Abstract
Oligomerization of human Islet Amyloid Polypeptide (IAPP) has been increasingly considered a primary pathogenic process in Type II Diabetes. Here structural features of the IAPP monomer have been probed using a combination of Ion Mobility Mass Spectrometry (IMS-MS) and all-atom Replica Exchange Molecular Dynamics (REMD) simulations. Three distinct conformational families of human IAPP monomer are observed in IMS experiments and two of them are identified as dehydrated solution structures based on our simulation results: one is an extended β-hairpin structural family and the second is a compact helix-coil structural family. The extended β-hairpin family is topologically similar to the peptide conformation in the solid state NMR fibril structure published by Tycko and coworkers. It is absent in both experiments and simulations performed on the non-amyloidogenic rat IAPP suggesting it may play an important role in the fibrillation pathway of human IAPP. In addition, pH dependence studies show the relative abundance of the β-hairpin structural family is significantly enhanced at pH 8.0. This observation is consistent with the increased rate of fibrillation at high pH in vitro and offers a possible explanation of the pH dependent fibrillation in vivo. This paper, to the best of our knowledge, presents the first experimental evidence of a significant population of β-hairpin conformers for the IAPP peptide. It is consistent with a previous suggestion in the literature that β-sheet rich oligomers are assembled from ordered β-hairpins, rather than from coiled structures.
Keywords: IAPP, Amylin, β-hairpin, Ion Mobility, Replica Exchange
Introduction
Aggregation of Islet Amyloid Polypeptide (IAPP) has been increasingly identified as the primary pathogenic process in Type II Diabetes,1–3 a disease afflicting more than 150 million people world wide.4 Like a number of other amyloid peptides5 human IAPP forms highly ordered β-sheet rich fibrils. In contrast rat IAPP does not form fibrils6–14 even though it differs from the human form by only 6 amino acids in the “core mutation region” (residues 18–29), indicating the disease related aggregation process is highly sequence dependent. While atomistic characterization of the fibril form of this amyloid peptide has been emerging,15, 16 the structures of the early aggregation species, including monomer and small oligomers, remain poorly understood. These early oligomeric species have been strongly implicated as the proximate toxic agent in Type II Diabetes.17–21 Clearly, to better our understanding of the oligomerization process more detailed structural descriptions of the human and rat IAPP monomers would be very useful, as differences in monomer structure may well lead to the differences observed in oligomerization, amyloid fibrillation and β-cell toxicity.22
The most detailed structural information available for human IAPP is from the fibril form due to its homogeneity. Luca et al.15 published a single fibril structure derived from solid state NMR data. They report a structure in which a cross-β subunit consists of stacked β-sheets formed from the parallel in-registry assembly of a U-shaped β-strand-loop-β-strand motif. The loop is located at residues 18–27, straddled by two β-strands comprising residues 8–17 and 28–37 respectively. Recently Wiltzius et al. 16 published a fibril model of the full length human IAPP extrapolated from x-ray diffraction data of cross-β spine structures of two segments of human IAPP. The topology of this latest model is similar to that reported by Luca et al.
Circular dichroism (CD) studies found the IAPP monomer adopts a primarily random coil structure.23, 24 NMR studies on human IAPP by Yonemoto et al.25 concluded that IAPP populates conformers with some N-terminal helix content and did not find any significant contribution of β-sheet conformers. However, recent two-dimensional IR results suggest early oligomers may assemble with a β-hairpin motif and β-sheet coupling between the hairpins.22 A study by Kayed et al.26 suggested a more complex view of the IAPP monomer. They followed the thermal denaturing pathway of IAPP with CD and proposed that the IAPP monomer adopts two distinct conformers: one non-amyloidogenic and another amyloidogenic, with the latter having higher contributions of both β-sheet and α-helix. The study suggested that at room temperature the IAPP monomer would populate both conformers at equilibrium. This view of the IAPP monomer parallels the studies on Amyloid-β peptide fragments where both experimental27–29 and theoretical30, 31 work find that the peptides adopt a number of conformers at equilibrium including β-strands and random coils.
One challenge of solution phase experiments is that the measurements provide information about the “average” structure in solution. In the case of IAPP the monomers may exist in several structural families that are in equilibrium both with themselves and with the soluble early oligomer distribution.21, 32 The monomers may very well develop a somewhat different structure in the oligomers than they have on their own. The fact that conventional solution techniques currently can not resolve different peptide oligomeric states and consequently cannot resolve the structural characteristics of isolated monomers and monomer units in oligomers remains a major experimental challenge.
In this paper we present a combination of ion mobility mass spectrometry (IMS/MS) experiments33 and replica exchange molecular dynamics (REMD)34–37 simulations to investigate monomeric structural differences between human and rat IAPP. In recent years, with the development of electrospray ionization (ESI) it has become possible to introduce biological molecules into the gas phase while minimizing the impact on their native solution structures. By combining this soft ionization source with ion mobility mass spectrometry33, 38 and state-of-the-art computational methods, multiple conformers of the same molecular ion can be resolved. In addition, oligomers with the same mass to charge ratio can be resolved for oligomer specific experiments. This development has led to significant progress in examining the solution structure of biological molecules,39 including in our group, the structures of oligonucleotides and their complexes with drug candidates,40–47 hormone-metal ion interactions,48 and amyloid forming systems including Aβ32, 49–53 and α-synuclein.54, 55 In this paper we describe IMS/MS studies combined with all-atom ab initio REMD simulations to investigate the structural details of monomeric IAPP.
Materials and Methods
Peptide Sample Preparation
All samples of human IAPP and rat IAPP were purchased from Bachem Inc. (Torrance, CA) and used without further purification. Stock solutions of the peptides were prepared at 1mM in 100% hexafluoroisopropanol (HFIP) purchased from Sigma (St. Louis). Aliquots of the stock were dried and the peptides were re-suspended in 50mM ammonium acetate buffer at pH 6.4 for final peptide concentrations of 20 µM. For pH dependent studies glacial acetic acid or ammonium hydroxide were added to the ammonium acetate buffers until the desired pH was achieved and then the peptide stock solution was added. To verify that the buffer and concentration conditions were appropriate for amyloid fibril formation, the presence of IAPP fibrils was confirmed using atomic force microscopy (AFM) to obtain fibril images from the aged human IAPP solution (Appendix 2 in the supporting material).
DC Ion Mobility Experiments
The instrument and operational parameters of the IMS/MS experiments have been described in detail elsewhere.38 Briefly, ion mobility experiments were conducted on a home built instrument comprised of a nano-electrospray ionization (nano-ESI) source, an ion funnel, a temperature controlled drift cell, a quadrupole mass analyzer and an electron multiplier detector. Samples were loaded into gold coated borosilicate capillaries that had been pulled to a fine point on an in-house tip puller (Sutter Instrument Co., Novato, CA). A positive voltage was applied to the tip and ions were extracted and entered the instrument through a small (0.010” id) orifice. The ions were collected, desolvated and focused in the ion funnel where they were stored immediately preceding the drift cell. A 10 µs pulse of ions was injected into the drift cell filled with ~5 torr of He gas where they gently drift through under the influence of a weak electric field (5–20 V/cm). The injection energy was usually kept as low as possible to minimize thermal heating of the ions during the injection process. Following the drift cell the ions were mass analyzed with a quadrupole mass filter and detected with a traditional conversion dynode/electron multiplier arrangement. Total analysis time of this instrument is between 0.5 and 2 ms.
Ions that are injected into the drift cell experience a constant force from the electric field, E, which is balanced by a retarding frictional force due to collisions with the buffer gas. Under these conditions the velocity, υd, at which the ions move through the buffer gas is proportional to the magnitude of the electric field where the mobility, K is the proportionality constant:
| (1) |
The absolute mobility of the ion packet is dependent on the temperature and the pressure of the buffer gas so it is customary to use the reduced mobility K0.
| (2) |
After the ions exit the drift cell they are mass analyzed and detected as a function of the arrival time, tA. The reduced mobility can be determined from instrumental parameters by converting equation (1) to equation (3) and plotting tA versus p/V.
| (3) |
In equation 3, l is the length of the drift cell, V is the voltage across the cell, and t0 is the time the ions spend outside the drift cell before hitting the detector.
