K, Gates KS. Entering the leinamycin rearrangement via 2-(trimethylsilyl)ethyl sulfoxides. Org. Biomol. Chem. 2007;5:1595–1600. doi: 10.1039/b701179b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zang H, Gates KS. Sequence specificity of DNA alkylation by the antitumor natural product leinamycin. Chem. Res. Toxicol. 2003;16:1539–1546. doi: 10.1021/tx0341658. [DOI] [PubMed] [Google Scholar]
- 18.Breydo L, Zang H, Gates KS. Synthesis and noncovalent DNA-binding properties of thiazole derivatives related to leinamycin. Tetrahedron Lett. 2004;45:5711–5716. [Google Scholar]
- 19.Nooner T, Dutta S, Gates KS. Chemical properties of the leinamycin 2'-deoxyguanosine adduct. Chem. Res. Toxicol. 2004;17:942–949. doi: 10.1021/tx049964k. [DOI] [PubMed] [Google Scholar]
- 20.Gates KS, Nooner T, Dutta S. Biologically relevant chemical reactions of N7-alkyl-2'-deoxyguanosine adducts in DNA. Chem. Res. Toxicol. 2004;17:839–856. doi: 10.1021/tx049965c. [DOI] [PubMed] [Google Scholar]
- 21.Shipova K, Gates KS. A fluorimetric assay for the spontaneous release of an N7- alkylguanine residue from duplex DNA. Bioorganic Med. Chem. Lett. 2005;15:2111–2113. doi: 10.1016/j.bmcl.2005.02.058. [DOI] [PubMed] [Google Scholar]
- 22.Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Ann. Rev. Genet. 1986;20:201–230. doi: 10.1146/annurev.ge.20.120186.001221. [DOI] [PubMed] [Google Scholar]
- 23.Boiteux, S, Guillet, M. Abasic sites in DNA: repair and biological consequences in Saccaromyces cerevisiae. DNA Repair. 2004;3:1–12. doi: 10.1016/j.dnarep.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Wilson DM, III, Barsky, D. The major human abasic endonuclease: formation, consequences and repair of abasic sites in DNA. Mutation Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 25.Kelley, MR, Parsons, SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref1. Antioxidants Redox Signalling. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- 26.Lhomme J, Constant J-F, Demeunynck M. Abasic DNA structure, reactivity, and recognition. Biopolymers. 2000;52:65–83. doi: 10.1002/1097-0282(1999)52:2<65::AID-BIP1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 27.Bailly V, Verly WG. Possible roles of beta-elimination and gamma-elimination reactions in the repair of DNA containing AP (apurinic/apyrimidinic) sites in mammalian cells. Biochem. J. 1988;253:553–559. doi: 10.1042/bj2530553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lindahl T, Andersson A. Rate of chain breakage at apurinic sites in double-stranded deoxyribonucleic acid. Biochemistry. 1972;11:3618–3623. doi: 10.1021/bi00769a019. [DOI] [PubMed] [Google Scholar]
- 29.Dutta S, Chowdhury G, Gates KS. Interstrand crosslinks generated by abasic sites in duplex DNA. J. Am. Chem. Soc. 2007;129:1852–1853. doi: 10.1021/ja067294u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutation Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 31.Wilson DM, III, Barsky D. The major human abasic endonuclease: formation,consequences and repair of abasic sites in DNA. Mutation Res. 2001;485:283–307. doi: 10.1016/s0921-8777(01)00063-5. [DOI] [PubMed] [Google Scholar]
