Abstract
Protein phosphorylation serves as a primary mechanism of signal transduction in the cells of biological organisms. Technical advancements over the last several years in mass spectrometry now allow for the large-scale identification and quantitation of in vivo phosphorylation at unprecedented levels. These developments have occurred in the areas of sample preparation, instrumentation, quantitative methodology, and informatics so that today, ten to twenty thousand phosphorylation sites can be identified and quantified within a few weeks. With the rapid development and widespread availability of such data, its translation into biological insight and knowledge is a current obstacle. Here we present an overview of how this technology came to be and is currently applied, as well as future challenges for the field.
Keywords: Protein phosphorylation; Post translational modification that can occur on proteins in which a phosphoryl group is added; Phosphoproteomics; The global analysis of protein phosphorylation on a given proteome; Shotgun proteomics; High-throughput sequencing method where peptides from digested protein samples are subjected to liquid chromatography and subsequent analysis with tandem mass spectrometry. After the mass-to-charge ratio (m/z) and intensity of eluting peptide precursors is recorded by an initial MS1 scan, the m/z values for peaks with high intensity are automatically selected for dissociation and determination of the m/z of the fragment ions in a subsequent MS2 scan; Collisional dissociation; Several similar methods (CAD, HCD, PQD) for dissociation of peptide cations during tandem MS analysis via collsions with inert gas molecules; Electron-based dissociation; Methods of peptide ion dissociation based on the capture (ECD) or transfer (ETD) of an electron, which are particularly effective for peptides with labile post-translational modifications (e.g., phosphorylation, glycoslyation, etc.); Phosphopeptide enrichment; Separation of phosphopeptides from more abundant non-modified peptides prior to tandem MS analysis. Commonly used approaches include chelating acidic phosphoryl groups with metals (e.g., IMAC, MOAC), immunoaffinity purification with phosphotyrosine-specific antibodies, and separation based on isoelectric point (e.g., SCX) or polarity (HILIC); Isotope labeling; Strategies in which stable isotopes are incorporated into peptides from different samples, allowing for quantitative comparison between conditions during MS; False discovery rate; The standard measure of error rate for proteomic datasets. It is defined as the number of incorrect matches (false positives) over the total number of matches (true positives plus false positives), and is commonly estimated by target-decoy database searching
PHOSPHOPROTEOMICS APPLICATIONS
Protein phosphorylation is a central mechanism of signal transduction across species, with kinases and phosphatases accounting for 2–4% of eukaryotic proteomes (1–2). Current estimates suggest that one-third of eukaryotic proteins are phosphorylated (3–4) – determining the sites, abundances, and roles of each of these modifications in a biological sample is a critical challenge. Traditional biochemical techniques are chiefly limited to testing how phosphoryl modifications at specific sites affect a single protein of interest (5). Identification of unknown in vivo phosphoryl modification sites on a broad scale, however, is simply not possible with these approaches. As few as ten years ago standard methods for protein phosphorylation site identification were limited to 32Pi labeling and two-dimensional phosphopeptide mapping (4, 6), which are time-intensive, low-throughput, limited to cell culture, and carry safety concerns. Since that time, advances in mass spectrometry have rapidly evolved so that today thousands of in vivo phosphorylation sites can be detected and quantified in just a few weeks (7–9). Application of these cutting-edge MS technologies have allowed for investigating phosphoproteins in a wide variety of biological contexts. Here we highlight recent studies having aims ranging from broad to narrow. Next, we detail these MS methods, recent technological breakthroughs, and speculate about future directions of the technology.
Whole-tissue physiology
A number of recent studies have performed comprehensive tissue phosphoproteome analysis, revealing insight into cell signaling in the context of whole organisms. In one example, 5,635 non-redundant phosphorylation sites were identified on proteins in fresh mouse liver (10). From these large-scale data the authors concluded that the C-terminus of a protein was more frequently a target of kinases than all other regions of proteins. This discovery adds new insight to determining the substrate preference for mammalian kinases. Another example of tissue work focused on Alzheimer’s disease (AD), which is associated with dysfunctional protein maintenance, including aberrant protein phosphorylation. Here, similar MS-based methods were employed to map 466 phosphorylation sites in a 20 hours post-mortem human AD brain (11). The identification of phosphorylation sites on protein Tau, and other novel substrates, provides candidate targets for future investigation into the physiology of AD. Recent work by our own group identified 3,404 non-redundant sites of in vivo phosphorylation in roots of the model legume Medicago truncatula (12). The identification of novel phosphorylation sites on proteins involved in initiation of symbiosis with nitrogen-fixing rhizobia bacteria will lead to a better understanding of how a eukaryotic host associates with microbes. Furthermore, analysis of the phosphorylation sites observed revealed phosphorylation motifs not previously observed in other species of plants, providing insight into the specificity of kinases in the plant kingdom.