Collision theory56 relates K0 to σ, the collision cross section of the ion of interest:
| (4) |
In this relationship, N is the buffer gas number density, μ is the reduced mass of the collision system (ion + He) and kb is Boltzmann’s constant. The collision cross sections are reported as the average of multiple measurements, typically more than three, and all measurements are reproducible within 1%. The measured reduced mobility (K0) and subsequently the collision cross section (σ) contain information about the three dimensional configuration of the ion. In the case of peptide ions, secondary and tertiary structural characteristics can be identified by comparison with computational models.
The flux of ions exiting the drift tube can be calculated56 and is used to fit the experimental arrival time distributions. It is assumed that the ion packet takes the form of a periodic delta function and the flux is given by equation 5.
| (5) |
Here z is the ion charge, r0 is the radius of the initial ion packet, a is the area of the exit aperture, DL and DT are the longitudinal and transverse diffusion coefficients, s is the initial ion density and α is the loss of ions due to reactions in the drift tube.
Molecular Dynamics Simulation
The AMBER 857 simulation suite was used in replica exchange molecular dynamics (REMD)34 simulations. All three peptides were modeled using the AMBER all-atom point-charge protein force field, ff96.58 Recent studies59, 60 have shown that AMBER ff96 combined with a recent generalized-born model (IGB=5)61 plus the surface term (gbsa=1) predicts reasonable structures for small α, β and α/β proteins, and thus AMBER ff96 is fairly well-balanced between α-helix and β-sheet secondary structures.
In order to improve conformational sampling, REMD protocols were used to generate both solution and solvent free structure ensembles. Solvent free simulations were carried out in vacuum and solution phase structures were generated using the recent implicit solvent model (IGB=5)61 plus the surface term (GBSA=1, 0.005 kcal/Å2/mol) to represent water solvent effects with an effective salt concentration of 0.2 M. For each peptide simulation an initial energy minimization was performed on an extended conformation and the minimized structure was used as the input for REMD. 16 replicas were set up with initial temperatures exponentially spaced from 270 to 465 K for solution phase calculations (i.e 270.0 278.9 289.3 300.0 311.2 322.7 334.7 347.2 360.1 373.4 387.3 401.7 416.6 432.1 448.2 465.0, see reference53 for the algorithm used to optimize them), and 300–2200 K for gas phase calculations (i.e 300.0 342.6 391.3 446.9 510.4 582.8 665.6 760.2 868.2 991.5 1132.4 1293.2 1476.9 1686.7 1926.4 2200.0 K, see reference53 for the algorithm used to optimize them). Extreme high temperature (i.e. up to 2200 K) was used to overcome slow structural relaxation in vacuum. We confirmed that the high temperatures applied to our simulations did not lead to any notable structural artifacts such as trans to cis isomerizations of the peptide bonds within the duration of the simulations (40.0 ns). Initial velocities for each replica system were generated according to the Maxwell-Boltzmann distribution according to that replica’s initial temperature. The first 1.0 ns of molecular dynamics simulation for each replica was performed without replica exchanges to equilibrate the system at its target temperature. After the equilibrium phase, exchanges between neighboring replicas were attempted every 2000 MD steps (2.0 ps) and the exchange rate among replicas was ~20% in the production phase. SHAKE62 was applied to constrain all bonds linking hydrogen atoms and a shorter time step of 1.0 fs rather than the typical 2.0 fs was used to circumvent the SHAKE failure probably caused by large atomic displacements at the high temperatures used in our simulations (up to 2200 K leading to a higher kinetic velocity). In order to cut computation time, non-bonded forces were calculated using a two-stage RESPA (reference system propagator algorithm approach)63 where the fast varying forces within a 12 Å radius were frequently updated (e.g. every step) and those beyond 12 Å were updated every two steps. Temperature was regulated using Berendsen’s algorithm64 with a coupling constant of 1.0 ps. The center of mass translation and rotation were removed every 500 MD steps (0.5 ps). Each solution phase replica was run for 200.0 ns giving a cumulative simulation time of 3.2 µs for each peptide system and each gas phase replica was run for 40.0 ns, resulting in 640.0 ns of cumulative simulation time for each peptide system. The snapshots in the replica trajectories were saved at 2.0 ps intervals for further analysis.
Secondary structure assignment and Tertiary structure clustering
The STRIDE program of Frishman and Argos65 was used to analyze secondary structure propensities. For tertiary structure analysis, the snapshots of the last 20.0 and 100.0 ns at 300 K respectively for solvent free and solution simulations were classified into structural families by the GROMACS clustering protocol66, in which the structure similarity metric is based on pair wise RMSD over Cα atoms of the peptide. A larger Cα rmsd (root mean square deviation) cutoff (3.0 Å), rather than the 2.0 Å typically used for ordered peptides, was used to capture major structural features. The centroid structure that has the largest number of neighboring structures within the cutoff, was selected as the representative structure of the structure family, and the total number of member snapshots within the family over the total ensemble indicates the stability of the structure.
The 10/5 (water solvent / solvent-free) most populated structural families of each peptide system (human IAPP(+3), human IAPP(+4) and rat IAPP(+4)) were obtained by applying the clustering method to the last 100ns/20ns (water solvent / solvent-free) trajectory at 300K. The structural families were further merged into a few super structural families to capture major features. The abundance of a supper family was just the sum of the abundance of its member families; the mean and the standard error of the cross section for a supper family were the weighted average from those of the representative structures of its member families.
Collision Cross Section Calculation
The representative structure of the structure family was used to calculate the cross section by a trajectory method67, 68 for solvent free-simulations. In order to better correlate with the solvent free experiments, solution phase structures were converted to ‘dehydrated’ structures via an energy minimization (500000 steps) in vacuum prior to cross section calculations. This “dehydration” reduces the overall size of the structures, while maintaining their solution structural features and in this paper these structures are referred to as “dehydrated solution structures”. For comparison with experimental collision cross sections structures are typically considered in good agreement if the calculated collision cross section falls within 2–3% of the experimental value.
Convergence of the REMD Simulations
Convergence was rigorously verified by a block analysis: the total sampling at 300 K was equally divided into four blocks, and for each block the secondary structure and tertiary structure clustering analysis detailed above was performed to see when convergence was reached. For all 6 sets of REMD simulations good convergence was found during the last half of the trajectory (see, for example, the data for human IAPP(+4) in Figure S2 of the supporting material). Thus, the structural results presented in this paper were calculated from the last half trajectory (the last 100.0 ns in implicit water solvent and the last 20.0 ns in solvent free at 300 K).
Results
Ion Mobility Mass Spectrometry
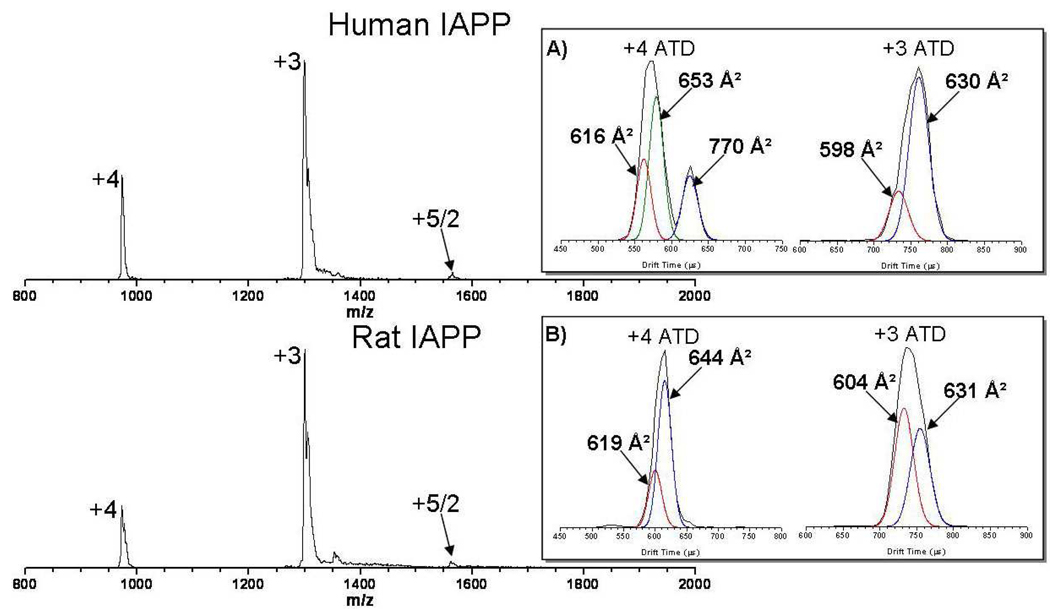
The mass spectrum for both human IAPP and rat IAPP are shown in Figure 2. Rat IAPP shows dominant +3 and +4 charge states. The +3 charge state likely corresponds to protonation of the Lys1, Arg11 and Arg18 residues and the +4 charge state arises from the subsequent protonation of the N-terminus.
