Abstract
Background
The role of the innate immune system in the pathophysiology of chronic rhinosinusitis (CRS) is poorly understood. In this study, we compared sinonasal expression of toll-like receptors (TLRs), complement components, serum amyloid A, and inflammatory genes (chemokines and cytokines) in control subjects and patients undergoing sinus surgery for CRS.
Methods
Eleven control subjects and 30 subjects with CRS unresponsive to medical management were enrolled prospectively before undergoing endoscopic sinus surgery. Ethmoid mucosal specimens were obtained surgically and processed for RNA extraction. Real-time polymerase chain reaction was used to quantitate the level of expression of messenger RNA (mRNA) for TLR, acute phase proteins, and cytokine genes. Subjects were followed for a minimum of 6 months postoperatively with nasal endoscopy to assess for recurrence of polyps.
Results
mRNA for all target genes was detected in the ethmoid mucosa of both control and CRS subjects. The level of gene expression was normalized to the housekeeping genes 18s RNA and glyceraldehyde-3-phosphate dehydrogenase. As compared with controls, CRS was associated with significantly higher expression of TLR2 and the inflammatory genes macrophage-inflammatory protein α, RANTES, and granulocyte-macrophage colony-stimulating factor. Patients with early recurrence of polyps after surgery had significantly decreased expression of TLR2, 9, and serum amyloid A and increased expression of macrophage-inflammatory protein α compared with surgery-responsive patients.
Conclusion
This study shows the increased levels of expression of TLR2 and a variety of inflammatory genes in sinonasal mucosa of CRS patients compared with controls. Whether these differences play a role in pathogenesis or are merely manifestations of disease activity is worthy of investigation.
Chronic rhinosinusitis (CRS) is a widely prevalent health condition that may severely impact the quality of life of affected individuals. When the disease process responds poorly to medical and surgical therapy, CRS can be a frustrating disorder for the otolaryngologist to manage. Although a number of contributing factors are recognized, the underlying pathogenesis of CRS remains largely unknown. CRS with nasal polyps (CRSwNP) is characterized by persistent eosin-ophilic inflammation, edematous mucosa with polyps, and thickened sinonasal secretions.1 Although it has been proposed that CRSwNP is fundamentally a disease of inflammation, rather than infection, it also has been postulated that microbes often found in the nasal cavity, such as fungi, bacteria, viruses, or their associated products, may play a role in initiating or perpetuating mucosal inflammation.2–8 It is clear that these microorganisms can be found in patients both with and without sinus disease, and evidence that eradication of these organisms cures CRS is inconsistent. However, an attractive theory of CRS is that it represents a dysregulation of normal mucosal immune mechanisms that act to protect or repair the sinonasal epithelium.
Situated at the boundary of the body and the outside world, the sinonasal mucosa has many strategies to cope with the threat of infection. In broad terms, immune mechanisms can be divided into those that are innate and those that are adaptive. The adaptive mechanisms, including the generation and trafficking of antigen-specific lymphocyte populations, are well studied and are unquestionably active in CRS.9,10 The more evolutionarily ancient innate pathways of defense are less emphasized but are beginning to gain attention as important players in many inflammatory processes. Epithelial cells contribute to the defense of the mucosa by acting as a mechanical barrier and by keeping the mucus blanket in motion through the movement of their cilia. In addition, epithelial cells are able to elaborate antimicrobial products that assist in inhibiting growth of microorganisms at the mucosal surface. 11–13 These defensive strategies are nonspecific and often constitutively active. More recently, it has been discovered that human sinonasal epithelial cells express a series of pattern recognition proteins known as toll-like receptors (TLRs).14 These receptors allow epithelial cells to recognize pathogen-associated molecular patterns in the outside environment. On activation of TLRs, epithelial cells may initiate production of defensive molecules specific to a particular pathogen and perhaps transmit a “danger” signal to alert the appropriate elements of the adaptive immune system.15–18 Through an immediate and aggressive offensive response, the sinonasal epithelium can ward off pathogens before they have the opportunity to threaten the health of the host. Numerous other cell types can participate in innate immune responses of the upper airways, including secretory cells, macrophages, dendritic cells, infiltrating leukocytes, and other tissue cells.
Although the mechanisms underlying the persistent inflammation in CRS are unknown, innate immune processes may play a role. The early recruitment of immune effector cells to sites of inflammation is dictated by the local production of specific signaling mediators and cell surface proteins. It is critical to understand how events occurring at the sinonasal mucosal surface are translated to the immune system through the epithelium, to learn how dysfunction of this process might result in persistent immune activation. A starting point for such an analysis is the documentation of the pattern of sinonasal TLR and innate immune effector gene expression in health and sinus disease. This may implicate specific TLRs or suggest an innate immune pathway that is abnormally regulated in CRS. In this study, we investigated the hypothesis that CRS is associated with altered expression of TLRs and innate immune mediators in the sinonasal mucosa and that the resistant form of CRSwNP has a different pattern of expression than the form more amenable to medical and surgical control.