Cell differentiation status
In addition to influencing the physiology of entire tissues, phosphorylation events can play roles in determining the fate of an individual cell. Pluripotent cells, such as embryonic stem cells, have the potential to differentiate into any cell type in the adult body. However, little is understood of the cellular signaling events that both maintain pluripotency and direct differentiation down a specific cell lineage. To date, only a handful of phosphoproteomic analyses have been reported on such cells (13–16). Our large-scale characterization of pluripotent human embryonic stem cells yielded the identification of 10,844 phosphorylation sites, including sites on two transcription factors, OCT4 and SOX2, both known to regulate pluripotency (13). This work was followed by two reports which quantitatively monitored human embryonic stem cell phosphorylation as a function of differentiation (14–15). Cell signaling events were detected via the quantitative change of half of the observed phosphorylation events within an hour of BMP4-induced differentiation (15). Using the algorithm NetworKIN (17), which predicts the kinases responsible for phosphorylating sites in phosphoproteomic datasets, CDK1/2 was implicated as playing a central role in both differentiation and self-renewal (15). These findings are notable first steps in clarifying our understanding of embryonic stem cell differentiation, and illustrate the need for further investigation of the role of phosphorylation in such cellular events.
Signal transduction cascades
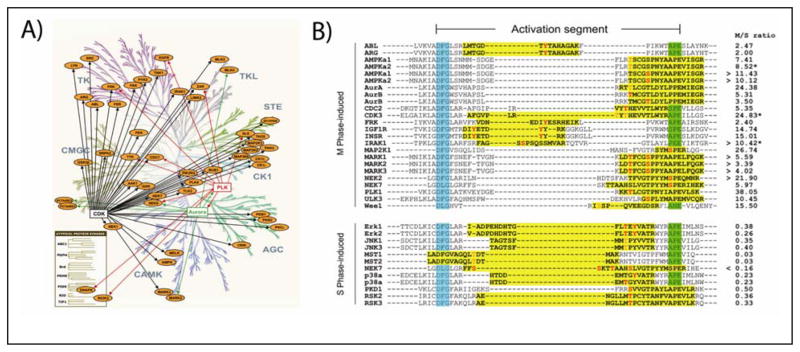
Individual protein phosphorylation events often have roles in broad signaling networks within a cell. Recent advancements have made MS-based phosphoproteomics the ideal way to study signal transduction within seconds of stimulation (18). Quantitative phosphoproteomic analysis of cultured cells treated with activators or inhibitors of the insulin/IGF-1 and MAP kinase pathways have expanded our understanding of two of the most well-studied signaling pathways in all of biology (19–20). While phosphorylation of kinases frequently regulates their own activity, they are typically underrepresented in phosphoproteomic studies, at least in part due to their low expression. Kinase affinity purification has proved to be a viable solution to this problem. In one example, the approach was applied to quantitatively map the ‘phosphokinome’ of human cancer cell lines arrested in different phases of the cell cycle (21). Data from the study of Daub et al., highlighted in Figure 1, illustrates the cell cycle regulation of 1,000 phosphorylation sites on 219 protein kinases from cells arrested in S and M phase.
Figure 1. Cell-cycle-regulated phosphorylation of the kinome.
(A) Protein kinase networks in mitosis are depicted within the context of the human kinome, represented as a dendrogram. Protein kinases for which the identified phosphopeptides were more than 4-fold up-regulated in M phase and contain consensus phosphorylation sites for CDK, PLK, or Aurora kinases are included. (B) An alignment of kinases having activation loops which contained phosphorylation sites that changed in abundance with progression of the cell cycle. The identified phosphopeptides and phosphorylation sites are indicated with yellow highlighting and red lettering, respectively. The ratios of relative abundances in M and S phases (M/S) observed are indicated. M/S ratios which could not be normalized for protein expression are marked by an asterisk (*). All panels reprinted with permission from (21), with the left panel originally adapted from www.cellsignaling.com.
Phosphatases can play equally important roles in regulating signaling pathways through the removal of phosphoryl groups from proteins. Depleting cells of specific protein phosphatases and employing quantitative phosphoproteomics can be used to determine which proteins are regulated by the phosphatase of interest, either directly or downstream. Recent work has taken this approach to investigate protein tyrosine phosphatase 1B (PTP1B) and its Drosophila ortholog Ptp61F (22–23). The Drosophila study detected changes in 288 of 6,478 phosphorylation sites without observing significant alterations at the proteome level.
Kinase/substrate specificity
While monitoring the phosphorylation of individual substrates by specific kinases is a challenging task (24), recent technologies have allowed for significant advancements when combined with the appropriate sample preparation. In the analog-sensitive (as) kinase approach, a kinase of interest is mutated by a single amino acid substitution, allowing it to accommodate the bulky sulfur-containing ATP analog N6-(benzyl)ATP-γ–S (25). When extracts from cells expressing the as kinase are incubated with the ATP analog, the thiophosphate group is transferred specifically to direct substrates of the as kinase. After quenching activity, digesting proteins, and performing thiopeptide purification to capture phosphopeptides containing an unnatural thiophosphoryl group, MS analysis is used for phosphopeptide sequencing. This approach has been applied to cultured human cells to identify 72 phosphorylation events on 68 protein substrates of CDK1/cyclin B (25). A variation of the as kinase approach which renders a specific kinase susceptible to the chemical inhibitor 1-NM-PP1, has been utilized for identifying 547 phosphorylation sites on 308 Cdk1 substrates in budding yeast by quantifying changes in phosphorylation of proteins bearing Cdk1 phosphorylation motifs in response to Cdk1 inhibition (26). This approach allows for investigation of in vivo phosphorylation events; however, the trade-off is that no signature tag is retained on a substrate protein, as in the N6-(benzyl)ATP-γ–S approach. Hence, rigorous validation is required to link kinases to the putative substrates.
