Abstract
Background
Mutations of PCSK9 are associated cross-sectionally with plasma LDL cholesterol (LDL-C) levels, but, little is known about their longitudinal association with LDL-C levels from young adulthood to middle age.
Methods and Results
We investigated the associations of 6 PCSK9 variants with LDL-C over 20 years in 1750 African Americans and 1828 whites from the CARDIA study. Generalized estimating equations were used to assess longitudinal differences in LDL-C levels between genotype categories. For African Americans, LDL-C levels at age 18 were significantly lower (p<0.001) among those with 3 genetic variants (L253F, C679X, and Y142X; 81.5 mg/dL) and A443T (95.5 mg/dL) compared with non-carriers (109.6 mg/dL). The difference in LDL-C levels from non-carriers tended to widen for those with the 3 variants only, by 0.24 mg/dl per year of age (p=0.14). For whites with the R46L variant, compared with non-carriers, LDL-C levels at age 18 were significantly lower (84.4 vs 100.9 mg/dL, p<0.001), and the increase in LDL-C with age was similar to non-carriers. The 3 genetic variants and the A443T variant in African American men were associated with lower carotid intima-media thickness and lower prevalence of coronary calcification measured at ages 38~50.
Conclusions
Our results suggest that participants with several genetic variants of PCSK9 have persistently lower serum LDL-C levels than non-carriers from ages 18–50. Such long-term reduction in LDL-C levels is associated with reduced subclinical atherosclerosis burden in African-American men.
Keywords: PCSK9, LDL-C, genetic variant, longitudinal study
Introduction
Elevated LDL cholesterol (LDL-C) is a major causal risk factor for atherosclerosis and its most common clinical manifestation, coronary heart disease (CHD). In addition to lifestyle, variants in genes coding for proteins such as apolipoprotein E (APOE), low-density lipoprotein receptor (LDLR) and apolipoprotein B (APOB) have been shown to play an important role in the regulation of low density lipoprotein (LDL) homeostasis1. The recent discovery of several variants of proprotein convertase subtilisin kexin type 9 (PCSK9) has suggested its role in LDL metabolism2–6. Two independent microarray studies7, 8 support a role for PCSK9 in sterol metabolism, and adenoviral-mediated over-expression of PCSK9 in mouse liver depletes hepatic LDL-receptor protein. A landmark epidemiological study9 by Cohen et al. also demonstrated that individuals with two nonsense mutations in PCSK9, Y142X and C679X, in African Americans and one nonsense mutation, R46L, in whites have significantly lower LDL-C levels in middle-age, and dramatically lower subsequent incidence of CHD. As a result, PCSK9 has emerged as a promising therapeutic target for lowering plasma LDL-C levels10.
Novel variants associated with LDL-C levels have been reported in various studies from ethnically diverse populations11–16. However, several of these variants have not been validated in large epidemiological studies. Moreover, little is known about the long-term trends of LDL-C levels associated with various variants of PCSK9 gene. In this study, we examined the association between 6 genetic variants of PCSK9 (R46L (rs11591147), A443T (rs28362263), L253F, C679X (rs28362286), Y142X, and E6709G (rs505151)) and longitudinal change with aging of LDL-C from examinations in year 0 to year 20 among 1,750 African American and 1,828 white healthy men and women enrolled in the Coronary Artery Risk Development In Young Adults (CARDIA) study. In order to determine whether variant-associated differences in LDL-C levels through young adulthood to middle age may also be associated with differences in development of subclinical atherosclerosis, we examined the association of these 6 genetic variants with carotid intimal-medial thickness (IMT) and coronary artery calcium (CAC) in middle age.
Methods
Study sample
The CARDIA Study is a multicenter longitudinal study examining cardiovascular risk factor development in young adults. The purpose of CARDIA is to study factors related to the evolution of cardiovascular disease risk factors by following a cohort from young adulthood to middle age. Details of participant enrollment and study procedures have been published elsewhere.17 Briefly, 5,115 African-American and white men and women aged 18 to 30 years were enrolled in 1985–1986 at four centers in Birmingham, AL (University of Alabama at Birmingham), Chicago, IL (Northwestern University), Minneapolis, Minnesota (University of Minnesota), and Oakland, CA (Kaiser Permanente Oakland). Participant recruitment was approximately balanced on age, sex, race, and education status at each center. Participants were examined at baseline (Year 0) and at follow-up examinations in years 2 (1987–1988), 5 (1990–1991), 7 (1992–1993), 10 (1995–1996), 15 (2000–2001), and 20 (2005–2006). At these follow-up visits, 90, 86, 81, 79, 74, and 72%, respectively, completed the examination. All participants have signed informed consent at each examination, and all study protocols have been approved by the institutional review boards at each site. Each participant’s age, race, and sex were self-reported during the recruitment phase and verified during the baseline clinic visit. Blood samples for measurement of blood lipids were collected according to standardized CARDIA protocols,17 after participants had fasted for 12 hours, and were processed at central laboratories as described previously.17, 18
Use of LDL-lowering medication was also self-reported and only available for Year 5, 7, 10, 15, and 20 examinations. For those on lipid-lowering medication, the fasting blood sample was collected before they took their medications that day. No special treatment was performed on blood samples of participants who used lipid-lowering medication. At each examination, weight and height were measured with subjects in light clothing and without shoes. Body weight was measured to the nearest 0.1 kg, using a calibrated scale. Height was measured to the nearest 0.5 cm with a vertical ruler. Body mass index (BMI) was calculated as the weight in kilograms divided by the square of height in meters. Carotid IMT was measured at the Year 20 examination in CARDIA using high-resolution B-mode ultrasound. Images were acquired bilaterally for both the common and internal carotid arteries in accordance with a previous published protocol19. The maximal IMT of the common carotid artery was defined as the mean of the maximal IMT of the near and far wall on both the left and right sides, measured 10 mm proximal to the common carotid bulb. The internal carotid IMT included the mean of the maximal IMT of the near and far wall on both the left and right sides in the internal artery exclusive of the carotid bulb. Coronary artery calcium was assessed by two computed tomography (CT) scans for each participant using electron beam CT with a standardized protocol20. For each scan, 40 consecutive images from the root of the aorta to the apex of the heart were obtained. Scans were read centrally by a trained reader who examined each participant’s scan independently of the other. The reader identified a region of interest for each potential focus of CAC, defined as 4 or more adjacent pixels (1.87 mm2) with a CT number > 130 HU (field of view = 35 cm). Agatston scores were adjusted for between-center differences using a standard calcium phantom scanned underneath each participant, and summed across the 4 major coronary arteries to compute a total calcium score. The presence of CAC was defined as having a positive, non-zero Agatston score, using the average of 2 scans.