METHODS
Patients
Thirty patients with CRS and 10 control subjects were enrolled in the study. The research protocol was approved through the Johns Hopkins Institutional Review process, and all subjects gave signed informed consent. The CRS patients were defined by historical, endoscopic and radiographic criteria and by meeting the definition of the American Academy of Otolaryngology–Head and Neck Surgery Chronic Rhinosinusitis Task Force.19 Specifically, the patients had continuous symptoms of rhinosinusitis as defined by the Task Force report for >12 consecutive weeks, associated with computed tomography of the sinuses revealing isolated or diffuse sinus mucosal thickening or air-fluid levels. Twenty of the patients met criteria for CRS with polyps (CRSwNP), and the other 10 patients were classified as CRS without nasal polyps.1
Surgery was performed only if a patient’s symptoms and radiographic findings failed to resolve despite at least 6 weeks of treatment with oral antibiotics, topical corticosteroids, decongestants, and mucolytic agents, as well as 4 weeks of systemic corticosteroid. The 10 control subjects had no evidence of sinus disease and were undergoing endoscopic approaches for orbital decompression, cerebrospinal fluid leak repair, or sphenoidotomy for biopsy of an isolated, noninflammatory sphenoid sinus process. Before surgery, the patients with CRS received 1 week of oral methylprednisolone and oral antibiotics. All tissue specimens were taken from the resected uncinate process (Fig. 1) or the anterior ethmoid sinus. The CRS specimens were carefully selected to avoid polyps or tissue that was grossly polypoid in appearance. The specimens were immediately placed in RNAlater (chloride, pH 7.4; Ambion, Austin, TX)). The samples were refrigerated until further processing was performed. All CRS patients received identical perioperative medications, including systemic corticosteroids, and they underwent weekly sinonasal endoscopy for the 1st month after surgery. The number and frequency of subsequent visits was determined by the needs of the individual patients. All patients were followed for a minimum of 9 months after surgery and assessed for recurrence or persistence of polyps.
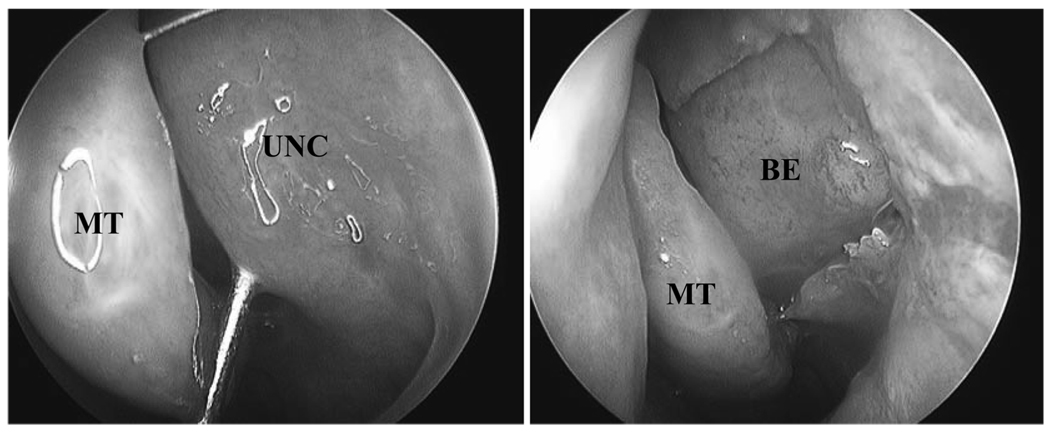
Figure 1.
Standard procedure for obtaining uncinate mucosa endoscopic sinus surgery. The uncinate process was resected in an atraumatic fashion and immediately preserved in RNAlater solution. UNC, uncinate process; MT, middle turbinate; BE, bulla ethmoidalis.
RNA Extraction/Reverse Transcription
Total messenger RNA (mRNA) was isolated from the stored mucosa using the TRIzol (0.8 mL) reagent (Invitrogen, Carlsbad, CA) and DNA was removed using DNAfree (Ambion) per the manufacturer’s protocol. Isolated mRNA was reverse transcribed using a poly(dT)15 primer (Roche, Indianapolis, IN) and Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA) using conditions provided by the manufacturer. Samples were visualized using agarose gel electrophoresis.
Real-Time Polymerase Chain Reaction
Real-time polymerase chain reaction (RT-PCR) was performed in an ABI PRISM 7700 Sequence Detection System thermal cycler (PE Applied Biosystems, Foster City, CA) to quantify target mRNA, as well as mRNA for the housekeeping genes glyceraldehyde-3-phosphate dehydrogenase (GAPDH), β-actin, 18s RNA, and hypoxanthine phosphoribosyltransferase (HPRT). Primers and probes were designed using Primer Express 1.5 software (PE Applied Biosystems) from sequences stored in the GenBank sequence database. Target genes, primers, and probes used are listed in Table 1. Primer and probe sets for β-actin, HPRT, and 18s RNA were purchased as Taqman Assays on Demand kits from Applied Biosystems (PE Applied Biosystems).
Table 1.