Kinase activity assay for kinome profiling (KAYAK) is an alternative approach to monitor site-specific activity of kinases. Here, peptides from a library of known kinase substrates are individually subjected to phosphorylation by endogenous kinases in cell extracts (27). After activity is quenched, isotopically labeled phosphopeptide standards are added and, after sample pooling, quantitative MS-based analysis determines the relative amount of phosphorylation on each library substrate peptide. These data then provide a direct means to monitor activities of specific kinases (or kinase families) under various conditions. KAYAK can also identify novel in vivo kinase substrates after separating cell lysates chromatographically to bin the cell’s kinases into different fractions (28). Each fraction is then subjected to KAYAK to monitor kinase activity and traditional shotgun proteomics to obtain a profile of the relative abundances of the kinases (as well as all other detectable proteins) which may be responsible for the activity observed. This approach successfully identified substrates of the Cdc2/cyclin B1 complex, AMPK1, and the tyrosine kinase Lck in a variety of cell lines.
SAMPLE PREPARATION
The transformation we have observed in large-scale phosphoproteomic analyses over the past decade has largely been driven by a determined collective effort to isolate phosphorylated peptides from complex mixtures. Phosphopeptide enrichment is critical for two primary reasons: (1) phosphopeptides exist at low stoichiometric abundances and (2) they can be suppressed during ionization. Enrichment strategies have, and continue to, evolve. Today, a collection of metal-based affinity methods are among the most common, with variations on loading, column format, and elution conditions. Below we provide a brief summary of these and other popular approaches. For more detail we direct readers elsewhere (9, 29).
Affinity-based approaches
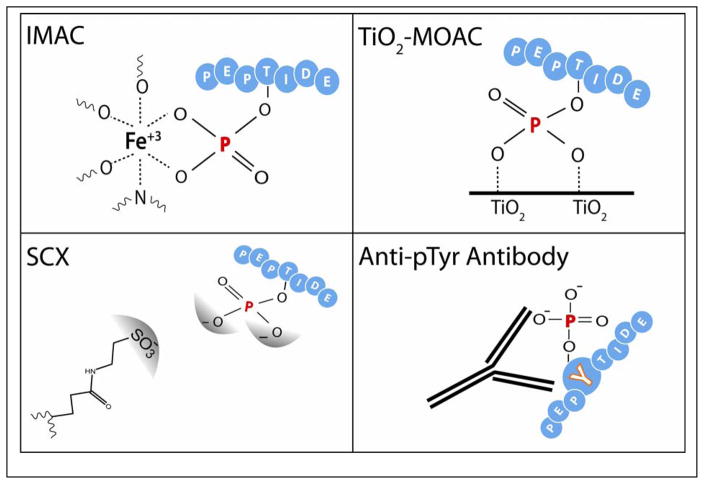
The most frequently used strategies for phosphopeptide enrichment are affinity-based, as illustrated in Figure 2. These methods include immobilized metal affinity chromatography (IMAC), metal oxide affinity chromatography (MOAC), and strong cation exchange (SCX). Note the significantly smaller proportion of phosphorylated tyrosine residues are generally directly targeted by an anti-pTyr immunoaffinity method (30–31).
Figure 2. Phosphopeptide enrichment.
The first challenge in phosphoproteomics is enrichment of low-abundance phosphopeptides or phosphoproteins. The most commonly used enrichment techniques exploit the chemical characteristics of the phosphate group in affinity capture. The figure panels have been modified with permission from previous publications (124–125).
IMAC (32), the classic phosphopeptide enrichment technique, is traditionally performed offline in a column-based format. Here, positively charged metal ions, such as Fe(III) (13, 33–35) or Ga(III) (36), are chelated onto a solid phase nitrilotriacetic/iminodiacetic acid resin and presented for interaction with negatively charged phosphate groups. The most persistent complication with both IMAC and MOAC has been specificity. At moderate pH, carboxylic acid groups are negatively charged, and can compete with phosphoryl groups in binding. To mitigate these issues, binding and washing conditions are often performed at low pH, such that carboxylic acid groups carry no charge. Elution is subsequently accomplished at a more basic pH. Some protocols utilize derivatization to convert carboxylic acids to their corresponding methyl esters prior to IMAC to prevent nonspecific binding (33). Caveats of this chemical conversion include incomplete labeling and increased search complexity during data analysis. Great success has recently been found in a less labor-intensive magnetic bead-based IMAC protocol, which does not call for O-methyl ester formation (16). Phosphate metal affinity chromatography (PMAC) is a similar magnetic bead-based metal affinity strategy applied more specifically to the enrichment of intact phosphoproteins (37).