Genotype data
Participants eligible for the current study included 4,119 individuals who consented to isolation of genomic DNA from a blood sample obtained at the year 10 or year 15 examinations. Each of the 6 PCSK9 polymorphisms was genotyped using the TaqMan assay (Applied Biosystems, Foster City, CA) as previously described21. Primers and probe sequences are available upon request. Polymorphism genotyping in the CARDIA study adheres to a rigorous quality control (QC) program, which includes barcode identification of samples, robotic sample handling, and blind replicate genotype assessment on 5% of the total sample (n=195). The genotyping success rate was 94%, 94%, 93.4%, 95%, 96%, 96%, and 94% for E670G, A443T, L253F, C697X, Y142X, and R46L, respectively. As a result, the number of participants with missing genotyping data for each of the 6 PCSK9 polymorphisms ranged from 162 to 271.
Statistical analysis
To assess the longitudinal association of PCSK9 genetic variants with LDL-C levels, generalized estimating equations (GEEs) were utilized with PCSK9 genetic variants, time-dependent age, and interaction term of PCSK9 genetic variants and time-dependent age as the independent variables, adjusting for gender, lipid medication use, time-dependent BMI, and examinations represented by 6 dummy variables. Separate analyses were performed for African American and white participants. Participants without any of these 6 genetic variants (“non-carriers”) were considered as the reference group for their race. LDL-C at age 18 and LDL-C change per year were computed for E670G, A443T, and the 3 combined low-prevalence genetic variants (L253F, C679X, and Y142X) and compared with non-carriers among African Americans; only the R46L genetic variant was used for comparison among whites, given very low/absent prevalence of the other variants in whites. The same GEE procedure with exclusion of lipid medication use as an independent variable was also used to assess the association of PCSK9 genetic variants with HDL-C levels as a comparison, given that PCSK9 is not known to have any direct association with HDL-C levels in other studies9, 15. Finally, multiple linear regression models and multiple logistic regression models were performed to assess the association of these genetic variants with carotid artery IMT measures and CAC prevalence at the year 20 examination, respectively. Data analysis was performed using SAS 9.1 and R 2.4.0.
Results
Of the final 1,750 African American participants for data analysis, 46 men and 78 women were homozygous for the E670G allele, 148 and 169, respectively, were heterozygous or homozygous for the A443T allele, and 18 and 29 had at least one of the 3 genetic variants (L253F, C679X, and Y142X); there were 534 male and 728 female non-carriers. For the 1,828 white participants included, 30 men and 30 women were heterozygous or homozygous for the R46L allele, with 852 male and 916 female non-carriers. Baseline anthropometric characteristics and lipid profiles from the examination are shown in Table 1. Total cholesterol and LDL-C levels are significantly lower among those with A443T, 3 genetic variants, or R46L when compared with non-carriers. The minor allele frequency was 24.8% for E670G, 10% for A443T, 0.66% for C679X, 0.46% for L253F, 0.28% for Y142X, and 1.64% for R46L. There was no significant association between variants of PCSK9 and gender in both race groups (p=0.25 for African Americans, p=0.89 for whites, χ2 test). Changes in BMI over the 20 year period of time were similar among genetic variant groups and non-carriers within each race (data not shown). No deviation from Hardy-Weinberg equilibrium was observed among the PCSK9 genetic variants except for E670G (p=0.04, χ2 test).
Table 1.
Baseline characteristics among PCSK9 variants.
| African American | White | |||||||
|---|---|---|---|---|---|---|---|---|
| Non-carriers (n=1262) |
E670G (n=124) |
A443T (n=317) |
3 variants† (n=47) |
Non-carriers (n=1768) |
R46L (n=60) |
|||
| Male (%) | 42.3 | 37.1 | 46.7 | 38. | 48.2 | 50.0 | ||
| Age | 24.5±3.8 | 24.2±4.1 | 24.1±3.6 | 24.1±4.0 | 25.5±3.4 | 26.0±3.7 | ||
| BMI | 25.2±5.6 | 25.6±6.5 | 25.8±6.5 | 25.1±5.9 | 23.8±4.1 | 23.4±3.5 | ||
| SBP (mm Hg) | 111.4±11.0 | 111.0±10.7 | 111.7±10.5 | 110.4±9.6 | 109.5±10.9 | 108.7±11.3 | ||
| DBP (mm Hg) | 68.8±10.5 | 69.1±10.0 | 68.8±9.5 | 66.9±9.5 | 68.5±9.1 | 68.9±9.0 | ||