Primer/Probe Combinations for TLRs and Other Gene Targets
| Target | Forward Primer | Reverse Primer | Probe |
|---|---|---|---|
| TLR1 | TCAAGACTGTAGCAAATCTGGAACTATC | TTTCAAAAACCGTGTCTGTTAAGAGA | TGCCCATCCAAAATTAGCCCGTTCCTA |
| TLR2 | GGCCAGCAAATTACCTGTGTG | AGGCGGACATCCTGAACCT | CCATCCCATGTGCGTGG |
| TLR3 | CCTGGTTTGTTAATTGGATTAACGA | GATTGGTCTTCCTTTTCCATTGAA | CATACCAACATCCCTGAGCT |
| TLR4 | GCAGTGAGGATGATGCCAGGAT | GCCATGGCTGGGATCAGAGT | TGTCTGCCTCGCGCC |
| TLR5 | TGCCTTGAAGCCTTCAGTTATG | CCAACCACCACCATGATGAG | CCAGGGCAGGTGCTTATCTGACCTTAACA |
| TLR6 | GAAGAAGAACAACCCTTTAGGATAGC | AGGCAAACAAAATGGAAGCTT | TGCAACATCATGACCAAAGACAAAGAACCTA |
| TLR7 | TTACCTGGATGGAAACCAGCTAC | TCAAGGCTGAGAAGCTGTAAGCTAG | AGATACCGCAGGGCCTCCCGC |
| TLR8 | AGCGGATCTGTAAGAGCTCCATC | CCGTGAATCATTTTCAGTCAAGAC | CCTGACAACCCGAAGGCAGAAGGC |
| TLR9 | GCAGTCAATGGCTCCCAGTTC | GCGGTAGCTCCGTGAATGAGTG | CCCGCAATAAGCTG |
| TLR10 | TTATGACAGCAGAGGGTGATGC | CTGGAGTTGAAAAAGGAGGTTATAGG | TTGACCCCAGCCACAACGACACTG |
| Factor P | ACCCACATCTGCAACACAGCT | CCCGCGTGACTGGTGG | CTGTCAAGAAATCC |
| Factor B | CCCTACTACAATGTGAGTGATGAGATC | GTCTGCCCACTCCATCGG | CTCTGCCAATCGCAC |
| SAA | CTTGGCGAGCCTTTTGATG | TAGTTCCCCCGAGCATGG | ACATGAGAGAAGCCAATTA |
| C3 | AGAAAGACATGGCCCTCACG | TCGCAAATATCTTTAGCCTCCTG | CTTTGTTCTCATCTCGC |
| MIP-1α | CTACACCTCCCGGCAGATTC | CCGGCTTCGCTTGGTTAG | CAGTGCTCCAAGCCCGGTGTCATC |
| RANTES | TCGCTGTCATCCTCATTGCTA | GCACTTGCCACTGGTGTAGAAA | CTGGGACACCACACCCTGCTGC |
| GM-CSF | AGGGCCCCTTGACCATGA | CAAAGGTGATAGTCTGGGTTGCA | CAGCACTGCCCTCCAACCCCG |
| TNF-α | Applied Biosystems Assay on Demand Kit | ||
| β-Actin | CTGGCCGGGACCTGAT | GCAGCCGTGGCCATCTC | CACCACCACGGCCA |
| HPRT | Applied Biosystems Assay on Demand Kit | ||
| 18s RNA | Applied Biosystems Assay on Demand Kit |
One microliter of each complementary DNA (cDNA) preparation, corresponding to 25 µg of mRNA, was diluted 1:10 and 5 µL of this preparation was placed in a total volume of 25 µL with the following components: 1X TaqMan PCR buffer, 5.5 mM of MgCl2, 0.25 mM of dNTP (dATP, dCTP, dUTP, and dGTP), 0.25 U of AmpErase UNG (uracil-N-glycosylase, Roche, Indianapolis, IN), 0.75 U of Ampli Taq Gold (PE Applied Biosystems, Foster City, CA), 0.4 µM of each primer, and 0.2 µM of the Taqman probe. Cycle parameters used were 50°C for 2 minutes to activate UNG and 95°C for 10 minutes to activate Taq, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The level of expression of target mRNA was determined as the ΔCT. The ΔCT method uses the difference in CT value obtained between a normalizing housekeeping gene and the target gene to calculate relative quantification (ΔCT is the difference in threshold cycles for target and housekeeping gene). This normalization reduces sample-to-sample variations in signal strength. A decrease in the ΔCT by 1 U equals a doubling of the level of the target gene.
Data Analysis
Raw data from RT-PCR was entered into a Microsoft Excel (Microsoft Corp., Redmond, WA) spreadsheet and statistical analysis was performed. Data are expressed as mean values ± SEM. Statistical significance of differences was determined by Student’s t-test. Differences were considered statistically significant at p < 0.05.
RESULTS
Patients
The 10 control subjects were confirmed to have normal sinus mucosa at the time of surgery and did not have any evidence of inflammatory sinonasal mucosal disease in the postoperative follow-up period. Of the 20 CRSwNP patients, 11 developed recurrence or persistence of polyps or diffuse polypoid mucosa within 6 months after surgery. In 10 of those patients, the sinonasal mucosa never completely normalized after surgery. There was a single patient in whom the nasal polyps returned in a delayed fashion after a period of apparent resolution. The remaining 9 CRSwNP patients did not have regrowth of polyps over the follow-up period (at least 6 months) after the surgery. Figure 2 shows representative endoscopic images from the nasal cavities of a responsive and a recalcitrant CRSwNP subject. In the CRS without nasal polyps group, 8 out of 10 patients had complete resolution of their mucosal disease after surgery. The remaining two patients developed diffuse polypoid mucosal changes associated with persistent symptoms.