Recently evolved MOAC protocols are often staged around micro-stage tip columns that take advantage of titania or zirconia as metal oxide chromatography modifiers (38). At present, titanium dioxide (TiO2) remains the most commonly used modifier. Again, phosphopeptides are loaded onto the metal oxide at acidic pH and eluted at basic pH. Instead of relying on prior O-methyl ester formation, MOAC uses various acids, including DHB (39), glycolic acid, and lactic acid (38), to increase phosphopeptide specificity. In contrast to IMAC, which can have greater affinity for multiply phosphorylated peptides, an LC–ESI–MS/MS-based study using TiO2–MOAC reported preferential detection of singly phosphorylated peptides (39). There are many variations of the metal oxide presentation and staging for which varied success has been reported from lab to lab (40–42).
SCX is characteristically presented as a component of the multidimensional protein identification technology (MudPIT) common to shotgun proteomics (43). Peptides are retained on a column through the interaction of positively charged peptide side chains with negatively charged column resin. Peptides are eluted from the column in order of increasing isoelectric point (pI) over a salt concentration gradient. The negatively charged nature of the phosphoryl group at low pH causes phosphopeptides to have lower affinity for the negatively charged resin than a corresponding non-phosphorylated peptide of the same sequence; hence, phosphopeptides are enriched in the earlier-eluted fractions. Many phosphoproteomic methodologies involve SCX followed by either IMAC or MOAC for maximum enrichment (44). A modified low-pH SCX methodology can produce fractions consisting almost entirely of phosphopeptides, eliminating the need for further enrichment (45). As an alternative to SCX, hydrophilic interaction chromatography (HILIC) has been used for pre-enrichment fractionation (46).
Recent efforts have aimed to completely automate the multistep, tedious task of phosphopeptide enrichment by conducting IMAC (47), TiO2 (48–49), or SCX (50) online with MS. Some contend that online enrichment should increase reproducibility; that aside, labor could obviously be reduced by use of an automated format (48). Another benefit of such analyses is the elimination of phosphopeptide storage – e.g., offline strategies require eluted phosphopeptides to be acidified and stored below −20 °C to prevent degradation and conversion of phosphoserine or phosphotheronine residues to dehydroalanine or methyl-dehydroalanine respectively via β-elimination.
Alternative phosphopeptide enrichment strategies
A variety of chemical methodologies have likewise appeared. BEMA (β-elimination/Michael addition), takes advantage of the ease of β-elimination of phosphorylated serine and threonine residues at basic pH and the ability to subject the resulting dehydroalanine/methyl-dehydroalanine products to Michael addition with a desired tag for affinity purification (51–53). Calcium phosphate precipitation has proven to be a fast, economical, and simple enrichment technique (11) in exchange for diminished specificity. Phosphoramidate chemistry (PAC) is another approach in which phosphopeptides are coupled to a solid-phase support such as an amino-derivatized dendrimer or controlled-pore glass derivatized with maleimide for selection (29, 54). Phosphopeptides are de-protected and collected under acidic conditions.
Note that the methods described above are not readily compatible with phosophohistidine-containing proteins and peptides. A detailed description of method development specific for phosphohistidine analysis is found in the following references (55–56).
TANDEM MS METHODOLOGY
During the MS-based experiments referred to above, a phosphopeptide mixture is separated using capillary liquid chromatography. A typical separation column is 25 to 100 microns in diameter and 5 to 30 cm in length. The eluent is concurrently introduced into the mass spectrometer via electrospray ionization (ESI), a process that generates multiply protonated gas-phase peptide cations. The mass-to-charge ratio (m/z) and intensity of the intact peptide precursors are recorded by an initial MS scan – commonly referred to as a full scan MS or MS1. Next, m/z values for peaks with high intensity (i.e., abundant peptide cations) are automatically selected in order of decreasing abundance for sequencing by tandem MS (MS/MS). This process of precursor selection, dissociation, and fragment ion mass analysis is repetitively performed on analyte species as they elute from the LC column. Ideally, MS/MS interrogation of a phosphorylated peptide generates a series of fragment ions that differ in mass by a single amino acid, such that the peptide primary sequence and position of the phosphoryl modification(s) can be determined. This necessitates peptide bond cleavage that is not only specific to the peptide backbone, but is robust enough to elucidate differences in peptides whose primary amino acid sequence are the same, yet vary in the site of phosphorylation (i.e., positional isomers).