| HDL (mg/dL) | 54.8±13.4 | 55.4±15.2 | 54.5±12.3 | 52.7±11.0 | 51.9±13.2 | 54.7±12.9 | ||
| LDL (mg/dL) | 112.6±31.7 | 117.0±30.5 | 103.4±31.2§ | 78.9±22.9§ | 109.2±29.5 | 92.2±26.3§ | ||
| Cholesterol (mg/dL) | 180.7±33.7 | 184.6±32.7 | 170.4±33.4§ | 142.4±25.8§ | 176.8±32.2 | 160.9±26.1§ | ||
| Triglyceride (mg/dL) | 66.9±37.1 | 65.2±34.3 | 63.5±38.6 | 56.2±27.4‡ | 79.4±57.8 | 69.9±40.2 | ||
| Smoker (%) | 34.0 | 33.9 | 29.6 | 32.6 | 25.4 | 28.3 | ||
BMI: body mass index; SBP: systolic blood pressure; DBP: diastolic blood pressure; LDL: low-density lipoprotein; HDL: high-density lipoprotein
L253F, C679X, and Y142X
p<0.05 when compared with non-carriers;
p<0.01 when compared with non-carriers
Baseline and longitudinal differences in LDL-C levels
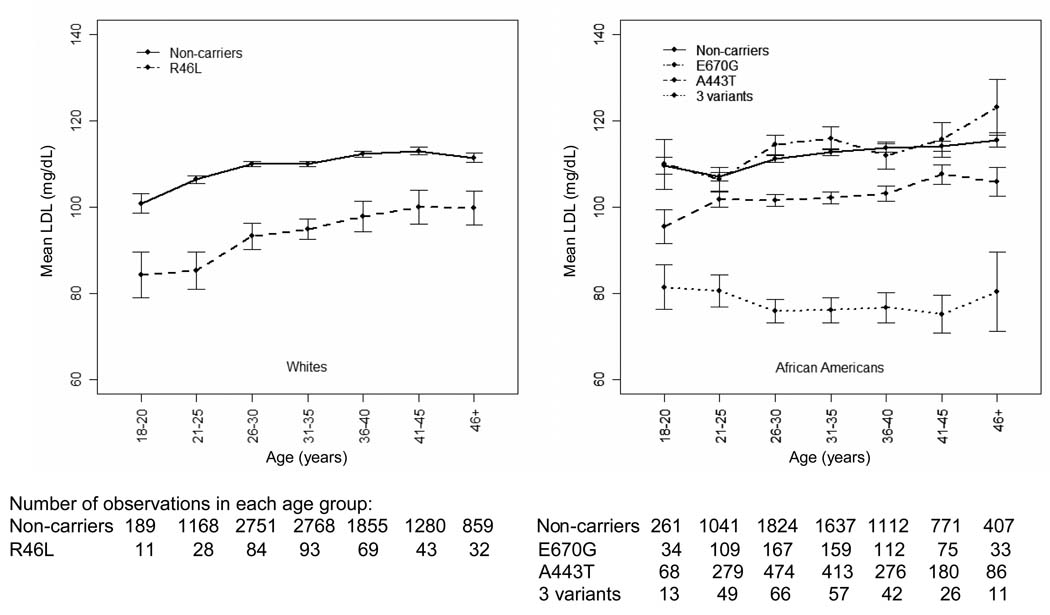
To illustrate the change of LDL-C levels over the 20 years of follow-up, we pooled all the data from the seven CARDIA examinations and calculated the unadjusted mean LDL-C levels in participants at seven age groups over 20 years of follow-up as shown in Figure 1 for African Americans and whites separately, by genotype. For whites, mean LDL-C levels in the R46L variant group (mean±SD=84.4±17.5 mg/dL) were significantly lower (p<0.01) than in non-carriers (100.9±30.3 mg/dL) at ages 18 to 20. This difference diminished slightly over the 20 year period of follow-up, because the R46L variant group had a greater increase in LDL-C levels than non-carriers (99.8±21.9 vs 111.4±31.5 mg/dL at age 46 or above, p=0.039). For African Americans, the E670G variant group had similar mean LDL-C levels as non-carriers in all ages. Conversely, the mean LDL-C levels in both the 3 genetic variants group (81.5±18.5 mg/dL) and the A443T variant group (95.5±32.1 mg/dL) were significantly lower than for non-carriers (109.6±32.0 mg/dL) at ages 18 to 20 years (p<0.01 for both), and the mean difference between the 3 gene variants and non-carriers widened through age 46 or above (80.5±30.3 vs 115.5±33.5 mg/dL, p<0.01). The Y142X variant was associated with the lowest mean LDL-C levels among the 3 variants in all age groups (supplementary Figure 1). The mean differences between the A443T variant and the non-carriers remained similar over the 20 year period of time (p<0.05 for all 7 age groups). Notably, mean LDL-C levels in the 3 genetic variants group remained approximately the same (i.e., did not increase) over the 20 years, a pattern unique to participants with these 3 genetic variants.
Figure 1. Unadjusted mean LDL-C levels of PCSK9 genetic variants in different age groups among whites and African Americans.
•3 variants: L253F, C679X, and Y142X
Results of the GEE models examining the longitudinal changes of LDL-C with age in both race groups, adjusted for sex, time-dependent lipid medication use, time-dependent BMI, and examination are shown in Table 2. White non-carriers had a mean 0.83 mg/dL LDL-C increase per year (p<0.001). When compared to non-carriers, participants with the R46L variant had a mean 16.2 mg/dl lower LDL-C at age 18 (p<0.001), and a non-significant increase in LDL-C level over the 20 years was observed (0.02 mg/dL per year; p=0.90 compared with non-carriers). As expected, mean LDL-C levels in men were higher than women (by 8.22 mg/dL, p<0.001). Furthermore, each one unit of BMI higher was associated with a mean 1.33 mg/dL higher LDL-C level (p<0.001).
Table 2.
Adjusted longitudinal association of LDL-C levels with PCSK9 genetic variants among African Americans and whites.
| African American | ||||
|---|---|---|---|---|
| Variable | Difference in LDL-C (mg/dL) at age 18 |
P | Annual change in LDL-C s(mg/dL) |
P† |
| Non-carrier | Reference | 0.73 (0.41, 1.06) | ||
| E670G | 2.74† (−3.65, 9.13) | 0.40 | 0.63 (0.21, 1.05) | 0.47 |
| A443T | −9.69† (−13.52, −5.86) | <0.001 | 0.70 (0.35, 1.05) | 0.69 |
| 3 variants‡ | −30.63† (−37.71, −23.55) | <0.001 | 0.49 (0.04, 0.94) | 0.14 |
| Male (vs female) | 4.72 (2.12, 7.32) | <0.001 | ||
| Lipid medication (yes vs no) | −33.58 (−40.34, −26.81) | <0.001 | ||
| BMI (per 1 unit increase) | 1.27 (1.11, 1.43) | <0.001 | ||
| White | ||||
| Non-carrier | Reference | 0.83 (0.50, 1.16) | ||
| R46L | −16.20† (−23.29, −9.11) | <0.001 | 0.85 (0.41, 1.28) | 0.90 |
| Male (vs female) | 8.22 (5.76, 10.68) | <0.001 | ||
| Lipid medication (yes vs no) | −39.22 (−44.68, −33.75) | <0.001 | ||
| BMI (per 1 unit increase) | 1.33 (0.75, 1.91) | <0.001 | ||
Compared with non-carriers and adjusted for year of examination.