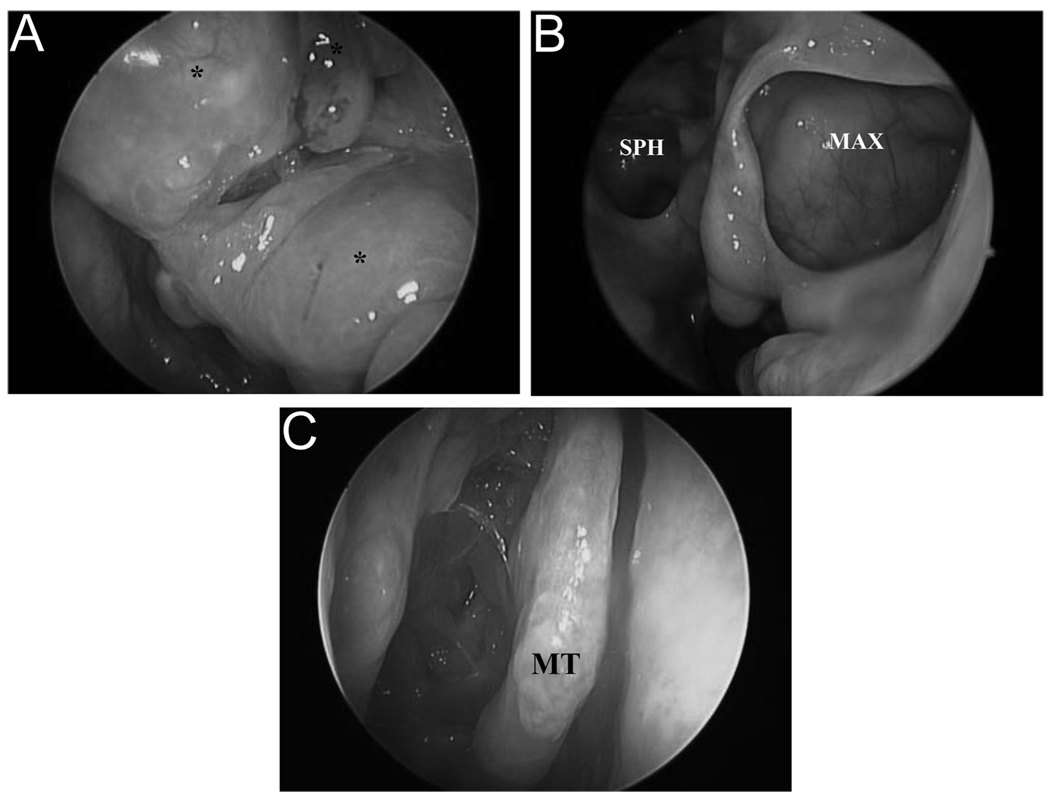
Figure 2.
Representative examples of responsive and recalcitrant CRS. (A) Responsive CRSwNP prior to surgical management. Left nasal cavity is shown (*nasal polyps). (B) Same sinonasal cavity 6 months postsurgery (MAX, maxillary sinus; SPH, sphenoid sinus). (C) Recalcitrant CRSwNP 9 months postsurgery. The right nasal cavity is shown (MT, middle turbinate).
RT-PCR
CRS Compared with Controls
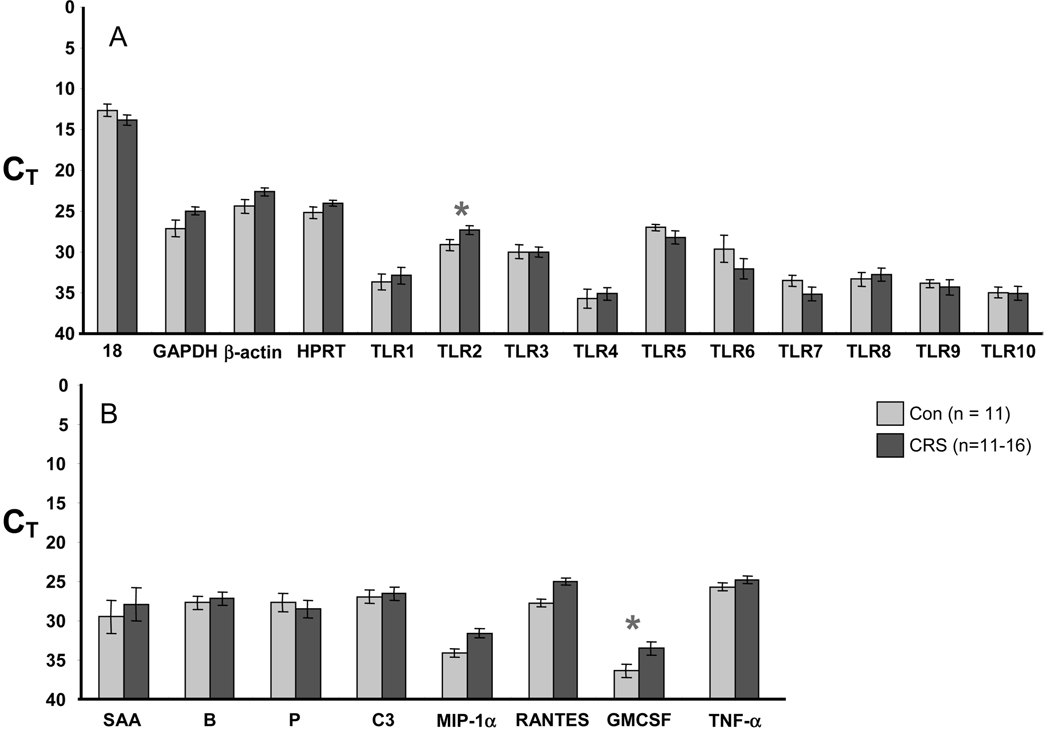
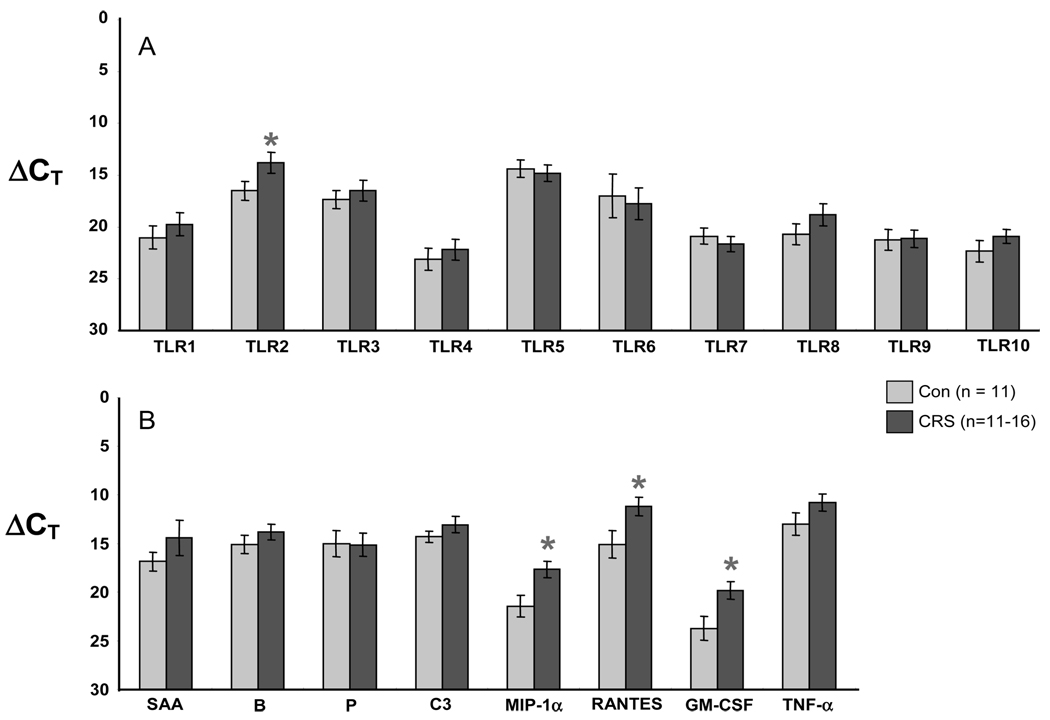
RT-PCR showed that all 10 TLR genes were expressed in both controls and CRS patients. Figure 3 depicts the level of expression for the 10 TLR genes as well as five acute phase-like proteins: serum amyloid A (SAA), complement factor B, complement factor P, and C3. Values in panels A and B are expressed as raw CT; a lower value corresponds to a higher level of expression and each unit represents a twofold change. CRS patients had statistically lower CT for TLR2 and granulocyte-macrophage colony-stimulating factor (GM-CSF; and thus higher levels of expression) when compared with the controls (p < 0.05). Figure 4 depicts ΔCT, which is the raw PCR cycle threshold CT for each target gene normalized to the CT for the housekeeping gene 18s RNA. There were statistically significant increases in expression of TLR2 (Fig. 4A) and in the inflammatory genes macrophage-inflammatory protein α (MIP-1α), RANTES, and GM-CSF (Fig. 4B) in CRS relative to controls (p < 0.05). We chose 18s RNA for normalization because the raw CT values for the housekeeping genes GAPDH, β-actin, and HPRT were decreased in the CRS samples as compared with controls (i.e., increased levels in CRS, see left side of Fig. 3A). It is important to note that each CT unit represents a twofold change. Thus, levels of mRNA for TLR2, MIP-1α, RANTES, and GM-CSF were all elevated approximately 5- to 10-fold.