Collisional dissociation
The most prevalent method of peptide fragmentation is collision-activated dissociation (CAD). During CAD, a precursor ion population undergoes multiple collisions with inert gas atoms (e.g., helium), causing the internal energy of a peptide cation to progressively increase. This energy is distributed throughout the peptide cation until the threshold of dissociation is reached (57). The dominant cleavage location is the protonated amide bonds, resulting in the formation of fragments carrying either the N- terminus (b-type ions) or C-terminus (y-type ions). Ideally, the site of cleavage is distributed along the entire peptide backbone among the population of precursor ions, such that a series of homologous product ions are produced and the peptide precursor sequence may be deciphered. However, in the presence of phosphorylated serine or threonine residues, the phosphoryl group is often the preferred site of protonation, resulting in cleavage of the bonds anchoring these modifications to the peptide. Thus, CAD MS/MS spectra are often dominated by the neutral loss of phosphoric acid (H3PO4). For many phosphorylated peptide cations, this pathway is preferred such that only low-level b- and y-type ions are observed. Of course, these ions are critical to both peptide sequence identification and localization of the phosphoryl group to a specific amino acid residue.
The exact implementation of CAD can be varied to impact the energy of collisions and consequently the resulting MS/MS spectrum. CAD performed within a collision cell (beam-type CAD) results in more energetic collisions and often generates more sequence-informative b- and y-type ions of greater intensity than that of lower-energy implementations, such as resonant-excitation CAD, typically performed in ion trapping systems (58–59). More recent work has called attention to another possible problem during CAD – phosphoryl group rearrangement. Here, 45% of synthetic phosphopeptides interrogated via resonant-excitation CAD displayed evidence of rearrangement of the phosphoryl group to an alternate hydroxyl-containing amino acid (60). Rearrangement was not observed under beam-type CAD conditions or with electron transfer dissociation (vide infra).
Electron-based dissociation
Fundamentally different methods of peptide fragmentation based on the capture or transfer of an electron have also been developed. Electron capture dissociation (ECD) involves the capture of a low-energy electron by a multiply charged precursor cation, while in electron transfer dissociation (ETD) an electron is transferred from a radical anion with low electron affinity to the peptide precursor cation (61–64). Both are radical-driven dissociation methods which induce cleavage of the N–Cα bond to produce predominantly c- and z-type product ions. The result is a dissociation method highly complementary to CAD, as ECD/ETD cleaves the peptide backbone without cleaving labile PTMs, such as phosphorylation. These fragmentation methods frequently facilitate the determination of the peptide primary sequence and exact site of modification and are consequently becoming widespread in their application.
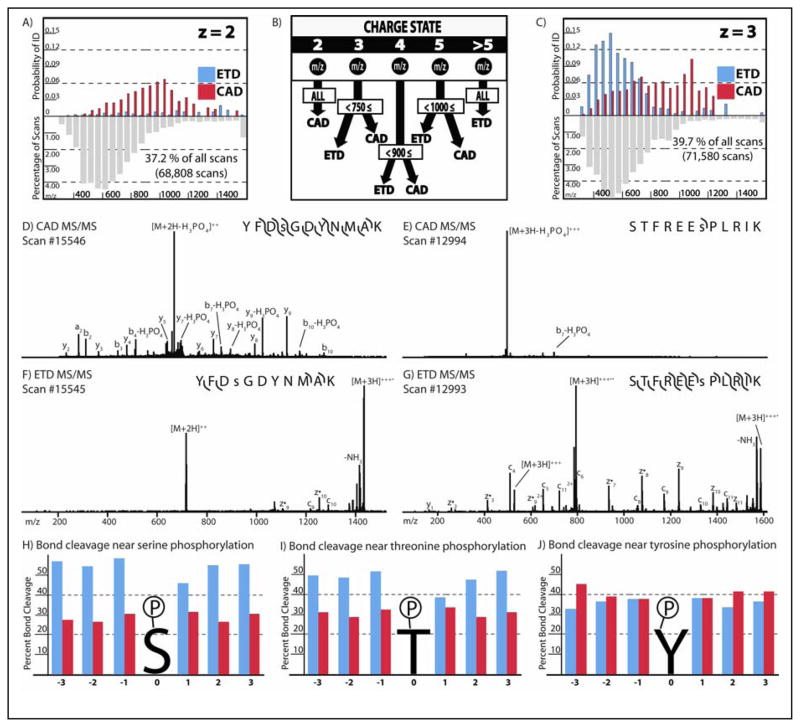
Recent work has focused on quantifying the exact benefit of ETD for phosphopeptide analysis. This work, summarized in Figure 3, shows that ETD has the greatest probability for successful phosphopeptide identification for precursors of low m/z and charge states > 2, while CAD is more successful for doubly charged peptides and those of high m/z (65). Additionally, ETD and CAD are highly complementary, with only 18% of peptides being identified via both CAD and ETD (65). The generation of sequence ions surrounding sites of phosphorylation is critical to site localization. As displayed in Figure 3, the percent of bond cleavage via CAD surrounding phosphorylated serine and threonine residues is less than that of ETD, likely a result of the favored neutral loss of H3PO4 rather than the generation of sequence-informative ions. In contrast, bond cleavage via CAD and ETD are equivalent surrounding phosphorylated tyrosine residues. Overall, the few studies to date which have compared CAD and ETD suggest that the most comprehensive analysis of phosphorylation can be achieved when utilizing both methods of fragmentation (13, 65–67). Another recent review article discusses the fundamentals behind different tandem MS methods for sequencing phosphopeptides in greater detail (68).