L253F, C679X, and Y142X
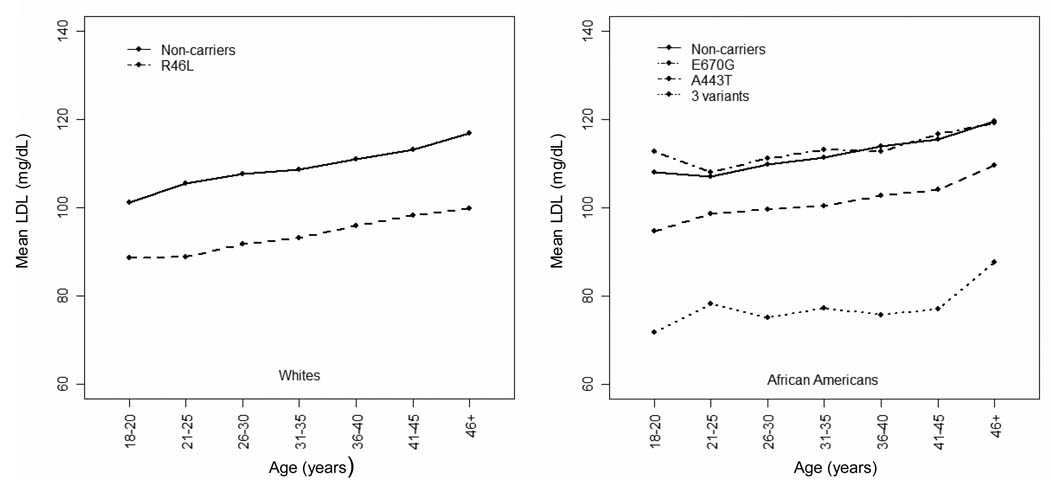
For African Americans, when compared with African American non-carriers, participants with the 3 genetic variants had a substantially lower mean LDL-C level (by 30.63 mg/dL, p<0.001) at age 18. The A443T variant group also had significantly lower LDL-C at age 18 (by 9.69 mg/dL, p<0.001), whereas there was no statistically significant difference in those with E670G (p=0.40). Notably, the mean difference in LDL-C levels tended to widen by 0.24 mg/dL per year of age between the 3 genetic variants and non-carriers (p=0.14). No significant differences in changes of LDL-C per year of age were observed over the 20-year period of time between the E670G or A443T variant groups and non-carriers. Similarly to whites, mean LDL-C levels in women were lower than men by 4.72 mg/dL (p<0.001). Each one unit of BMI higher was associated with a mean 1.27 mg/dL greater LDL-C levels (p<0.001). To illustrate the trend for adjusted mean LDL-C levels wit age groups, we used the same GEE procedure but replaced the continuous age variable with 7 age groups as shown in Figure 1. The adjusted mean LDL-C levels for PCSK9 variants by race are shown in Figure 2.
Figure 2. Adjusted* mean LDL-C levels of PCSK9 genetic variants in different age groups among whites and African Americans.
*Adjusted for: sex, time-dependent lipid medication use, time-dependent body mass index, and year of examination.
•3 variants: L253F, C679X, and Y142X
Additional gender-specific analyses were performed using the same GEE model for both African Americans and whites. For African Americans, the effects of PCSK9 variants on LDL-C levels were similar between men and women. However, the mean difference in longitudinal changes in LDL-C levels between the 3 genetic variants and non-carriers became less significant (p=0.22 and 0.25 for men and women, respectively), possibly due to smaller sample sizes. For whites, compared to non-carriers, the R46L variant group had similarly lower LDL-C levels at age 18 by 18.51 mg/dL (p<0.001) and 13.47 mg/dL (p=0.004) for men and women, respectively. The mean LDL-C levels widened by 0.33 mg/dL per year for women (p=0.04, compared with non-carriers), while a non-significant increase in LDL-C level over the 20 years was observed for men (0.32 mg/dL per year, p=0.21).
Acknowledging that LDL-lowering medication use has significant impact on the LDL-C level, we repeated the above analysis by excluding participants with LDL-lowering medication use in the GEE models. The numbers of participants on LDL-lowering medication at the Year 5, 7, 10 examinations were only <1%. At the Year 15 examination, the use of LDL-lowering medication were 1.7%, 4.5%, and 1% among African American non-carriers, E670G, and LDL-lowering variants, respectively, and 3.1% and 0% among white non-carriers and R46L variant, respectively. At the Year 20 examination, there were 7.4%, 10.7%, and 5.5% of participants with LDL-lowering medication use among African American non-carriers, E670G, and LDL-lowering variants, respectively, and 9.9% and 5.8% among white non-carriers and R46L variant, respectively. After excluding those with LDL-lowering medications use in our multivariate GEE models, our results remained similar except for the mean difference in LDL-C levels with age between the 3 variants and non-carriers among African Americans; the difference has now widened to 0.37 mg/dL per year of age and has become statistically significant (p=0.005). This significant difference persists even in the gender-specific analysis.
Due to the possibility that LDL-C levels may be influenced by lifestyle and nutrient intake patterns, we further considered fat intake and physical activity as potentially confounding factors. However, after adjusting for the Keys score and total/heavy physical activity in our GEE procedure. There were no substantive difference in the association between PCSK9 variants and LDL-C levels.
Longitudinal changes of HDL-C levels
We also investigated the association of PCSK9 variants with longitudinal changes in HDL-C levels. No association has been reported so far between variants of PCSK9 and variation in HDL-C. The same GEE models as for the LDL analysis were used. In whites, when compared to non-carriers, participants with the R46L variant tended to have a higher mean HDL-C level, by 3.5 mg/dL, at age 18 (p=0.05), and this disparity was maintained with aging. In African Americans, after adjusting for gender, age, BMI, and 6 dummy variables for examination, no significant differences in mean HDL-C levels were observed in any variant groups when compared with non-carriers at age 18. Participants with the A443T variant had an additional increase of 0.09 mg/dL (p=0.048) mean HDL-C levels per year of age relative to non-carriers. Additional analyses were performed by adjusting for LDL-lowering medication use and excluding participants with any LDL-lowering medication. The results remain similar to those reported above.