Figure 3.
RT-PCR comparison of (A) housekeeping genes and TLR or (B) host defense and inflammatory genes in control (light bars) and CRS patients (dark bars). Values are CT; a lower value is a higher level of expression and each unit represents a twofold change. Figure shows increased expression of TLR2 and GM-CSF in CRS patients (*p < 0.05; TLR2, p < 0.04; GM-CSF, p = 0.04).
Figure 4.
RT-PCR comparison (A) of the expression of TLR and (B) acute phase proteins and inflammatory genes in control (light bars) and CRS patients (dark bars). Values are ΔCT, normalized to the housekeeping gene 18s RNA; a lower ΔCT value corresponds to a higher level of expression. Figure shows increased expression of TLR2, MIP-1α, RANTES, and GM-CSF in sinonasal tissue from CRS patients (*p < 0.05; TLR2, p = 0.05; MIP-1α, p = 0.01; RANTES, p = 0.03; GM-CSF, p = 0.02).
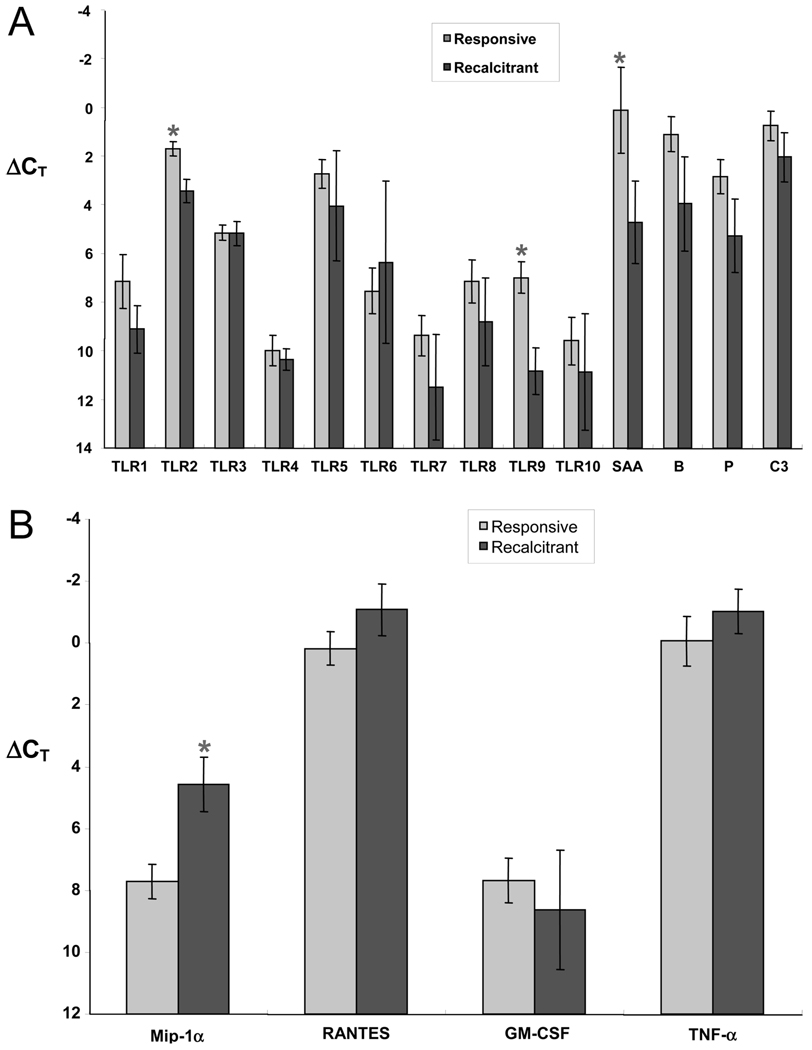
Recalcitrant CRS Compared with Treatment-Responsive CRS
Based on the postoperative course of the CRS patients over the next 9 months, the CRS samples were subdivided into recalcitrant (13 patients) and treatment-responsive (17 patients) groups. These samples were then compared for their level of expression of the innate immunity gene targets and cytokine genes normalized to GAPDH (Fig. 5). There was no statistical difference between GAPDH CT for the responsive (24.4 ± 0.8) and recalcitrant (24.3 ± 0.4) patients. The results indicate that TLR2, TLR9, and SAA were significantly different between groups, with lower gene expression in the recalcitrant CRS group than the responsive CRS group (Fig. 5A). On the other hand, MIP-1α gene expression was significantly increased in recalcitrant CRS patients compared with the treatment-responsive CRS patients (Fig. 5B).