Figure 3. Phosphopeptide fragmentation.
For all panels, ETD results are shown in blue and CAD in red. (A, C) The probability of a high confidence phosphopeptide identification via either ETD or CAD MS/MS for peptide cations having various charge (z) as a function of precursor m/z ratio is indicated (65). To evaluate the importance of any one m/z ratio bin, the percentage of all precursors observed having the specific z and m/z ratio are given below. Note, that for dications, CAD is the most successful method; however, for triply charge cations, ETD is the best method for peptides below 750 m/z. (B) A probabilistic decision tree for using ETD and CAD together for phosphopeptide sequencing was generated from the data represented in panels A and C, as well as those for other charge states indicated. (D–G) Comparison of CAD and ETD tandem mass spectra for representative doubly and triply-charged phosphopeptides. (H–J) The percentage of all backbone bonds cleaved via ETD or CAD for the 3 backbone bonds to the N-terminal side (−3 through −1) and the C-terminal side (+1 through +3) of a phosphorylated serine (H), threonine (I) or tyrosine (J) (13). Panels A–C from the Supporting Information from (65) and panels H–J reprinted with permission from (13).
QUANTITATION
Systems biologists are rightly enthusiastic about the capabilities of modern MS approaches for large-scale phosphorylation site discovery; that said, imparting the measurements with quantitative data can add a whole new level of information that is likely key for most inquiries. Significant progress has been achieved in this regard so that today comparing several biological samples simultaneously is routine for many laboratories. Figure 4 presents an overview of the various quantitation strategies. Most methods rely on the incorporation of heavy stable isotopes to produce identically-behaving peptides that vary slightly in mass. The intensity of these mass spectral peaks is then used to determine the relative amount of the peptide in one condition versus the other. Label-free approaches have also been utilized for phosphopeptide quantitation (69–70); see the following articles for more detailed discussions (71–72).
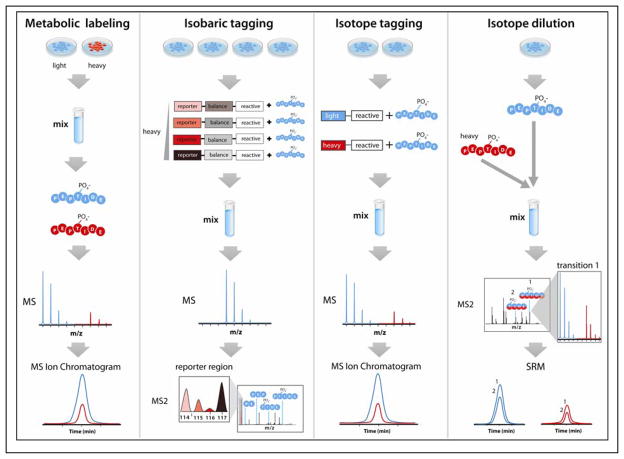
Figure 4. Isotope labeling strategies for phosphopeptide quantitation.
Metabolic labeling introduces heavy isotopes into proteins synthesized in living cells, using heavy isotope-containing amino acids (SILAC) or nutrients (15N or 13C). Peptides analyzed by MS exhibit an m/z difference between light- and heavy-labeled peptides, allowing for relative quantitation by monitoring extracted ion chromatograms of eluting peptides. Isobaric tagging strategies (e.g., iTRAQ and TMT) label peptides from different samples after protein digestion is performed. As co-eluting peptides modified with tags having the same nominal mass are isolated and fragmented, the MS2 scan provides assessment of relative abundance through analysis of the intensity of low m/z reporters. Isotope tagging strategies label digested peptides with heavy and light reactive tags (e.g., ICAT), which allow for determining relative abundance from extracted ion chromatograms. Absolute quantitation (AQUA) of phosphopeptides can be achieved using isotope dilution, in which a known amount of a synthetic heavy isotope-labeled phosphopeptide is spiked into a sample after enzymatic digestion is performed to produce the corresponding endogenous phosphopeptide of interest. Quantitation is achieved by selected reaction monitoring (SRM), typically performed on a triple quadrupole mass spectrometer. For all strategies, the step in the workflow which incorporates heavy isotopes is indicated, with blue representing samples with naturally occurring light isotopes and red representing samples containing stable heavy isotopes. Figure panels adapted with permission from the following references (96, 126).