Effect of PCSK9 and APOE on LDL-C levels
Gene-gene interactions could possibly affect the LDL-C levels. Therefore, we assessed the interaction between APOE and PCSK9. As shown in the supplementary Table 1, participants with the APOE ε2/ε2 or ε3/ε2 genotypes have the lowest mean LDL-C levels in each of the PCSK9 variant groups and non-carriers. On the other hand, among those with the APOE ε2/ε2 or ε3/ε2 genotypes, mean LDL-C levels were still lower among those carrying PCSK9 LDL-C lowering variants when compared to non-carriers. Therefore, an additive effect seems to exist on LDL-C levels between PCSK9 and APOE. However, when testing the differences in mean LDL-C levels across APOE genotypes within each PCSK9 LDL-C variant, only APOE ε2/ε2 or ε3/ε2 genotypes within the PCSK9 A443T variant showed significantly lower mean LDL-C levels (p<0.01) compared to other APOE genotypes. The non-significant result for other PCSK9 variants is likely due to small sample sizes in several PCSK9/APOE genotypes.
Association of PCSK9 genetic variants with carotid IMT and CAC
The mean IMT measures and CAC prevalence in each of the 6 PCSK9 variants, stratified by race and sex, at the Year 20 examination are shown in Table 3. Of note, among African American men, only 13% (12/92) in the A443T variant group and none (0/9) in the 3 genetic variants group had CAC > 0 as compared to 28% (89/318) in non-carriers. Mean IMT measures were also significantly lower among those with A443T variant when compared with non-carriers. A similar association was not observed among African American women. For whites, CAC prevalence was lower in both genders and IMT measure was also lower in female among those with R46L variant compared to non-carriers although none of these differences reached statistical significance. Further analysis for the association of PCSK9 variants with IMT measures using multiple linear regression and CAC prevalence using multiple logistic regression were performed separately for whites and African Americans by sex. The descriptive statistics of cardiovascular risk factors included in the regression model are shown in supplementary Table 2. After adjusting for diabetic status, smoking status, systolic blood pressure, sex, BMI, and age at the Year 20 examination, no significant difference in mean IMT measures was observed for any of the 6 variant groups when compared with non-carriers of the same race. However, significantly lower CAC prevalence among the A443T variant group remained among African American men (p=0.03, odds ratio 0.45, 95% CI [0.22, 0.93]). When combining the A443T variant and the 3 genetic variants in African American men, these 4 LDL-lowering variants had lower CAC prevalence (p=0.01, odds ratio 0.4, 95% CI [0.20,0.82]) as shown in Table 4. Further examination of CAC scores among African Americans with CAC>0 reveals similar distributions between non-carriers and those with these 4 LDL-lowering variants. As shown in Table 5, within both gender groups, the median CAC scores for those with the LDL-lowering variants were lower than the non-carriers. The 75th percentile CAC scores were lower among males, but not females, with the LDL-lowering variants; the 90th percentile CAC scores, however, were much lower for both males and females with the variants than for the non-carriers.
Table 3.
Carotid artery IMT (mm) and CAC prevalence (%) among PCSK9 genetic variants at the Year 20 examination.
| IMT*(mm) | CAC*(%) | |||
|---|---|---|---|---|
| Male | Female | Male | Female | |
| African American | ||||
| E670G | 0.85±0.13 (34) | 0.82±0.11 (49) | 20 (30) | 9 (44) |
| A443T | 0.83±0.13†(99) | 0.82±0.12 (115) | 13†(92) | 9 (108) |
| 3 variants | 0.84±0.11 (10) | 0.81±0.11 (19) | 0 (9) | 12 (17) |
| Non-carriers | 0.87±0.16 (318) | 0.81±0.12 (529) | 28 (318) | 11 (510) |
| White | ||||
| R46L | 0.80±0.12 (27) | 0.73±0.12 (24) | 24 (25) | 4.6 (22) |
| Non-carriers | 0.80±0.12 (677) | 0.74±0.10 (723) | 33 (669) | 11 (715) |
IMT: intima-medial thickness, mm; CAC: coronary artery calcium, % with positive, non-zero Agatston score; 3 variants: L253F, C679X, and Y142X
Number in the parenthesis is the number of participants for which IMT or CAC measures were available.
p<0.05 when compared with non-carriers. The t-test was used to calculate the p value for IMT, and the chi-square test for CAC prevalence.
Table 4.
Multiple logistic regression for the association of coronary artery calcium with PCSK9 LDL-lowering variants among African American men at the Year 20 examination.
| Variable | Odds Ratio† | 95% C.I. | P-value |
|---|---|---|---|
| 4 Variants‡ | 0.40 | (0.20, 0.82) | 0.01 |
| BMI (per 1 unit increase) | 1.09 | (1.03, 1.14) | 0.001 |
| Age (per 1 year increase) | 1.15 | (1.07, 1.23) | <0.001 |
| SBP (per 1 mm Hg increase) | 1.01 | (0.99, 1.03) | 0.20 |
| Diabetes | 1.16 | (0.53, 2.55) | 0.72 |
| Never Smoker | 0.34 | (0.19, 0.61) | <0.001 |
| Former Smoker | 0.37 | (0.16, 0.87) | 0.02 |
Compared with non-carriers.
L253F, C679X, Y142X, and A443T combined
Note: the outcome (coronary artery calcium) is binary, i.e., present versus absent.
Table 5.
Distribution of coronary artery calcium (CAC) score among African Americans with CAC>0 at the Year 20 examination.