Figure 5.
(A) RT-PCR comparison of the expression of TLR and acute phase protein genes in treatment-responsive (light bars) and recalcitrant CRS (dark bars) samples. Values are ΔCT, normalized to the housekeeping gene GAPDH; a lower ΔCT value corresponds to a higher level of expression. Figure shows decreased expression of TLR2, TLR9, and SAA in sinonasal tissue from CRS patients (*p < 0.05; TLR2, p = 0.003; TLR9, p = 0.002; SAA, p = 0.04). (B) RT-PCR comparison of expression of inflammatory genes in treatment-responsive (light bars) and recalcitrant CRS (dark bars) samples. Values are ΔCT, normalized to the housekeeping gene GAPDH; a lower ΔCT value corresponds to a higher level of expression. Figure shows increased expression of MIP-1α in sinonasal tissue from CRS patients with recalcitrant disease (*p < 0.05, MIP-1α, p < 0.05).
DISCUSSION
The sinonasal mucosa often is the first site of contact for airborne microorganisms and viruses. Multiple innate and adaptive immune strategies of defense have evolved to prevent these agents from colonizing or infecting the airway. It is hypothesized that dysregulation of these pathways could contribute to the persistent inflammatory state seen in CRS. In this study, we begin to explore the potential role of the innate immune system in the pathogenesis of CRS. We have shown that the expression of TLR2 and a number of inflammatory genes is higher in patients with sinus disease. Moreover, the most recalcitrant cases of CRS were found to be associated with significantly lower expression of TLR2, TLR9, and acute phase proteins, and higher expression of the inflammatory gene MIP-1α, when compared with CRS that is responsive to combined medical and surgical management. These findings suggest that the role of innate immune processes in the pathogenesis of CRS merits additional investigation.
In light of the association of fungal elements or fungal colonization with certain forms of CRS, it is interesting to note that, in drosophila, Toll protein is a molecule critical to resistance against fungal infection. When drosophila Toll binds to fungal components, it activates a signal transduction pathway and transcription factor that up-regulates expression of drosomycin, an antifungal peptide.18,20 Because insects lack an adaptive immune system, Toll acts as the central agent of the fly’s pathogen defense against fungus. Using the Toll gene sequence as a model, a series of 11 homologous TLRs have been identified in the mammalian genome. Unlike the role of Toll in flies, TLR genes in humans and other mammals are not restricted to fungal immunity. A diverse set of bacterial and viral products, such as unmethylated bacterial DNA motifs, lipopolysaccharide, viral double-stranded RNA, mycobacterial lipoproteins, and flagellin are recognized by the different TLRs.21,22 Two TLRs in particular, TLR2 and TLR4, have been implicated as being involved in fungal recognition in mammalian cells, although these receptors are activated also by products of both Gram-positive and Gram-negative bacteria. 20,23 Increased TLR2 gene expression in CRS patients therefore could be of relevance whether bacteria or fungi are involved in the pathogenesis of CRS. Binding of TLRs induces signaling cascades and changes in gene expression that are being elucidated rapidly. When TLRs are made constitutively active in human cell cultures, there is activation of NF-κB pathways commonly associated with inflammation.24 In human epithelial cell lines, activation of TLR by double-stranded RNA and other agonists has been shown to result in the increased expression of innate and adaptive proinflammatory genes.25
A common element in the existing competing theories of CRS pathogenesis is the concept that there is an abnormal immune response to a trigger at the mucosal surface. Whether the inciting factor is a fungus, a staphylococcal antigen, a biofilm of Gram-negative bacteria, or any other microbial agent, there is ultimately a common pathway of immune activation that characterizes CRS. The role of the epithelium in regulating the sinonasal mucosal immune response to these agents has not been explored fully. The increased expression of TLR2 in CRS may either reflect an underlying pathophysiological mechanism in the disease or may occur secondarily as a consequence of the ongoing inflammatory process. We have not established whether increased TLR2 mRNA is associated with either increased TLR2 protein or increased signaling. Increased TLR2 responses could contribute to the inflammatory responses in CRS. TLR2 has been shown to be involved in the response of airway epithelial cells to air pollution particulates, as well as to fungi and Gram-positive bacteria.26 TLR9 is activated by viral products as well as by bacterial CpG DNA motifs.27,28 As expression of both of these TLRs seems to be expressed at higher levels in patients with surgery-responsive CRS; this may reflect a greater response to colonization by bacteria or fungi or infection by viruses. Systemic corticosteroids, which are potent anti-inflammatory agents in CRS, not only decrease expression of inflammatory cytokines, but also tend to increase expression of innate immune genes.29 Additional studies will be necessary to determine the influence of corticosteroids on expression of inflammatory genes, TLR, and host defense molecules in CRS.