Figure 2.
Positive ion ESI mass spectra showing +3, +4 and +5/2 charge states of human IAPP (top) and rat IAPP (bottom). Arrival time distributions for the +3 and +4 charge states are given in the insets. The +5/2 charge state is due to a minor amount of oligomer formation (see text). Individual fitted features in the ATDs are shown in color. Cross sections for the major features are noted.
The human IAPP mass spectrum shows a charge state distribution nearly identical to the rat form. The +3 charge state of human IAPP comes from protonation of the N-terminus, Lys1 and Arg11. The +4 charge state then arises from the protonation of the His18. Both of the human and rat IAPP mass spectra show a small component of dimer in the +5 charge state. The intensity of oligomer states increases dramatically either at long times (hours/days) or at higher concentrations. For the purposes of this manuscript we will limit our discussion to the structures of the monomer states, saving the discussion of the early oligomer state distributions, structures and formation mechanisms for future publications.
To gain insight into the peptide conformation we turn to the ion mobility experiment. Arrival time distributions (ATDs) are shown as insets in Figure 2 for the +3 and +4 charge states. The experimental cross sections for each peptide and charge state are listed in Table 1. The ATDs for the +3 charge state of both human and rat IAPP are quite similar. Both appear to have one dominant conformer. However, according to collision theory56, the ATDs are wider than a single conformer and require at least two features to fit the data. The best fit +3 rat IAPP features have cross sections of 604 Å2 and 631 Å2 and the +3 human IAPP features have cross sections of 598 Å2 and 630 Å2.
Table 1.
Experimental collision cross sections at room temperature.
| IAPP Cross Sections (Å2)1 | ||
|---|---|---|
| Charge State | Human IAPP | Rat IAPP |
| +3 | 630, 598 | 631, 604 |
| +4 | 770, 653, 616 | 644, 619 |
Experimental error ± 1% for all cross sections.
Likewise the rat IAPP +4 ATD looks like a single feature; however, it again is too wide to contain a single conformer. Two features with cross sections of 619 Å2 and 644 Å2 provide an optimal fit. These larger cross sections, compared to the +3 charge state, are consistent with increased charge repulsion in the +4 charge state which is a common trend observed in multiply charged ions in the gas phase54. The +4 ATD for human IAPP is different than any of the other ATDs recorded, having three distinct features. There are two conformers in the more intense, shorter time peak, with measured cross sections of 616 Å2 and 653 Å2 (they are more apparent under ion funnel conditions that favor annealing to solvent free conformers (see Figure S1 in the supporting material)), and a peak composed of only one feature at much longer time with a cross section of 770 Å2. To verify that that the ATD of the +4 charge state corresponds to dominantly monomer species, mass spectra were obtained for the individual ATD features. Both the compact and extended features show 0.25 amu 13C spacing indicating that they are both monomers (Figure S2). The features with cross sections of 616 Å2 and 653 Å2 likely correlate to the compact structural families in the human IAPP +3 charge state (604 Å2 and 631 Å2) where the slightly larger cross sections again are due to differences in the charge repulsion as observed between the +3 and +4 charge states in rat IAPP.
Interestingly, the extended conformer with a cross section of 770 Å2 in the human +4 ATD is ~18% larger in cross section than the compact solution conformer. For the experiments shown in Figure 2 the source ion funnel conditions were kept as soft as possible to minimize the energy input into the ions. This procedure maximizes the preservation of solution structures, suggesting the long time feature in the human +4 charge state ATD correlates to an IAPP structural family found in solution.
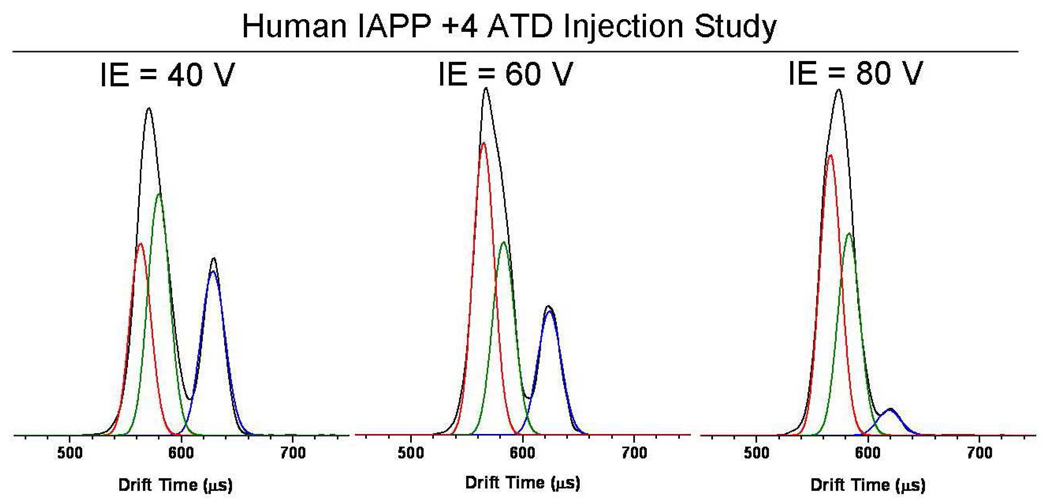
Injection Energy Study of +4 Human IAPP
To examine the relative stability of the structural families (the extended vs. the compact) in the +4 ATD of human IAPP, the energy with which ions were injected into the drift cell was increased systematically. ATDs at varying injection energies are shown in Figure 3. Increasing the injection energy is analogous to an annealing experiment where energy is input into the ion (via buffer gas collisions), allowing it to overcome energy barriers and refold into the lowest energy solvent free conformation. A detailed description is given elsewhere52. At injection energy 40 V the compact conformers are present with about twice the abundance as the extended conformer. With increased injection energy (60 V) the intensity of the extended conformer decreases until at the highest injection energy (80 V) it is nearly gone. Figure S1 shows the same human IAPP +4 ATD at elevated ion funnel DC and RF voltages which favor conversion of solution conformers to solvent free conformers. In this ATD the most compact structural family with a cross section of 616 Å2 is significantly enhanced while the extended family at 770 Å2 is reduced. These results indicate that the extended conformer is metastable in the gas phase and is likely due to a solution phase structural family that is preserved during the ESI process. The extended solution structure is stable in the absence of solvent on the time scale of our experiment (10−3 seconds) as long as ion energies are minimized. These results are also consistent with the shortest time feature (616 Å2 for human IAPP +4 charge state) being associated with the solvent free structure of the peptide.
Figure 3.
Arrival time distributions of the +4 charge state of human IAPP monomer at varying injection energies.