Metabolic labeling
Metabolic labeling strategies incorporate heavy stable isotopes directly into proteins of living cells. One approach involves the use of isotopically labeled amino acids that are incorporated into proteins during translation, a method commonly referred to as stable isotope labeling with amino acids in cell culture (SILAC) (73–74). During MS analysis, the eluting peptide cations are detected as pairs that are spaced by the number of heavy isotopes in each heavy amino acid the peptide carries. These two signals enable quantitation through the integration of extracted ion chromatograms. The utility of this method for phosphoproteomics was examined by culturing yeast in normal media and media containing 13C6–arginine and 13C6–lysine, to produce peptide pairs differing by multiples of 6 Da upon tryptic digestion (75). From yeast treated with mating pheromone, 139 out of over 700 phosphopeptides detected were quantified to change by at least two-fold. A more recent use of SILAC for phosphoproteomics compared three conditions by labeling HeLa cells with three distinct isotopic signatures by incorporating different combinations of 2H, 13C, and 15N into arginine and lysine (76). Of the 6,600 phosphorylation sites on 2,244 proteins identified, 14% were found to change by at least twofold over a three-point time course after treatment with EGF. Metabolic labeling with light and heavy isotope-containing non-amino acid nutrients provides an alternative to SILAC in which heavy isotopes are incorporated into all amino acids synthesized by the cell (77–78). One such application of quantitative phosphoproteomics was achieved by culturing HeLa cells in media containing 14N and 15N ammonium sulfate to monitor TNF signaling (79). Metabolic labeling with 15N was also used to monitor phosphorylation dynamics of a plasma membrane H-ATPase in Saccharomyces cerevisiae, demonstrating an 11-fold change in phosphorylation at two adjacent residues on the C-terminus in response to glucose stimulation (80). Still, these strategies are fundamentally limited to two, or at most three, quantitative comparisons. Additionally, while metabolic labeling of mammals is possible (81–83), large sample requirements make it cost prohibitive for most phosphoproteomics studies, making quantitative tissue phosphoproteomics particularly challenging (84).
Isobaric tagging
Commercially available isobaric tags (iTRAQ or TMT) provide an alternative to metabolic labeling that can be utilized without growing cells or organisms in special conditions. These stable isotope-containing amine-reactive molecules are used to label peptides from different samples with tags having the same nominal mass. When interrogated via MS/MS, however, each tag dissociates from the peptide to produce low m/z reporter ions that differ by 1 Da. A recent study reported a dynamic range of two orders of magnitude for iTRAQ-based phosphopeptide quantification using beam-type CAD on an orbitrap mass analyzer (59). Further instrumentation advancements are likely to improve this dynamic range. At present, up to eight different labels can be used simultaneously to compare up to eight different conditions. Note while these tags were designed for cleavage by beam-type CAD (85), ETD fragmentation can be applied, but produces reporter ions of lower intensity which are not ideal for accurate quantitation (86). To circumvent this issue, parallel fragmentation of a precursor with ETD for phosphopeptide identification with a subsequent scan using collision-based dissociation for quantitation can be performed (87–88).
Isotope tagging
Several differential isotope-labeling strategies provide the sample growth flexibility afforded by isobaric tags with the ability to monitor co-eluting peptides pairs with characteristic m/z shifts as in metabolic labeling. Isotope-coded affinity tags (ICAT), thiol-reactive molecules containing light or heavy isotopes, represent one of the earliest-developed strategies (89), and have been successfully utilized for phosphopeptide quantitation (90). Another recent labeling strategy utilized normal or deuterated methanol to convert the carboxylic acid moiety of the C-terminus of phosphopeptides to light or heavy O-methyl esters (91). A similar strategy utilizes dimethyl labeling of amines with normal or deuterated formaldehyde (92). However, subtle changes in chromatographic elution can occur from deuterium incorporation – an issue that can be problematic. Another strategy combines digesting with trypsin in 16O or 18O water followed by O-methyl ester formation using 16O or 18O methanol (93). For labeling phosphopeptides specifically, converting peptidyl phosphates to phosphoramidates and incorporating 16O or 18O during subsequent acid-catalyzed hydrolysis has been shown to result in isotope incorporation which is stable under LC separation conditions (94).
Isotope dilution
While all the above methods provide relative quantitation, recent advancements have also been made in methodology for determining absolute abundance of phosphopeptides. One commonly used strategy for determining absolute quantitation of a peptide involves isotope dilution with a synthetic heavy amino acid-containing peptide standard (95), an approach that has long been used for small molecule quantitation. More recent implementations of isotope dilution for proteomics, termed absolute quantification of proteins (AQUA), uses selected reaction monitoring (SRM) on triple quadrupole mass spectrometers to compare the chromatograms for individual fragment ions, or transitions, from an isotopically labeled synthetic peptide to those from the corresponding endogenous peptide (96). For phosphoproteomics, the absolute abundance of an in vivo phosphorylation event at a specific site can be monitored and compared between as many samples as desired when the appropriate heavy phosphopeptide standard is used (96). This approach is typically limited to targeted analysis of specific proteins, such as the recent characterization of the site-specific de-phosphorylation of the cell polarity protein Par 3 by protein phosphatase 1α (97).
INFORMATICS
With the amount of information generated from the approaches described above, data analysis challenges are numerous in the field of phosphoproteomics. Fortunately, most of the strategies developed for conventional shotgun proteomics are likewise applicable to phosphopeptide datasets. Chief among these is the use of target–decoy database searching at both the peptide and protein level, a relatively simple approach that provides the ability to control the false discovery rate (FDR) of a dataset (98). This has proven to be a critical advance in contemporary proteomics, giving empirical statistical validation to peptide and protein identifications made by tandem mass spectrometry, which previously relied upon disparate and somewhat uncertain scoring models.