| Sex | Variant | N | Median | 75th percentile | 90th percentile | Max | Mean |
|---|---|---|---|---|---|---|---|
| Male | 4 variants* | 12 | 23.8 | 77.0 | 117.1 | 121.5 | 43.7 |
| non-carriers | 89 | 32.7 | 99.3 | 271.2 | 6057.9 | 168.6 | |
| Female | 4 variants | 11 | 6.8 | 91.1 | 155.4 | 156.1 | 41.0 |
| non-carriers | 57 | 33.7 | 75.9 | 259.4 | 2025.3 | 108.6 | |
L253F, C679X, Y142X, and A443T combined
Discussion
In addition to LDLR and APOB, PCSK9 was recently identified as a novel gene involved in autosomal dominant hypercholesterolemia (ADH) from several familial studies22–24. Recent studies have also described several variants of PCSK9 among specific populations that are associated with particularly high or low plasma LDL-C levels13, 15, 22, 25, 26. In this study, we investigated six genetic variants of PCSK9 and their longitudinal association with LDL-C levels in CARDIA. Consistent with other studies, the R46L variant was more common in whites, whereas the other 5 variants (L253F, C679X, Y142X, A443T, and E670G) were more common in African Americans. We observed that the 3 low-prevalence genetic variants (L235, C679X, and Y142X) and the A443T variant among African Americans, and the R46L variant among whites, were associated with significantly lower LDL-C levels in young adulthood when compared with non-carriers. Such differences were preserved over 20 years of follow-up, from young adulthood to middle age. Furthermore, the mean LDL-C levels of those carrying the 3 genetic variants stayed approximately the same over the 20 years of follow-up; the difference in mean LDL-C levels between those carrying the 3 genetic variants and non-carriers increased by 0.25 mg/dL per year of age, implying that these 3 genetic variants of PCSK9 may provide strong regulation of plasma LDL-C levels.
Chen et el.13 reported an association between LDL-C levels and E670G among an African American population. Evans and Beil12 found the E670G SNP associated with hypercholesterolemia in men. However, such an association was not confirmed in our study population or in the study by Kotowski et al15. This discrepancy is most likely due to race and ethnic differences among these 4 studies. The study sample of Evans and Beil was men of European origin, and the majority (84%) of Chen’s sample was American whites. In contrast, our results regarding the E670G polymorphism are restricted to African Americans (124/1750=7%) since only 3 whites were found to have this PCSK9 genetic variant. Our study sample is comparable to that of Kotowski et al.15, which included 1,822 African Americans, 1,045 whites, and 601 Hispanics from the Dallas Heart Study.
In a recent landmark study, Cohen et al9 studied PCSK9 variants among 3363 African American and 9524 white participants of the Atherosclerosis Risk in Communities (ARIC) study. They observed that African American participants with the uncommon nonsense mutations C679X and Y142X (two of the 3 genetic variants group in the present study) had 28% lower mean plasma LDL-C levels (100 mg/dL vs. 138 mg/dL; P<0.001) at a mean age of 54 years. In a 15-year follow-up, African American participants with C679X or Y142X had an 88% lower rate of clinical CHD events compared with non-carriers (1.2% vs. 9.7%; P=0.008), despite similar levels of other risk factors except for hypertension, which was less prevalent at baseline among carriers (37% vs. 55%). Of note, African American participants who carried these mutations had significant but only modestly lower carotid IMT compared with non-carriers (0.70 mm vs, 0.73 mm; P=0.04). In Cohen’s study, whites with the R46L variant (3.2% of the study sample) had a mean LDL-C level that was 15% lower than non-carriers (116 mg/dL vs. 137 mg/dL), similar levels of other risk factors, and significant but only modestly lower carotid IMT (0.71 mm vs. 0.73 mm; P=0.005) at the baseline age. Carriers of R46L had 47% lower incidence of CHD during follow-up (6.3% vs. 11.8%; P=0.003).
In the present study, we found differences in LDL-C levels between the LDL-lowering variants (the 3 genetic variants and the A443T variant) and non-carriers in African Americans, and between the R46L variant and non-carriers in whites that were similar to those reported by Cohen et al. Because of the young age of the CARDIA sample, there have been very few clinical CHD events to date. We therefore examined measures of subclinical atherosclerosis, carotid IMT and CAC, which were obtained at a mean age of 45 years, a decade earlier than the baseline age of the ARIC sample. We observed significantly lower prevalence of CAC and lower mean IMT among African American men with the LDL-lowering variants (the 3 genetic variants and the A443T variant). Lower CAC prevalence or mean IMT trends were also observed among white men and women and African American women although these associations did not reach statistical significance. The lack of significant differences in subclinical atherosclerosis measures in these race/gender groups may well be due to the younger age of the CARDIA cohort and the limited statistical power for detecting the associations. A recent study by Loria et al.27 shows that levels of modifiable risk factors including LDL-C levels in the early adulthood are more informative predictors of CAC than concurrent levels in mid-life. Our results are consistent with these findings, suggesting that a long-term reduction in LDL-C levels from young adulthood to middle age may be associated with subclinical atherosclerosis burden, especially for CAC. In addition, it is worth noting that whereas the relative differences in LDL-C were similar in the current CARDIA and previously studied ARIC samples, the absolute levels of LDL-C were substantially lower in the overall CARDIA sample (even among those aged 45 and above), compared with ARIC. A recent study revealed that among whites, elevated total cholesterol levels in younger adults (at age 40) have limited impact on the 10-year risk of CHD, but lead to substantially higher lifetime risks of CHD28. Thus, the differentiation of subclinical atherosclerosis burden between the carriers of LDL-lowering variants and non-carriers may increase with aging. Further follow-up to older ages, as was possible in the ARIC study, is needed to determine whether clinical CHD event rates will differ between carriers and non-carriers of PCSK9 variants in CARDIA. It is also worth noting that carotid IMT and CAC do not directly measure the burden or likelihood of rupture of vulnerable, lipid-laden plaques in the coronary circulation, and therefore they may be imperfect surrogates for future risk of clinical CHD events.
A limitation of our study is the loss of follow-up of a substantial number of CARDIA participants over two decades. Those who were lost to follow up were more likely to be men, African American, less educated, or smokers. The GEE procedure assumes an MCAR (missing complete at random) mechanism to handle the missing data. To guard against this strong assumption, we also used mixed model to examine the association of PCSK9 variants with LDL-C levels and found the results to be similar to that from the GEE procedure. We further compared the baseline characteristics between those with and without loss of follow-up. We found no significant difference in their mean LDL-C levels. In addition to loss of follow-up, we also excluded from our analyses those who provided blood for DNA isolation but for whom genotyping data were unavailable. These missing genotyping data were due to the unsuccessful genotyping of the samples and not a factor of participants’ LDL-C levels or any clinical features. Therefore, their exclusion from our analyses should not bias our results.