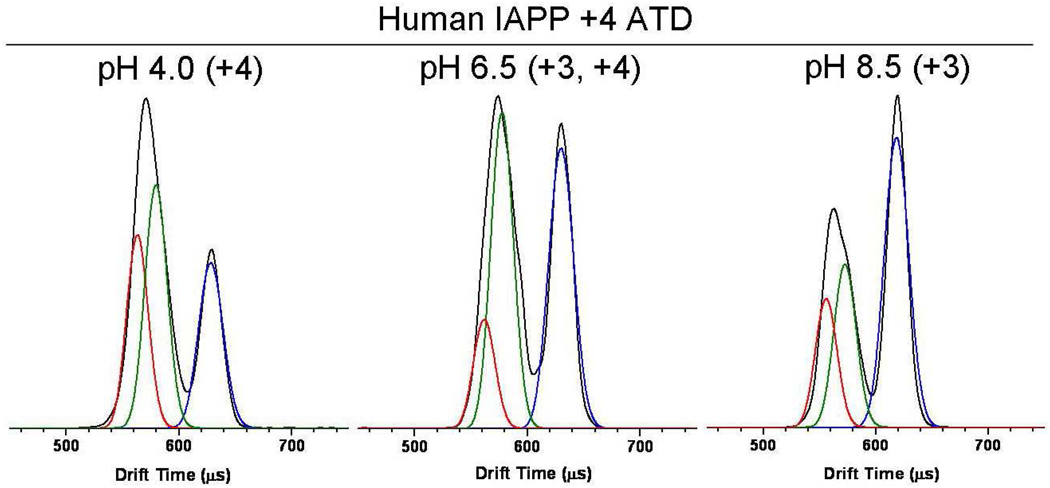
pH Study on +4 Human IAPP
Figure 4 shows the ATDs of the +4 charge state of human IAPP at solution pH’s 4.0, 6.5 and 8.5. Although the pKa of the histidine side chain at position 18 may vary with protein conformation, a pKa of 6.0 of the isolated histidine should be a reasonable estimate for most conformers of human IAPP. At pH 4.0 (<His pKa) all the peptide in solution is in the +4 charge state, resulting directly in the +4 ions observed in the experiment following electrospray. At this pH the shorter time ATD peak, containing the compact pair of conformers, comprises the largest ATD feature. At pH 6.5 (~His pKa) the compact and extended features have nearly equal intensities and at pH 8.5 the extended conformer is the most dominant feature. At pH 8.5 (>His pKa) all the peptide in solution is in the +3 charge state with the histidine deprotonated. Hence, at this pH the +4 ions observed in the experiment result from +3 ions in solution being protonated during the ESI process. We know from the injection energy studies the extended conformer is the energetically least favored conformer in the absence of solvent. Consequently, protonation during the ESI process appears to stabilize the extended +3 solution conformer so it can be detected in the IMS experiment. The extended conformer is not observed in the +3 ATD, suggesting rapid rearrangement to a more compact structure for this charge state in the absence of solvent. This point will be addressed again later in the paper.
Figure 4.
Arrival time distributions of the +4 charge state of human IAPP monomer at varying solution pHs. The charge states of IAPP in solution are listed in parenthesis next to the pH.
REMD Simulations
Detailed molecular structural features were explored with a goal of identifying conformers observed in the experiment and understanding structural motifs critical for initiation of oligomerization leading to fibrillation of human IAPP. REMD simulations started from an extended peptide chain were done on both the +3 and +4 charge states of human IAPP as well as the +4 charge state of the non-amyloidogenic rat IAPP. The +3 charge state of rat IAPP was not simulated as it is an unlikely state in solution. REMD simulations for each peptide system were performed with an implicit solvent model as well as under solvent free conditions.
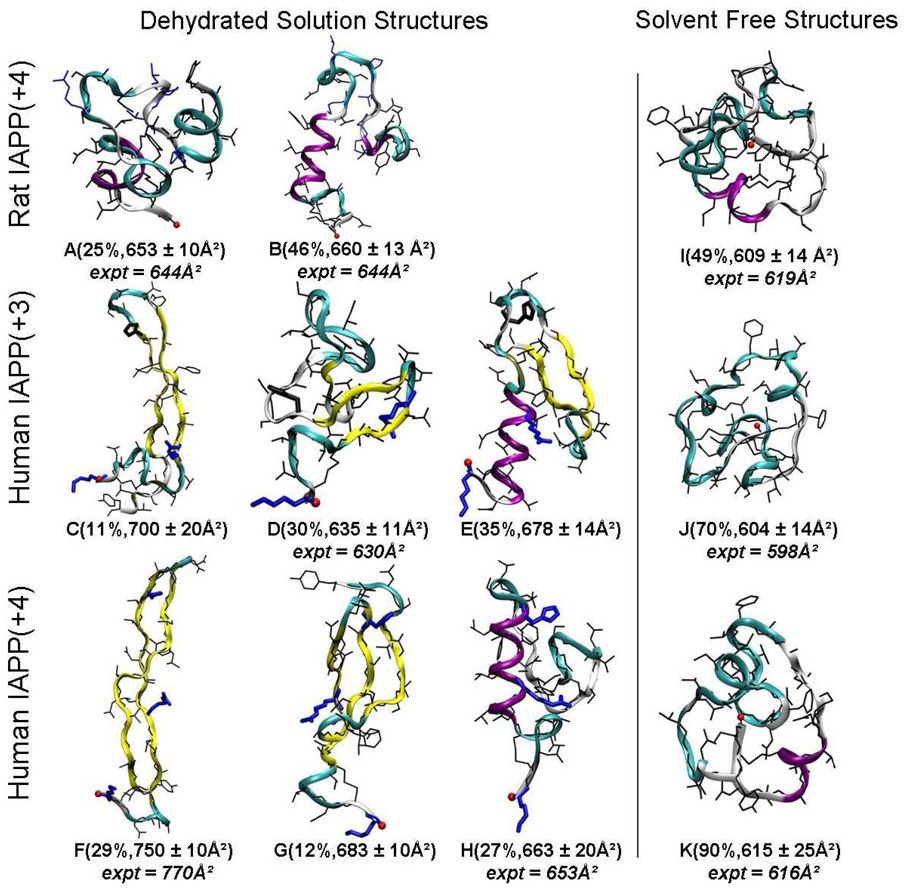
From our clustering analysis with a Cα root mean square deviation (RMSD) cutoff of 3.0 Ǻ, a large number of diversified structural families were identified and are presented in Figure S4 and S5 in the supplementary material. The most populated structural families were further merged into super structural families based on similarity in the molecular topology. Representative structure, abundance, calculated collisional cross section and possible assignment to experimental cross section for each super structural family are presented in Figure 5. Key features are stated below.
Figure 5.
Representative structures of super structural families for each peptide at a temperature of 300 K from REMD simulations. The abundance and the calculated cross sections with standard error for each super family are given in parenthesis (See Figures S4 and S5 for all structural families in each super family), the corresponding experimental cross section assignment is shown in italic font. The side chains of the protonated histidine at position 18 of human IAPP(+4) and the “mutated” residues (R18H, L23F, P25A, V28S, and P29S) of rat IAPP(+4) with respect to human IAPP are in blue. Positively charged residues Lys1, Arg11 and His18 are highlighted by blue bold lines in structures C–H; and neutral residue His18 in structures C–E by a bold black line. The backbone is shown in cartoon and the secondary structure is coded by color: coil in silver, β-sheet in yellow, isolated β-bridge in tan and turn in cyan. The N-terminus is indicated by a red ball.
The rat IAPP(+4) system has two dehydrated solution super structural families: a compact globular family A with mostly turn and coil secondary structure and a helix-coil family B containing a short turn-coil (residues 1–7), a short helix (residues 8–17) and a long turn-coil (residues 18–37). The two structural families have computed cross sections of 653 Å2 and 660 Å2 respectively, which are in reasonably good agreement with the experimental value of 644 Å2. The solvent free family I, with a computed cross section of 609 Å2 is significantly smaller and is a good match with the more compact feature in the rat +4 ATD with an experimental cross section of 619 Å2. Neither of the two super structural families has any significant β-sheet content and the “core mutation region” of rat IAPP(+4) adopts a turn-coil motif which is expected due to the presence of the turn-prone proline residues.