- 32.Lomax ME, Cunniffe S, O’Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 33.Rinne ML, He Y, Pachkowski BF, Nakamura J, Kelley MR. N-methylpurine DNA glycosylase overexpression increases alkylation sensitivity by rapidly removing non-toxic 7-methylguanine adducts. Nucleic Acids Res. 2005;33:2859–2867. doi: 10.1093/nar/gki601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fishel ML, He Y, Smith ML, Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin. Cancer Res. 2007;13:260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 35.Fishel ML, Seo YR, Smith ML, Kelley MR. Imbalancing the DNA base excision repair pathway in the mitochondria; targeting and overexpressing N-methylpurine DNA glycosylase in mitochondria leads to enhanced cell killing. Cancer Res. 2003;63:608–615. [PubMed] [Google Scholar]
- 36.Rinne ML, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol. Cancer Ther. 2004;3:955–967. [PubMed] [Google Scholar]
- 37.Behroozi SJ, Kim W, Dannaldson J, Gates KS. DNA Cleavage by 1,2-Dithiolan-3-one 1-Oxides: A Class of Thiol-Activated DNA Cleaving Agents Structurally Related to the Antitumor Antibiotic Leinamycin. Biochemistry. 1996;35:1768–1774. doi: 10.1021/bi952257t. [DOI] [PubMed] [Google Scholar]
- 38.Mitra K, Kim W, Daniels JS, Gates KS. Oxidative DNA cleavage by the antitumor antibiotic leinamycin and simple 1,2-dithiolan-3-one 1-oxides: Evidence for thiol-dependent conversion of molecular oxygen to DNA-cleaving oxygen radicals mediated by polysulfides. J. Am. Chem. Soc. 1997;119:11691–11692. [Google Scholar]
- 39.Chatterji T, Gates KS. DNA Cleavage by 7-Methylbenzopentathiepin: A Simple Analog of the Antitumor Agent Varacin. Bioorg. Med. Chem. Lett. 1998;8:535–538. doi: 10.1016/s0960-894x(98)00066-3. [DOI] [PubMed] [Google Scholar]
- 40.Chatterji T, Gates KS. Reaction of thiols with 7-methylbenzopentathiepin. Bioorganic Med. Chem. Lett. 2003;13:1349–1352. doi: 10.1016/s0960-894x(03)00103-3. [DOI] [PubMed] [Google Scholar]
- 41.Chatterji T, Keerthi K, Gates KS. Generation of reactive oxygen species by a persulfide (BnSSH) Bioorg. Med. Chem. Lett. 2005;15:3921–3924. doi: 10.1016/j.bmcl.2005.05.110. [DOI] [PubMed] [Google Scholar]
- 42.Breydo L, Gates KS. Thiol-dependent DNA cleavage by 3H-1,2-benzodithiol-3-one 1,1-dioxide. Bioorg. Med. Chem. Lett. 2000;10:4242–4246. doi: 10.1016/s0960-894x(00)00125-6. [DOI] [PubMed] [Google Scholar]
- 43.Sivaramakrishnan S, Gates KS. Possible mechanisms underlying the antitumor activity of S-deoxyleinamycin. Bioorg. Med. Chem. Lett. 2008;18:3076–3080. doi: 10.1016/j.bmcl.2007.11.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Halliwell B, Gutteridge JMC. Role of free radicals and catalytic metal ions in human disease: an overview. Methods Enzymol. 1990;186:1–85. doi: 10.1016/0076-6879(90)86093-b. [DOI] [PubMed] [Google Scholar]
- 45.Finkel T. Oxidant signals and oxidative stress. Curr. Opin. Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 46.Finkel T, Holbrook NJ. Oxidants, oxidative stress and the biology of ageing. Nature. 2000;408:239–247. doi: 10.1038/35041687. [DOI] [PubMed] [Google Scholar]
- 47.Winterbourn CC. Reconciling the chemistry and biology of reactive oxygen species. Nat. Chem. Biol. 2008;4:278–286. doi: 10.1038/nchembio.85. [DOI] [PubMed] [Google Scholar]
- 48.Ozben T. Oxidative stress and apoptosis: impact on cancer therapy. J. Pharm. Sci. 2007;96:2181–2196. doi: 10.1002/jps.20874. [DOI] [PubMed] [Google Scholar]
- 49.Bassett S, Urrabaz R, Sun D. Cellular response and molecular mechanism of antitumor activity by leinamycin in MiaPaCa human pancreatic cancer cells. Anti-Cancer Drugs. 2004;15:689–696. doi: 10.1097/01.cad.0000136886.72917.6f. [DOI] [PubMed] [Google Scholar]