A recent study by Claeyes et al. reported that expression of TLR2 and TLR4 was not different in patients with nasal polyposis compared with those with hypertrophic adenoids.30 In contrast, Pitzurra et al. showed the absence of TLR2, TLR3, TLR4, TLR5, and TLR9 in patients with confirmed colonization of the nasal vestibule with Staphylococcus aureus.31 In this study, we were careful to use nonpolypoid tissue from the ethmoid sinus of both CRS patients and controls. Increased TLR2 expression was observed when using both the raw CT values and values normalized to 18s RNA. The main disadvantages of analysis of whole tissues are variability in location from which the tissue was derived among subjects and variability of the cell types that occupy these surgical samples. Clearly, careful analysis of the expression of TLR, inflammatory genes, and host defense genes in single cell types (e.g., mucosal epithelial cells) will be necessary to resolve some of the variable results reported in the literature. Previous studies have shown increased expression of inflammatory genes in CRS including RANTES, GM-CSF, and tumor necrosis factor α.9,32,33 Our data showing higher levels of mRNA for these genes support the findings of these earlier studies. In this study we made the novel observation of an increase in the expression of mRNA for the chemokine MIP-1α in CRS patients when compared with the controls. Furthermore, this chemokine also was found to be increased in patients with recalcitrant disease compared with CRS patients responsive to surgery. MIP-1α is important for the recruitment of cells involved in both inflammation and host defense, including mononuclear cells and neutrophils. Additional studies will be necessary to determine whether MIP-1α protein levels are increased in CRS.
CRS is a heterogeneous disorder that is variable in its natural history and response to therapy. Recent efforts to define subcategories of CRS have identified two large divisions based on the presence or absence of polyps.1 CRSwNP, which often is associated with diffuse eosinophilic mucosal inflammation, can be difficult to control. However, even within the CRSwNP population, there is further variability in the degree of recalcitrance to medical and surgical therapy. Based on this study, differences in the level of expression of TLRs and other genes involved in innate immunity may be associated with disease reversibility. It would be beneficial to both the physician and the patient to identify additional clinical and laboratory parameters that might predict responsiveness to therapy. At the mucosal battlefield of the sinonasal cavity, the best defense is a good offense. The epithelium on the front line acts to detect pathogens, respond with an immediate counterattack, and alert the adaptive immune system. When these innate functions are impaired or suppressed, it may become possible for bacteria, fungi, or viruses to gain the advantage. Alternatively, overactivity of TLR and innate immune effectors could play an important role in the persistent inflammatory responses that characterize CRS. Additional research into the function of the innate immune system of the upper airway in health and disease is needed to better understand how these processes relate to CRS.
Footnotes
Presented at the spring meeting of the American Rhinologic Society, Boca Raton, Florida, May 2005
REFERENCES
- 1.Meltzer EO, Hamilos DL, Hadley JA, et al. Rhinosinusitis: Establishing definitions for clinical research and patient care. Otolaryngol Head Neck Surg. 2004;131 suppl 6:S1–S62. doi: 10.1016/j.otohns.2004.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duncavage J. Chronic rhinosinusitis is not always infectious sinusitis. Curr Opin Otolaryngol Head Neck Surg. 2005;13:1. [Google Scholar]
- 3.Bachert C, Gevaert P, van Cauwenberge P. Staphylococcus aureus enterotoxins: A key in airway disease? Allergy. 2002;57:480–487. doi: 10.1034/j.1398-9995.2002.02156.x. [DOI] [PubMed] [Google Scholar]
- 4.Shin SH, Ponikau JU, Sherris DA, et al. Chronic rhinosinusitis: An enhanced immune response to ubiquitous airborne fungi. J Allergy Clin Immunol. 2004;114:1369–1375. doi: 10.1016/j.jaci.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Conley DB, Tripathi A, Ditto AM, et al. Chronic sinusitis with nasal polyps: Staphylococcal exotoxin immunoglobulin E and cellular inflammation. Am J Rhinol. 2004;18:273–278. [PubMed] [Google Scholar]
- 6.Holtzman MJ, Shornick LP, Grayson MH, et al. “Hit-and-run” effects of paramyxoviruses as a basis for chronic respiratory disease. Pediatr Infect Dis J. 2004;23 suppl 11:S235–S245. doi: 10.1097/01.inf.0000144674.24802.c1. [DOI] [PubMed] [Google Scholar]
- 7.Cryer J, Schipor I, Perloff JR, Palmer JN. Evidence of bacterial biofilms in human chronic sinusitis. ORL J Otorhinolaryngol Relat Spec. 2004;66:155–158. doi: 10.1159/000079994. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein JM, Kansal R. Superantigen hypothesis for the early development of chronic hyperplastic sinusitis with massive nasal polyposis. Curr Opin Otolaryngol Head Neck Surg. 2005;13:39–44. doi: 10.1097/00020840-200502000-00010. [DOI] [PubMed] [Google Scholar]