The human IAPP(+3) has three dehydrated solution super structural families: (C) a long β-hairpin (β-strand 9–17, loop 18–23, and another β-strand 24–33), (D) a β-sheet rich compact structure and (E) a helix-loop-hairpin structure (helix 4–16, loop 17–23, and hairpin 24–36) with computed cross sections of 700 Å2, 635 Å2 and 678 Å2 respectively. There are no features in the experimental ATD with cross section near 700 Å2 indicating the β-hairpin family is not detected in this experiment. Family E, with significant β-hairpin like structure is also not observed, possibly due to the instability of the β-hairpin motif for the z/n = +3 system in the solvent free environment (more will be discussed later). The compact β-sheet rich structure D, with a computed cross section of 635 Å2 matches well with the experimental value of 630 Å2, while the solvent free structure J, with a computed cross section of 604 Å2, matches well with the small experimental shoulder feature at 598 Å2. The “core mutation region” of human IAPP(+3) tends to be part of the turn in the β-hairpin region in the first (C) and third (E) super families representing a high abundance of 46% of the total conformational ensemble in solution.
The human IAPP(+4) also has three dehydrated solution super structural families: (F) a long β-hairpin, similar to family C found in human IAPP(+3), (G) a three-stranded β-sheet and (H) a helix-coil family similar to the second rat IAPP(+4) family (B). The experimentally detected extended structure, with a cross section of 770 Å2, agrees very well with the extended β-hairpin family F, having a computed cross section 750 Å2. The compact feature in the ATD, with an experimental cross section of 653 Å2, is assigned as the helix-coil family H, having a computed cross section of 663 Å2. The solvent free family K, with a computed cross section of 615 Å2 matches well with the compact shoulder feature in the ATD at 616 Å2. (Again, this feature is more apparent under ion funnel conditions that favor annealing to solvent free conformers (Figure S1)). The “core mutation region” of human IAPP(+4) tends to be part of the β-hairpin turn region with a high abundance of 41% of the total conformational ensemble in solution.
Over all there is good agreement between experiment and the REMD simulations (Figure 5). For example, there is a quantitative match between the β-hairpin structure of human IAPP(+4) and the extended feature in the experiment (Figure 2). This indicates the solution structure for the hairpin is maintained in the solvent-free environment during IMS/MS analysis (i.e. for 0.5 and 2 ms). In contrast, some structures populated in the solution simulation (i.e. C and E of human IAPP(+3) and G of human IAPP (+4)), are not observed in experiment. There are two possible reasons for this. One, even though great care is taken to treat the ions as gently as possible (using electrospray ionization and low instrumental voltages) the IAPP ions are still perturbed to some degree. As a result, only structural families that are stable under our experimental conditions and in the absence of solvent will be detected. Solution conformers that do not survive these conditions refold to more stable solvent free conformations and will be detected as such (Figures 5I–K). Two, while there have been significant recent improvements in the force field we are using, the relative populations of some structures in the simulation may be overestimated and may not be readily observable in the experiment.
Secondary Structural Analysis
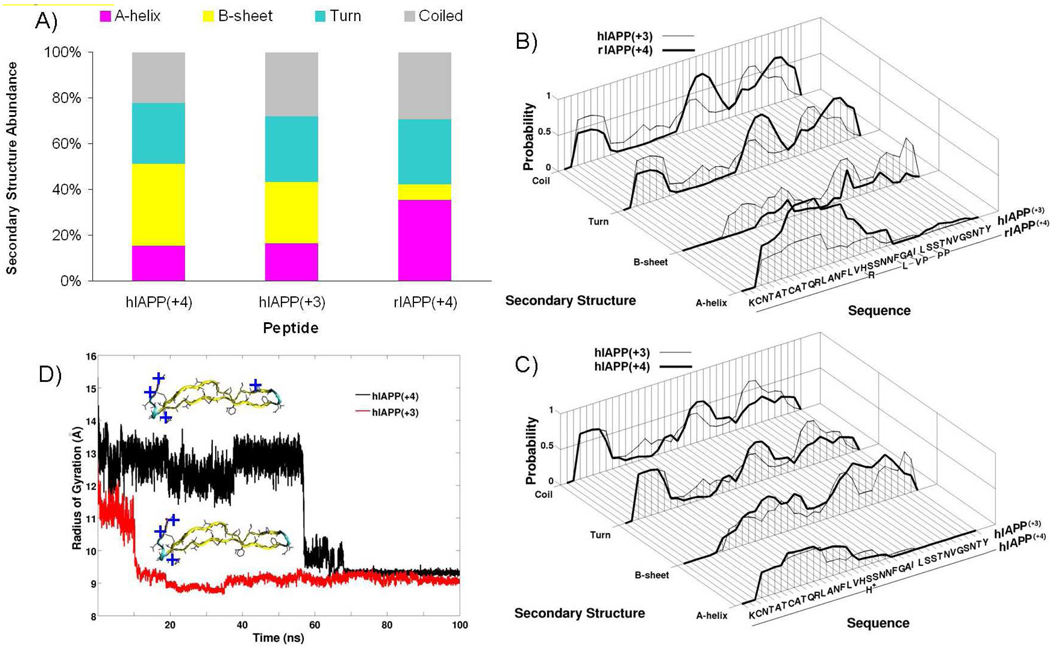
A detailed analysis of the secondary structure characteristics for each of the three peptides in solution is shown in Figure 6A. Rat IAPP(+4) has β-sheet and α-helical propensities of ~7% and ~36% respectively. Conversely human IAPP(+3) has β-sheet and α-helical propensities that are nearly the reverse at ~27% and ~6% respectively. Human IAPP(+4) is nearly identical to human IAPP(+3) but has a β-sheet content slightly enhanced at ~36%. These results follow the general trend that low propensities for α-helix formation and higher propensities for β-sheet formation favor amyloid fibril formation.
Figure 6.
Structural properties of IAPP monomer. A) Secondary structure propensity of human IAPP(+4), human IAPP(+3) and rat IAPP(+4) at 300 K. Standard error is less than 1% from the block analysis. B) Comparison of position-dependent secondary structure propensities for human IAPP(+3) and rat IAPP(+4). C) Comparison of position-dependent secondary structure propensities for human IAPP(+3) and human IAPP(+4). D) Gas phase stability simulations of β-hairpin structures of human IAPP at +3 and +4 charge states at 300 K. The locations of the positive charges in the two peptides are shown by blue plus signs (+).
The position dependent secondary structure analysis is shown in Figure 6B and 6C. A comparison between the +3 charge state of human IAPP and the +4 form of rat IAPP, which are the major forms at neutral pH for each peptide (Figure 6B), shows the residues 1–7 in both systems do not have any β-sheet propensity, likely due to the conformational restriction imposed by the 2–7 disulfide bridge. The α-helical propensity is localized in the N-terminal half of the peptide (residues 1–25) and is more pronounced in rat IAPP than human IAPP. Conversely human IAPP has a greater β-sheet propensity across residues 8–37; notably it is not limited to the “core mutation region” (residues 18– 29). Lastly the “core mutation region” in rat IAPP has greater components of turn and random coil than human IAPP, most likely contributed by the turn prone prolines at positions 25, 28 and 29. This analysis indicates that the sequence variations between the two forms of the peptide have a global effect on the peptide conformation and the β-sheet propensity of human IAPP(+3) is greatly enhanced in the “core mutation region”.
The same analysis was used to compare the +3 and +4 charge states of human IAPP (Figure 6C). The +4 charge state of human IAPP has an enhancement of turn and coil propensities localized in the “core mutation region” (residues 18–29). +4 human IAPP also has a greater β-sheet propensity in the two flanking regions: residues 9–18 and residues 29–37. These features are consistent with the tendency of +4 human IAPP to adopt the well defined β-hairpin structure shown in Figure 5F.