The mechanism of action of PCSK9 on cholesterol levels has not been fully elucidated. An in vitro study of human cells by Dubuc et al3 shows that PCSK9 mRNA expression is upregulated by statins and cholesterol depletion by twofold more than that of LDLR. Maxwell et al.8 found that PCSK9 mRNA levels were decreased by twofold in livers of mice fed a cholesterol-rich diet. Later studies further show that PCSK9 impairs low-density lipoprotein receptor, and in turn, inhibits LDL clearance29while a depletion of PCSK9 in mice results in accelerated LDL clearance5. Zhao et al. show that the three variants prevent the secretion of mature PCSK9 by disrupting synthesis (Y142X), autocatalytic cleavage (L253F), or folding (C697X) of proteins while R46 and A443T have no detectable effect on the processing or secretion of PCSK916. Moreover, circulating PCSK9 reduces LDL receptor levels, and knocking out PCSK9 increases LDL receptor levels and lowers plasma cholesterol levels. Therefore, the hypocholesterolemia associated with PCSK9 deficiency may be caused primarily by increased LDL clearance and not by reduced lipoprotein production. Taken together, these observations support the notion that PCSK9 affects cholesterol homeostasis by promoting LDLR degradation. Our work suggests that specific genetic variants of PCSK9 have a strong role in LDL metabolism. Data from Cohen’s study and ours show no increased triglycerides levels among those carrying LDL-lowering variants. Therefore, loss of function of PCSK9 does not appear to be harmful. Further understanding of the molecular regulation of these mutations may lead to a more effective therapeutic management of lipid disorders.
Elevated LDL cholesterol (LDL-C) is a major causal risk factor for atherosclerosis and its most common clinical manifestation, coronary heart disease (CHD). The recent discovery of several variants of proprotein convertase subtilisin kexin type 9 (PCSK9) has suggested its important role in the regulation of low density lipoprotein homeostasis. In this study, we investigated the associations of 6 PCSK9 variants with LDL-C over a follow-up period of 20 years in 1750 African Americans and 1828 whites from the CARDIA study. Our results suggest that participants with several genetic variants of PCSK9 (including R46L, A443T, L253F, C679X, and Y142X) have persistently lower serum LDL-C levels than non-carriers from age 18 to 50 years. Such long-term lower LDL-C levels is associated with less subclinical atherosclerosis burden in African-American men. The mechanism of action of PCSK9 on cholesterol levels has not been fully elucidated. Further understanding of the molecular regulation of these variants may lead to more effective therapeutic management of lipid disorders.
Acknowledgments
We thank Laura Colangelo for her assistance with data preparation. We also thank all the reviewers for their valuable comments to improve this manuscript.
Funding Sources: The CARDIA study was supported by contracts N01-HC-95095, N01-HC-48047, N01-HC-48048, N01-HC-48049, and N01-HC-48050 from the National Heart, Lung, and Blood Institute, National Institutes of Health.
Footnotes
Conflict of Interest Disclosures: None
REFERENCES
- 1.Rader DJ, Cohen J, Hobbs HH. Monogenic hypercholesterolemia: new insights in pathogenesis and treatment. The Journal of clinical investigation. 2003;111(12):1795–1803. doi: 10.1172/JCI18925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 3.Dubuc G, Chamberland A, Wassef H, Davignon J, Seidah NG, Bernier L, Prat A. Statins upregulate PCSK9, the gene encoding the proprotein convertase neural apoptosis-regulated convertase-1 implicated in familial hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2004;24(8):1454–1459. doi: 10.1161/01.ATV.0000134621.14315.43. [DOI] [PubMed] [Google Scholar]
- 4.Grozdanov PN, Petkov PM, Karagyozov LK, Dabeva MD. Expression and localization of PCSK9 in rat hepatic cells. Biochem Cell Biol. 2006;84(1):80–92. doi: 10.1139/o05-155. [DOI] [PubMed] [Google Scholar]
- 5.Rashid S, Curtis DE, Garuti R, Anderson NN, Bashmakov Y, Ho YK, Hammer RE, Moon YA, Horton JD. Decreased plasma cholesterol and hypersensitivity to statins in mice lacking Pcsk9; Proceedings of the National Academy of Sciences of the United States of America; 2005. pp. 5374–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun XM, Eden ER, Tosi I, Neuwirth CK, Wile D, Naoumova RP, Soutar AK. Evidence for effect of mutant PCSK9 on apolipoprotein B secretion as the cause of unusually severe dominant hypercholesterolaemia. Hum Mol Genet. 2005;14(9):1161–1169. doi: 10.1093/hmg/ddi128. [DOI] [PubMed] [Google Scholar]
- 7.Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes; Proceedings of the National Academy of Sciences of the United States of America; 2003. pp. 12027–12032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maxwell KN, Soccio RE, Duncan EM, Sehayek E, Breslow JL. Novel putative SREBP and LXR target genes identified by microarray analysis in liver of cholesterol-fed mice. Journal of lipid research. 2003;44(11):2109–2119. doi: 10.1194/jlr.M300203-JLR200. [DOI] [PubMed] [Google Scholar]
- 9.Cohen JC, Boerwinkle E, Mosley TH, Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 10.Lambert G. Unravelling the functional significance of PCSK9. Curr Opin Lipidol. 2007;18(3):304–309. doi: 10.1097/MOL.0b013e3281338531. [DOI] [PubMed] [Google Scholar]
- 11.Miyake Y, Kimura R, Kokubo Y, Okayama A, Tomoike H, Yamamura T, Miyata T. Genetic variants in PCSK9 in the Japanese population: Rare genetic variants in PCSK9 might collectively contribute to plasma LDL cholesterol levels in the general population. Atherosclerosis. 2007 doi: 10.1016/j.atherosclerosis.2006.12.035. [DOI] [PubMed] [Google Scholar]