- 9.Hamilos DL. Chronic sinusitis. J Allergy Clin Immunol. 2000;106:213–227. doi: 10.1067/mai.2000.109269. [DOI] [PubMed] [Google Scholar]
- 10.Pinto JM, Baroody FM. Chronic sinusitis and allergic rhinitis: At the nexus of sinonasal inflammatory disease. J Otolaryngol. 2002;31 suppl 1:S10–S17. doi: 10.2310/7070.2002.21310. [DOI] [PubMed] [Google Scholar]
- 11.Dauletbaev N, Gropp R, Frye M, et al. Expression of human beta defensin (HBD-1 and HBD-2) mRNA in nasal epithelia of adult cystic fibrosis patients, healthy individuals, and individuals with acute cold. Respiration. 2002;69:46–51. doi: 10.1159/000049369. [DOI] [PubMed] [Google Scholar]
- 12.Chen PH, Fang SY. The expression of human antimicrobial peptide LL-37 in the human nasal mucosa. Am J Rhinol. 2004;18:381–385. [PubMed] [Google Scholar]
- 13.Kaliner MA. Human nasal respiratory secretions and host defense. Am Rev Respir Dis. 1991;144:S52–S56. doi: 10.1164/ajrccm/144.3_pt_2.S52. [DOI] [PubMed] [Google Scholar]
- 14.Vandermeer J, Sha Q, Lane AP, Schleimer RP. Innate immunity of the sinonasal cavity: Expression of messenger RNA for complement cascade components and toll-like receptors. Arch Otolaryngol Head Neck Surg. 2004;130:1374–1380. doi: 10.1001/archotol.130.12.1374. [DOI] [PubMed] [Google Scholar]
- 15.Matzinger P. The danger model: A renewed sense of self. Science. 2002;296:301–305. doi: 10.1126/science.1071059. [DOI] [PubMed] [Google Scholar]
- 16.Pasare C, Medzhitov R. Toll-like receptors: Linking innate and adaptive immunity. Microbes Infect. 2004;6:1382–1387. doi: 10.1016/j.micinf.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 17.Seitz M. Toll-like receptors: Sensors of the innate immune system. Allergy. 2003;58:1247–1249. doi: 10.1046/j.1398-9995.2003.00225.x. [DOI] [PubMed] [Google Scholar]
- 18.Janssens S, Beyaert R. Role of toll-like receptors in pathogen recognition. Clin Microbiol Rev. 2003:16637–16646. doi: 10.1128/CMR.16.4.637-646.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lanza DC, Kennedy DW. Adult rhinosinusitis defined. Otolaryngol Head Neck Surg. 1997;117:S1–S7. doi: 10.1016/S0194-59989770001-9. [DOI] [PubMed] [Google Scholar]
- 20.Netea MG, Van der Graaf C, Van der Meer JW, Kullberg BJ. Recognition of fungal pathogens by toll-like receptors. Eur J Clin Microbiol Infect Dis. 2004;23:672–676. doi: 10.1007/s10096-004-1192-7. [DOI] [PubMed] [Google Scholar]
- 21.Akira S. Mammalian toll-like receptors. Curr Opin Immunol. 2003;15:5–11. doi: 10.1016/s0952-7915(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 22.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–376. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 23.Mambula SS, Sau K, Henneke P, et al. Toll-like receptor (TLR) signaling in response to Aspergillus fumigatus. J Biol Chem. 2002;277:39320–39326. doi: 10.1074/jbc.M201683200. [DOI] [PubMed] [Google Scholar]
- 24.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300:1524–1525. doi: 10.1126/science.1085536. [DOI] [PubMed] [Google Scholar]
- 25.Sha Q, Truong-Tran AQ, Plitt JR, et al. Activation of airway epithelial cells by toll-like receptor agonists. Am J Respir Cell Mol Biol. 2004;31:358–364. doi: 10.1165/rcmb.2003-0388OC. [DOI] [PubMed] [Google Scholar]
- 26.Becker S, Dailey L, Soukup JM, et al. TLR-2 is involved in airway epithelial cell response to air pollution particles. Toxicol Appl Pharmacol. 2005;203:45–52. doi: 10.1016/j.taap.2004.07.007. 15. [DOI] [PubMed] [Google Scholar]
- 27.Platz J, Beisswenger C, Dalpke A, et al. Microbial DNA induces a host defense reaction of human respiratory epithelial cells. J Immunol. 2004;173:1219–1223. doi: 10.4049/jimmunol.173.2.1219. [DOI] [PubMed] [Google Scholar]
- 28.Finberg RW, Kurt-Jones EA. Viruses and toll-like receptors. Microbes Infect. 2004;6:1356–1360. doi: 10.1016/j.micinf.2004.08.013. [DOI] [PubMed] [Google Scholar]
- 29.Schleimer R. Glucocorticoids suppress inflammation but spare innate immune responses in airway epithelium. Proc Am Thorac Soc. 2004;1:222–230. doi: 10.1513/pats.200402-018MS. [DOI] [PubMed] [Google Scholar]
- 30.Claeys S, de Belder T, Holtappels G, et al. Human beta-defensins and toll-like receptors in the upper airway. Allergy. 2003;58:748–753. doi: 10.1034/j.1398-9995.2003.00180.x. [DOI] [PubMed] [Google Scholar]
- 31.Pitzurra L, Bellocchio S, Nocentini A, et al. Antifungal immune reactivity in nasal polyposis. Infect Immun. 2004;72:7275–7281. doi: 10.1128/IAI.72.12.7275-7281.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamilos DL, Leung DYM, Huston DP, et al. GM-CSF, IL-5, and RANTES immunoreactivity and mRNA expression in chronic hyperplastic sinusitis with nasal polyposis (NP) Clin Exp Allergy. 1998;28:1145–1152. doi: 10.1046/j.1365-2222.1998.00380.x. [DOI] [PubMed] [Google Scholar]
- 33.Meltzer EO, Hamilos DL, Hadley JA, et al. Rhinosinusitis: Establishing definitions for clinical research and patient care. J Allergy Clin Immunol. 2004;114 suppl 6:155–212. doi: 10.1016/j.jaci.2004.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]