Solvent Free Stability Simulation
The relative stability of the extended β-hairpin conformers of human IAPP were compared for different charge states in the absence of solvent at 300 K. Injection energy experiments show that the extended solution like structures of the molecular ion collapse into the most compact experimentally observed conformer with significant input of energy. This process will also occur at 300 K if the ion is held in the gas phase for an extended period of time. Figure 6D shows two conventional molecular dynamics simulations for +3 and +4 human IAPP. The +4 β-hairpin conformer survives without solvent nearly six times longer than the +3 β-hairpin conformer. This is likely due to the increased charge repulsion interactions in the +4 species (Figures 5 C and F). In particular, the protonated His18 at the loop of the +4 β-hairpin appears to stabilize the fold in the solvent-free environment via electrostatic repulsion with the other three positive charges (at the N-terminus, Lys1 and Arg11). In contrast, this stabilizing effect does not exist for human IAPP(+3), as His18 is not protonated. Note the absolute time scale is undetermined due to the simple thermostat used in the simulations.
Discussion
Human IAPP readily forms highly ordered amyloid fibrils15, 16 via cytotoxic oligomeric intermediates,17, 18, 20, 21 while the rat form does not form fibrils and elicits no pathological response from β-cells in the pancreas. This stark difference in biological behavior is remarkable in the fact that the two peptides differ in primary sequence by only six amino acid residues (R18H, L23F, P25A, V26I, P28S and P29S). While the IAPP monomer structure has been studied with a number of experimental techniques,23–26 the detailed structural characteristics of human and rat IAPP that result in such different chemical morphologies and physiological interactions remain elusive. In this study we have observed two dehydrated solution structural families of human IAPP(+4) in ion mobility experiments and identified them as a β-hairpin structural super family (F) and a helix-coil structural super family (H) with our REMD simulations. In contrast, we observed only one dehydrated solution structural family in rat IAPP(+4) and identified it as a helix-coil structure family (B). Thus, the human IAPP β-hairpin family may be responsible for the observed differences in oligomerization and β-cell toxicity compared with rat IAPP.
Two CD studies on human IAPP23, 24 report that IAPP is primarily random coil. NMR studies on rat IAPP69 and human IAPP25 report some N-terminal helix content. Our clustering results show all three peptide simulations have super structural families (B, E and H in Figure 5) with significant N-terminal helices followed by random coil, the calculated cross sections of super families B and H match well with our experimental values. More important, we also identify super structural families with significant β-hairpin content (C–E, F–G in Figure 5), the calculated cross section of super family F agrees well with our experimental data. Put together, our combined data suggest IAPP adopts multiple stable partially ordered families rather than purely random-coils, or random-coils with some N-terminal helix.
Kayed et al.26 propose that IAPP populates two distinct conformers, one of which is more amyloidogenic than the other. This idea is also applicable to Amyloid-β peptide where both simulations30, 31 and experiments27–29 suggest the peptide significantly populates β-strand in addition to random coil conformation. In our experiments we observe two highly populated solution structural families of IAPP and by direct comparison with REMD simulations identify them as β-hairpin (F) and helix-coil (H). While these two structures appear to be in equilibrium in solution, the interconversion rate in the absence of solvent is sufficiently slow we can independently detect them on the millisecond time scale.
The absence of the β-hairpin structural family in the +3 charge state of human IAPP is surprising. However, the solvent free stability simulation (Figure 6D) shows that the +4 β-hairpin is stable 6 times longer than the +3 β-hairpin. This additional stability likely occurs due to charge repulsion between the positively charged His18 and the three positive charges in the N-terminal region. The His18 is located at the opposite end of the extended β-strand from the Arg11 and repulsion between these charged centers likely provides additional stabilization to the extended β-hairpin conformer for the +4 charge state in the absence of solvent. While the +3 β-hairpin is less stable than the +4 charge state in the absence of solvent, our pH study shows that at high pH, with His18 deprotonated, there is a large enhancement of the β-hairpin feature in the +4 ATD. In this case the peptides in solution are triply charged and the fourth charge is added in the electrospray process. Hence, the +3 β-hairpin is stable in solution and adding the fourth charge in the ESI process preserves it as solvent evaporates.
The discussion of charge state brings us back to the unresolved question of pH and its impact on fibrillation rate. The β-cell extracellular space has a pH of 7.4, where as the inside of the insulin secretory vesicles have low pH of 5.5. This pH difference is correlated with the observation that IAPP amyloid deposits are primarily found in the extracellular space and no amyloid deposits are found within the secretory vesicles. The observation is consistent with the finding that IAPP fibrillation is increased at high pH70, 71 and with our description of the IAPP monomer where the β-hairpin conformer is enhanced at high pH in the experiment. Yet the picture of fibrillation is more complex as Lin et al.18 report evidence that toxic oligomers form within the secretory vesicles. Our data may provide a structural explanation for this observation. In both experiments and simulations we found that the β-hairpin conformer is amply abundant (33%) under acidic conditions (see Figure 4). If β-sheet oligomers form inside the vesicles they have the opportunity to interfere with β-cell activities before they are released extracellularly; after release from the cell, early β-sheet rich oligomers could possibly act as seeds to accelerate fibril formation in the extracellular medium where neutral pH further promotes β-sheet propensity.
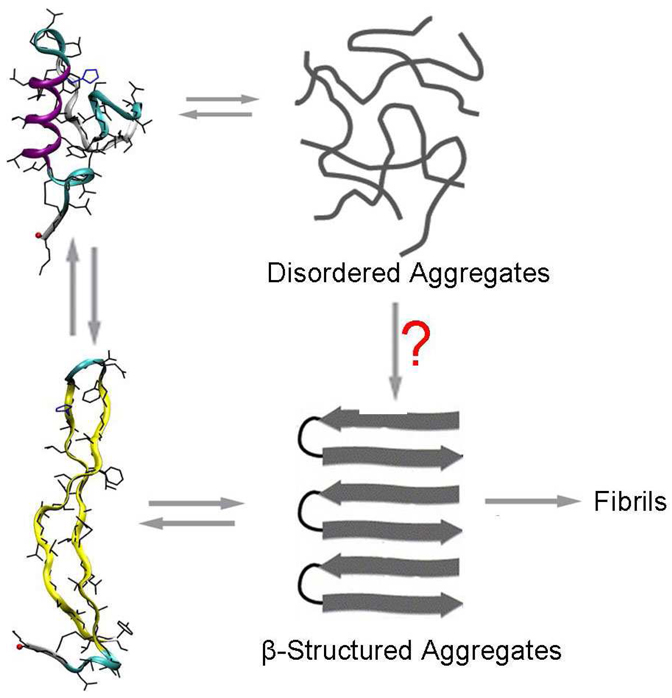
Lastly it is suggested that oligomers in most amyloid peptide systems go through a large scale conformational transition from a native disordered peptide oligomer to a highly ordered β-sheet oligomer during the late stages of amyloid fibril formation (not before nucleation).72 This idea is supported by the lack of monomer with β-sheet conformation in solution23, 24 based on experimental techniques that measure average structures. Yet, there are some theoretical studies on Amyloid-β peptide fragments (residues 12–28 and 10–35) that find large populations of random coil combined with significant contributions of β-hairpin like structural families.30, 31 Based on these structures, the authors propose a possible mechanism by which nucleation may occur from structured states and the conformation transition to β-structure may actually occur before nucleation (hereafter referred as the “early conformation transition” mechanism). The “early conformation transition” mechanism is further supported by theoretical evidence and recent 2-D IR experiments22 that these β-hairpins form multi-stranded β-sheets by side-to-side association.73, 74 Although this mechanism is theoretically sound, as it does not require a drastic, entropically unfavorable structural transition, there has been little supporting experimental evidence to date.22 Here, the data from our combined study suggest that the “early conformation transition” mechanism may be involved in IAPP fibrillation. Our simulation results show that the β-hairpin comprises a significant portion (~24–33%) of the conformation ensemble and our experiments confirm that the extended β-hairpin structural family does exist and in significant quantities. We argue that the monomer β-hairpin structure could be more aggregation prone than other conformers leading to β-sheet-type oligomers. Hence, the rate of IAPP fibrillation may well depend on the amount of time that IAPP spends in the β-hairpin conformation. It should be noted, however that conversion of disordered aggregates to fibrillar structures through a phase transition is not necessarily excluded by the data and analysis presented here. Consequently both options are pictured in Figure 7. A question mark (?) is associated with the latter mechanism since we currently have no data to support its presence.