- 50.Barkley LR, Ohmori H, Vaziri C. Integrating S-phase checkpoint signaling with trans-lesion synthesis of bulky DNA adducts. Cell Biochem. Biophys. 2007;47:392–408. doi: 10.1007/s12013-007-0032-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gates KS. Covalent Modification of DNA by Natural Products. In: Kool ET, editor. Comprehensive Natural Products Chemistry. New York: Pergamon; 1999. pp. 491–552. [Google Scholar]
- 52.Gates KS. Mechanisms of DNA Damage by Leinamycin. Chem. Res. Toxicol. 2000;13:953–956. doi: 10.1021/tx000089m. [DOI] [PubMed] [Google Scholar]
- 53.Kubo K, Ide H, Wallace SS, Kow YW. A novel, sensitive, and specific assay for abasic sites, the most commonly produced DNA lesion. Biochemistry. 1992;31:3703–3708. doi: 10.1021/bi00129a020. [DOI] [PubMed] [Google Scholar]
- 54.Atamna H, Cheung I, Ames BN. A method for detecting abasic sites in living cells: age-dependent changes in base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2000;97:686–691. doi: 10.1073/pnas.97.2.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collins AR. The comet assay for DNA damage and repair: principles, applications, and limitations. Mol. Biotechnol. 2004;26:249–261. doi: 10.1385/MB:26:3:249. [DOI] [PubMed] [Google Scholar]
- 56.Horvathova E, Slamenova D, Hlincikova L, Mandal TK, Gabelova A, Collins AR. The nature and origin of DNA single-strand breaks determined with the comet assay. Mutat. Res. 1998;409:163–171. doi: 10.1016/s0921-8777(98)00053-6. [DOI] [PubMed] [Google Scholar]
- 57.Buxser SE, Sawada G, Raub TJ. Analytical and numerical techniques for evaluation of free radical damage in cultured cells using imaging cytometry and fluorescent indicators. Methods Enzymol. 1999;300:256–275. doi: 10.1016/s0076-6879(99)00133-0. [DOI] [PubMed] [Google Scholar]
- 58.Gates KS. Chemical reactions of DNA damage and degradation. In: Platz MSM, R A, Jones M Jr, editors. Reviews of Reactive Intermediates. Hoboken: John Wiley and Sons, Inc; 2007. pp. 333–378. [Google Scholar]
- 59.Greenberg MM. Investigating nucleic acid damage processes via independent generation of reactive intermediates. Chem. Res. Toxicol. 1998;11:1235–1248. doi: 10.1021/tx980174i. [DOI] [PubMed] [Google Scholar]
- 60.Breen AP, Murphy JA. Reactions of oxyl radicals with DNA. Free Rad. Biol. Med. 1995;18:1033–1077. doi: 10.1016/0891-5849(94)00209-3. [DOI] [PubMed] [Google Scholar]
- 61.Greenberg MM. Elucidating DNA damage and repair processes by independently generating reactive and metastable intermediates. Org. Biomol. Chem. 2007;5:18–30. doi: 10.1039/b612729k. [DOI] [PubMed] [Google Scholar]
- 62.Pratviel G, Bernadou J, Meunier B. Carbon-hydrogen bonds of DNA sugar units as targets for chemical nucleases and drugs. Angew. Chem. Int. Ed. Eng. 1995;34:746–769. [Google Scholar]
- 63.Asaeda A, Ide H, Tano K, Takamori Y, Kubo K. Repair kinetics of abasic sites in mammalian cells selectively monitored by the aldehyde reactive probe (ARP) Nucleosides Nucleotides. 1998;17:503–513. doi: 10.1080/07328319808005194. [DOI] [PubMed] [Google Scholar]
- 64.Male R, Fosse VM, Kleppe K. Polyamine-induced hydrolysis of apurinic sites in DNA and nucleosomes. Nucleic Acid Res. 1982;10:6305–6318. doi: 10.1093/nar/10.20.6305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nakamura J, Swenberg JA. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999;59:2522–2526. [PubMed] [Google Scholar]