- 12.Evans D, Beil FU. The E670G SNP in the PCSK9 gene is associated with polygenic hypercholesterolemia in men but not in women. BMC medical genetics. 2006;7:66. doi: 10.1186/1471-2350-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen SN, Ballantyne CM, Gotto AM, Jr, Tan Y, Willerson JT, Marian AJ. A common PCSK9 haplotype, encompassing the E670G coding single nucleotide polymorphism, is a novel genetic marker for plasma low-density lipoprotein cholesterol levels and severity of coronary atherosclerosis. Journal of the American College of Cardiology. 2005;45(10):1611–1619. doi: 10.1016/j.jacc.2005.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Naoumova RP, Tosi I, Patel D, Neuwirth C, Horswell SD, Marais AD, van Heyningen C, Soutar AK. Severe hypercholesterolemia in four British families with the D374Y mutation in the PCSK9 gene: long-term follow-up and treatment response. Arteriosclerosis, thrombosis, and vascular biology. 2005;25(12):2654–2660. doi: 10.1161/01.ATV.0000190668.94752.ab. [DOI] [PubMed] [Google Scholar]
- 15.Kotowski IK, Pertsemlidis A, Luke A, Cooper RS, Vega GL, Cohen JC, Hobbs HH. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. American journal of human genetics. 2006;78(3):410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Z, Tuakli-Wosornu Y, Lagace TA, Kinch L, Grishin NV, Horton JD, Cohen JC, Hobbs HH. Molecular characterization of loss-of-function mutations in PCSK9 and identification of a compound heterozygote. American journal of human genetics. 2006;79(3):514–523. doi: 10.1086/507488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR, Jr, Liu K, Savage PJ. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41(11):1105–1116. doi: 10.1016/0895-4356(88)90080-7. [DOI] [PubMed] [Google Scholar]
- 18.Warnick GR, Mayfield C, Benderson J, Chen JS, Albers JJ. HDL cholesterol quantitation by phosphotungstate-Mg2+ and by dextran sulfate-Mn2+-polyethylene glycol precipitation, both with enzymic cholesterol assay compared with the lipid research method. American journal of clinical pathology. 1982;78(5):718–723. doi: 10.1093/ajcp/78.5.718. [DOI] [PubMed] [Google Scholar]
- 19.O'Leary DH, Polak JF, Kronmal RA, Savage PJ, Borhani NO, Kittner SJ, Tracy R, Gardin JM, Price TR, Furberg CD. Thickening of the carotid wall. A marker for atherosclerosis in the elderly? Cardiovascular Health Study Collaborative Research Group. Stroke. 1996;27(2):224–231. doi: 10.1161/01.str.27.2.224. [DOI] [PubMed] [Google Scholar]
- 20.Carr JJ, Nelson JC, Wong ND, McNitt-Gray M, Arad Y, Jacobs DR, Jr, Sidney S, Bild DE, Williams OD, Detrano RC. Calcified coronary artery plaque measurement with cardiac CT in population-based studies: standardized protocol of Multi-Ethnic Study of Atherosclerosis (MESA) and Coronary Artery Risk Development in Young Adults (CARDIA) study. Radiology. 2005;234(1):35–43. doi: 10.1148/radiol.2341040439. [DOI] [PubMed] [Google Scholar]
- 21.Fornage M, Doris PA. Single-nucleotide polymorphism genotyping for disease association studies. Methods Mol Med. 2005;108:159–172. doi: 10.1385/1-59259-850-1:159. [DOI] [PubMed] [Google Scholar]
- 22.Abifadel M, Varret M, Rabes JP, Allard D, Ouguerram K, Devillers M, Cruaud C, Benjannet S, Wickham L, Erlich D, Derre A, Villeger L, Farnier M, Beucler I, Bruckert E, Chambaz J, Chanu B, Lecerf JM, Luc G, Moulin P, Weissenbach J, Prat A, Krempf M, Junien C, Seidah NG, Boileau C. Mutations in PCSK9 cause autosomal dominant hypercholesterolemia. Nat Genet. 2003;34(2):154–156. doi: 10.1038/ng1161. [DOI] [PubMed] [Google Scholar]
- 23.Shioji K, Mannami T, Kokubo Y, Inamoto N, Takagi S, Goto Y, Nonogi H, Iwai N. Genetic variants in PCSK9 affect the cholesterol level in Japanese. J Hum Genet. 2004;49(2):109–114. doi: 10.1007/s10038-003-0114-3. [DOI] [PubMed] [Google Scholar]
- 24.Timms KM, Wagner S, Samuels ME, Forbey K, Goldfine H, Jammulapati S, Skolnick MH, Hopkins PN, Hunt SC, Shattuck DM. A mutation in PCSK9 causing autosomal-dominant hypercholesterolemia in a Utah pedigree. Hum Genet. 2004;114(4):349–353. doi: 10.1007/s00439-003-1071-9. [DOI] [PubMed] [Google Scholar]
- 25.Hooper AJ, Marais AD, Tanyanyiwa DM, Burnett JR. The C679X mutation in PCSK9 is present and lowers blood cholesterol in a Southern African population. Atherosclerosis. 2007;193(2):445–448. doi: 10.1016/j.atherosclerosis.2006.08.039. [DOI] [PubMed] [Google Scholar]
- 26.Hallman DM, Srinivasan SR, Chen W, Boerwinkle E, Berenson GS. Relation of PCSK9 mutations to serum low-density lipoprotein cholesterol in childhood and adulthood (from The Bogalusa Heart Study) The American journal of cardiology. 2007;100(1):69–72. doi: 10.1016/j.amjcard.2007.02.057. [DOI] [PubMed] [Google Scholar]
- 27.Loria CM, Liu K, Lewis CE, Hulley SB, Sidney S, Schreiner PJ, Williams OD, Bild DE, Detrano R. Early adult risk factor levels and subsequent coronary artery calcification: the CARDIA Study. J Am Coll Cardiol. 2007;49(20):2013–2020. doi: 10.1016/j.jacc.2007.03.009. [DOI] [PubMed] [Google Scholar]
- 28.Lloyd-Jones DM, Wilson PW, Larson MG, Leip E, Beiser A, D'Agostino RB, Cleeman JI, Levy D. Lifetime risk of coronary heart disease by cholesterol levels at selected ages. Arch Intern Med. 2003;163(16):1966–1972. doi: 10.1001/archinte.163.16.1966. [DOI] [PubMed] [Google Scholar]
- 29.Lalanne F, Lambert G, Amar MJ, Chetiveaux M, Zair Y, Jarnoux AL, Ouguerram K, Friburg J, Seidah NG, Brewer HB, Jr, Krempf M, Costet P. Wild-type PCSK9 inhibits LDL clearance but does not affect apoB-containing lipoprotein production in mouse and cultured cells. Journal of lipid research. 2005;46(6):1312–1319. doi: 10.1194/jlr.M400396-JLR200. [DOI] [PubMed] [Google Scholar]