Figure 7.
Schematic representation of a possible oligomerization mechanism consistent with the results presented in this paper. Among two interconverting structural families of human IAPP, β-hairpins are proposed to self-assemble into early ordered human IAPP oligomers by side-to-side association. The (?) symbol notes that we have no data pro or con for the occurrence of this mechanism and hence we include it for completeness.
Conclusion
We observe two distinct solution structural families of human IAPP in IMS/MS experiments and identify them as β-hairpin and helix-coil structural super families via REMD simulations. Previously observed increased rates of fibrillation at high pH70, 71 combined with our observed β-hairpin enhancement at high pH lead us to suggest that the β-hairpin structure may be an amyloidogenic precursor of human IAPP. This suggestion is supported by the absence of the β-hairpin conformer in both experiments and simulations of rat IAPP under identical conditions and with the topologic similarities of the β-hairpin to the solid state NMR structure.15 Similar suggestions of β-sheet assembly into ordered oligomers in simulations30, 31 and experiments27–29 have been made for fragments of the Amyloid-β peptide. As a consequence, we suggest one pathway to fibril formation in IAPP may be through direct assembly of β-hairpin like monomers.
Supplementary Material
Figure 1.
Primary sequences of human and rat IAPP. Positively charged residues are shown in blue and hydrophobic residues are shown in red. The mutations in rat IAPP are underlined (with respect to human IAPP).
Acknowledgment
The support of the National Institute of Health under grant IPOIAG027818(MTB), the David and Lucile Packard Foundation (JES), National Science Foundation grants CHE-0503728 and CHE-0909743 (MTB) and MCB-0642088 (JES). Simulations were performed on the Lonestar cluster at Texas Advanced Computing Center (LRAC MCA05S027 to JES). MTB also thanks Waters Corporation for donation a Synapt75 proto-type instrument used for part of the work presented here.
References
- 1.Hull RL, Westermark GT, Westermark P, Kahn SE. J Clin Endocr Metab. 2004;89:3629–3643. doi: 10.1210/jc.2004-0405. [DOI] [PubMed] [Google Scholar]
- 2.Hoppener JWM, Lips CJM. Int J Biochem Cell B. 2006;38:726–736. doi: 10.1016/j.biocel.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 3.Matveyenko AV, Butler PC. Ilar J. 2006;47:225–233. doi: 10.1093/ilar.47.3.225. [DOI] [PubMed] [Google Scholar]
- 4.Hossain P, Kawar B, Nahas ME. New Engl J Med. 2007;356:213–215. doi: 10.1056/NEJMp068177. [DOI] [PubMed] [Google Scholar]
- 5.Sipe JD. Amyloid proteins : the ?- sheet conformation and disease. Wiley- VCH: Weinheim; 2005. [Google Scholar]
- 6.Butler AE, Jang J, Gurlo T, Carty MD, Soeller WC, Butler PC. Diabetes. 2004;53:1509–1516. doi: 10.2337/diabetes.53.6.1509. [DOI] [PubMed] [Google Scholar]
- 7.Nishi M, Chan SJ, Nagamatsu S, Bell GI, Steiner DF. P Natl Acad Sci USA. 1989;86:5738–5742. doi: 10.1073/pnas.86.15.5738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaikaran ETAS, Higham CE, Serpell LC, Zurdo J, Gross M, Clark A, Fraser PE. J Mol Biol. 2001;308:515–525. doi: 10.1006/jmbi.2001.4593. [DOI] [PubMed] [Google Scholar]
- 9.Green J, Goldsbury C, Min T, Sunderji S, Frey P, Kistler J, Cooper G, Aebi U. J Mol Biol. 2003;326:1147–1156. doi: 10.1016/s0022-2836(02)01377-3. [DOI] [PubMed] [Google Scholar]
- 10.Abedini A, Raleigh DP. J Mol Biol. 2006;355:274–281. doi: 10.1016/j.jmb.2005.10.052. [DOI] [PubMed] [Google Scholar]
- 11.Abedini A, Meng FL, Raleigh DP. J Am Chem Soc. 2007;129:11300-+. doi: 10.1021/ja072157y. [DOI] [PubMed] [Google Scholar]
- 12.Padrick SB, Miranker AD. J Mol Biol. 2001;308:783–794. doi: 10.1006/jmbi.2001.4608. [DOI] [PubMed] [Google Scholar]
- 13.Betsholtz C, Christmansson L, Engstrom U, Rorsman F, Svensson V, Johnson KH, Westermark P. Febs Lett. 1989;251:261–264. doi: 10.1016/0014-5793(89)81467-x. [DOI] [PubMed] [Google Scholar]
- 14.Westermark P, Engstrom U, Johnson KH, Westermark GT, Betsholtz C. P Natl Acad Sci USA. 1990;87:5036–5040. doi: 10.1073/pnas.87.13.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luca S, Yau WM, Leapman R, Tycko R. Biochemistry-Us. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiltzius JJW, Sievers SA, Sawaya MR, Cascio D, Popov D, Riekel C, Eisenberg D. Protein Sci. 2008;17:1467–1474. doi: 10.1110/ps.036509.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ritzel RA, Meier JJ, Lin CY, Veldhuis JD, Butler PC. Diabetes. 2007;56:65–71. doi: 10.2337/db06-0734. [DOI] [PubMed] [Google Scholar]
- 18.Lin CY, Gurlo T, Kayed R, Butler AE, Haataja L, Glabe CG, Butler PC. Diabetes. 2007;56:1324–1332. doi: 10.2337/db06-1579. [DOI] [PubMed] [Google Scholar]
- 19.Haataja L, Gurlo T, Huang CJ, Butler PC. Endocr Rev. 2008;29:303–316. doi: 10.1210/er.2007-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Janson J, Ashley RH, Harrison D, McIntyre S, Butler PC. Diabetes. 1999;48:491–498. doi: 10.2337/diabetes.48.3.491. [DOI] [PubMed] [Google Scholar]
- 21.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, Glabe CG. Science. 2003;300:486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 22.Shim S-H, Gupta R, Ling YL, Strasfeld DB, Raleigh DP, Zanni MT. Proceedings of the National Academy of Sciences. 2009;106:6614–6619. doi: 10.1073/pnas.0805957106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higham CE, Jaikaran ETAS, Fraser PE, Gross M, Clark A. Febs Lett. 2000;470:55–60. doi: 10.1016/s0014-5793(00)01287-4. [DOI] [PubMed] [Google Scholar]
- 24.Goldsbury C, Goldie K, Pellaud J, Seelig J, Frey P, Muller SA, Kistler J, Cooper GJS, Aebi U. J Struct Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 25.Yonemoto IT, Kroon GJA, Dyson HJ, Balch WE, Kelly JW. Biochemistry-Us. 2008;47:9900–9910. doi: 10.1021/bi800828u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kayed R, Bernhagen J, Greenfield N, Sweimeh K, Brunner H, Voelter W, Kapurniotu A. J Mol Biol. 1999;287:781–796. doi: 10.1006/jmbi.1999.2646. [DOI] [PubMed] [Google Scholar]
- 27.Danielsson J, Jarvet J, Damberg P, Graslund A. Febs J. 2005;272:3938–3949. doi: 10.1111/j.1742-4658.2005.04812.x. [DOI] [PubMed] [Google Scholar]
- 28.Jarvet J, Damberg P, Danielsson J, Johansson I, Eriksson LEG, Graslund A. Febs Lett. 2003;555:371–374. doi: 10.1016/s0014-5793(03)01293-6. [DOI] [PubMed] [Google Scholar]
- 29.Jarvet J, Damberg P, Bodell K, Eriksson LEG, Graslund A. J Am Chem Soc. 2000;122:4261–4268. [Google Scholar]
- 30.Han W, Wu YD. J Am Chem Soc. 2005;127:15408–15416. doi: 10.1021/ja051699h. [DOI] [PubMed] [Google Scholar]
- 31.Baumketner A, Shea JE. J Mol Biol. 2006;362:567–579. doi: 10.1016/j.jmb.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 32.Teplow DB, Lazo ND, Bitan G, Bernstein S, Wyttenbach T, Bowers MT, Baumketner A, Shea JE, Urbanc B, Cruz L, Borreguero J, Stanley HE. Accounts Chem Res. 2006;39:635–645. doi: 10.1021/ar050063s. [DOI] [PubMed] [Google Scholar]
- 33.Wyttenbach T, Bowers MT. Top Curr Chem. 2003;225:207–232. [Google Scholar]
- 34.Sugita Y, Okamoto Y. Chem Phys Lett. 1999;314:141–151. [Google Scholar]
- 35.Swendsen RH, Wang JS. Phys Rev Lett. 1986;57:2607–2609. doi: 10.1103/PhysRevLett.57.2607. [DOI] [PubMed] [Google Scholar]
- 36.Hukushima K, Nemoto K. J Phys Soc Jpn. 1996;65:1604–1608. [Google Scholar]
- 37.Geyer CJ. Computing science and statistics: Proceedings of the 23rd Symposium on the Interface; 1991. pp. 156–163. [Google Scholar]
- 38.Wyttenbach T, Kemper PR, Bowers MT. Int J Mass Spectrom. 2001;212:13–23. [Google Scholar]
- 39.Ruotolo BT, Giles K, Campuzano I, Sandercock AM, Bateman RH, Robinson CV. Science. 2005;310:1658–1661. doi: 10.1126/science.1120177. [DOI] [PubMed] [Google Scholar]
- 40.Baker ES, Bernstein SL, Bowers MT. J Am Soc Mass Spectr. 2005;16:989–997. doi: 10.1016/j.jasms.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 41.Baker ES, Bernstein SL, Gabelica V, De Pauw E, Bowers MT. Int J Mass Spectrom. 2006;253:225–237. [Google Scholar]
- 42.Baker ES, Bowers MT. J Am Soc Mass Spectr. 2007;18:1188–1195. doi: 10.1016/j.jasms.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 43.Baker ES, Hong JW, Gaylord BS, Bazan GC, Bowers MT. J Am Chem Soc. 2006;128:8484–8492. doi: 10.1021/ja060069s. [DOI] [PubMed] [Google Scholar]
- 44.Baker ES, Lee JT, Sessler JL, Bowers MT. J Am Chem Soc. 2006;128:2641–2648. doi: 10.1021/ja0564968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gidden J, Baker ES, Ferzoco A, Bowers MT. Abstr Pap Am Chem S. 2004;227:U258–U258. [Google Scholar]
- 46.Gidden J, Baker ES, Ferzoco A, Bowers MT. Int J Mass Spectrom. 2005;240:183–193. [Google Scholar]
- 47.Gidden J, Ferzoco A, Baker ES, Bowers MT. J Am Chem Soc. 2004;126:15132–15140. doi: 10.1021/ja046433+. [DOI] [PubMed] [Google Scholar]
- 48.Wyttenbach T, Liu DF, Bowers MT. J Am Chem Soc. 2008;130:5993–6000. doi: 10.1021/ja8002342. [DOI] [PubMed] [Google Scholar]
- 49.Baumketner A, Bernstein SL, Wyttenbach T, Bitan G, Teplow DB, Bowers MT, Shea JE. Protein Sci. 2006;15:420–428. doi: 10.1110/ps.051762406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea JE. Protein Sci. 2006;15:1239–1247. doi: 10.1110/ps.062076806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Krone MG, Baumketner A, Bernstein SL, Wyttenbach T, Lazo ND, Teplow DB, Bowers MT, Shea JE. J Mol Biol. 2008;381:221–228. doi: 10.1016/j.jmb.2008.05.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bernstein SL, Wyttenbach T, Baumketner A, Shea JE, Bitan G, Teplow DB, Bowers MT. J Am Chem Soc. 2005;127:2075–2084. doi: 10.1021/ja044531p. [DOI] [PubMed] [Google Scholar]
- 53.Wu C, Murray MM, Bernstein SL, Condron MM, Bitan G, Shea JE, Bowers MT. J Mol Biol. 2009;387:492–501. doi: 10.1016/j.jmb.2009.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grabenauer M, Bernstein SL, Lee JC, Wyttenbach T, Dupuis NF, Gray HB, Winkler JR, Bowers MT. J Phys Chem B. 2008;112:11147–11154. doi: 10.1021/jp801175w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bernstein SL, Liu DF, Wyttenbach T, Bowers MT, Lee JC, Gray HB, Winkler JR. J Am Soc Mass Spectr. 2004;15:1435–1443. doi: 10.1016/j.jasms.2004.08.003. [DOI] [PubMed] [Google Scholar]
- 56.Mason EA, McDaniel EW. Transport properties of ions in gases. New York: Wiley; 1988. [Google Scholar]
- 57.Case DA, Cheatham TE, Darden T, Gohlke H, Luo R, Merz KM, Onufriev A, Simmerling C, Wang B, Woods RJ. J Comput Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gunsteren WFv, Weiner PK, Wilkinson T. Computer simulation of biomolecular systems. aordrecht ; Boston: Published under the KLUWER/ESCOM imprint by Kluwer Academic Publishers; 1997. [Google Scholar]
- 59.Shell MS, Ritterson R, Dill KA. J Phys Chem B. 2008;112:6878–6886. doi: 10.1021/jp800282x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ozkan SB, Wu GA, Chodera JD, Dill KA. P Natl Acad Sci USA. 2007;104:11987–11992. doi: 10.1073/pnas.0703700104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Onufriev A, Bashford D, Case DA. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 62.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23:327–341. [Google Scholar]
- 63.Procacci P, Berne BJ. Mol Phys. 1994;83:255–272. [Google Scholar]
- 64.Berendsen HJC, Postma JPM, Vangunsteren WF, Dinola A, Haak JR. J Chem Phys. 1984;81:3684–3690. [Google Scholar]
- 65.Frishman D, Argos P. Proteins-Structure Function and Genetics. 1995;23:566–579. doi: 10.1002/prot.340230412. [DOI] [PubMed] [Google Scholar]
- 66.Daura X, Gademann K, Jaun B, Seebach D, van Gunsteren WF, Mark AE. Angew Chem Int Edit. 1999;38:236–240. [Google Scholar]
- 67.Shvartsburg AA, Jarrold MF. Chem Phys Lett. 1996;261:86–91. [Google Scholar]
- 68.Mesleh MF, Hunter JM, Shvartsburg AA, Schatz GC, Jarrold MF. J Phys Chem-Us. 1996;100:16082–16086. [Google Scholar]
- 69.Williamson JA, Miranker AD. Protein Sci. 2007;16:110–117. doi: 10.1110/ps.062486907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Abedini A, Raleigh DP. Biochemistry-Us. 2005;44:16284–16291. doi: 10.1021/bi051432v. [DOI] [PubMed] [Google Scholar]
- 71.Charge SBP, Dekoning EJP, Clark A. Biochemistry-Us. 1995;34:14588–14593. doi: 10.1021/bi00044a038. [DOI] [PubMed] [Google Scholar]
- 72.Chiti F, Dobson CM. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 73.Jang S, Shin S. J Phys Chem B. 2008;112:3479–3484. doi: 10.1021/jp076450w. [DOI] [PubMed] [Google Scholar]
- 74.Jang S, Shin S. J Phys Chem B. 2006;110:1955–1958. doi: 10.1021/jp055568e. [DOI] [PubMed] [Google Scholar]
- 75.Pringle SD, Giles K, Wildgoose JL, Williams JP, Slade SE, Thalassinos K, Bateman RH, Bowers MT, Scrivens JH. Int J Mass Spectrom. 2007;261:1–12. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.