False discovery rate
Phosphopeptide searches are inherently more demanding than those of unmodified peptides due to the increased database size incurred with dynamic modifications (i.e., the possibility of phosphorylation at every serine, threonine, and tyrosine residue). As a result, the distribution of scores for correct target peptide–spectrum matches (PSMs) is typically less separated from that of incorrect PSMs when variable modifications are considered, placing a premium on the use of additional filtering criteria. The most popular of these is precursor mass error. When low part-per-million (ppm) precursor mass accuracy is required, correct PSMs are preferentially retained. Depending on how this information is applied, it can yield either a lower FDR while identifying roughly the same number of peptides, or a higher number of peptides at the same FDR. While high mass accuracy gives a significant improvement for complex peptide mixtures (up to 50% increases in peptides identified), it is vitally important for phosphoproteomic experiments, with gains of 100% or more (99).
Site localization
Another matter which requires attention is that since most database search algorithms do not explicitly evaluate different peptide isoforms with special consideration, the top PSM, regardless of score and mass accuracy, may not be the correct positional isomer. It is possible that only one or multiple peptide forms were present in the sample, or the data is inconclusive, and database search outputs usually do not provide the requisite evidence for the true situation. Therefore, localization algorithms are necessary to ascertain with greater confidence the likely modification form(s). There is a wide variety of such software available, the most well-known being Ascore, which uses probabilistic analysis of the occurrence of site-determining fragment ions (100). As Ascore was written for CAD and database searching with SEQUEST, additional software has recently been developed for other fragmentation methods and search algorithms, such as PhosphoScore (101), Phosphinator (13), and SLoMo (102).
Data sharing/mining
Due to the rapid growth of high-throughput phosphoproteomics, thousands of phosphorylation sites, and often the kinases responsible, are now known in a variety of organisms. This introduces challenges in terms of data accessibility and mining. Many websites exist for the storage and sharing of phosphorylation-related information, many of which accept external data contributions. Examples of such repositories are PhosphoSitePlus (http://www.phosphosite.org/), Phospho.ELM (103), PhosphoPep (104), and the Phosphorylation Site Database (PHOSIDA) (105). For mining this wealth of data, algorithms to extract sequence motifs associated with phosphorylation sites, such as motif-x, have been developed (106). These motifs make it possible for new phosphorylation sites to be hypothesized without direct observation, as demonstrated by the Scansite algorithm to augment existing and construct new signaling pathways (107). A related approach is the use of machine learning algorithms that recognize phosphorylation sites and the responsible kinases based on computational models trained with existing data. Users can then query their own protein sequences for potential phosphorylation sites using tools such as NetPhosK (108), PredPhospho (109), Group-based Phosphorylation Scoring (GPS) (110), KinasePhos (111), Prediction of pK-specific Phosphorylation site (PPSP) (112), and PhosphoMotif Finder (113) of the Human Protein Reference Database. Also of note is the NetworKIN algorithm, which was designed to predict the kinases responsible for known or newly discovered phosphorylation sites (17).
Sulfonation
Finally, one of the more interesting challenges in this field is the distinction between the isobaric modifications of phosphorylation (79.96633 Da) and sulfonation (79.95681 Da). Sulfonation occurs primarily on tyrosine residues, but recent studies have revealed its existence on serines and threonines as well (114)—causing concern for confusion with phosphorylation among these increasingly numerous large-scale reports. At a difference of only 9.5 mDa, or 9.5 ppm for a 1 kDa peptide and 3.2 ppm for a 3 kDa peptide, it pushes the limit of what is routinely achievable with high-throughput instrumentation and automated analysis (115). With sulfonation representing an extremely labile modification, the neutral loss of 80 Da (SO3) from sulfopeptides, as opposed to the more familiar 98 Da (H3PO4) loss characteristic of collisional dissociation of phosphopeptides, has been reported as a marker for serine and threonine sulfonation (114). Further studies have shown that such losses occur with electron-based fragmentation methods as well (116). As phosphoproteomics continues to advance, special consideration should be given to the confounding modification of sulfonation in data analysis software. The resolution of this matter should be more tractable given recent trends toward the use of hybrid instruments capable of high-resolving power mass analysis, particularly for phosphoproteomic experiments.
CHALLENGES AND FUTURE DIRECTIONS
The recent advancements in technology and methodology discussed here have allowed for new ways to address questions pertaining to protein phosphorylation, expanding our view of cell signaling in living organisms. While current instrumentation and experimental strategies for phosphoproteomics are significantly ahead of where they were just five years ago, limitations still exist. Enrichment is one area that could benefit from further development. All of the methods described above capture modestly overlapping subsets of phosphorylated peptides. We note some exceptional recent work utilizing antibodies for large-scale identification of tyrosine phosphorylation (117–118). To allow for complete phosphoproteome characterization, continued efforts to develop more robust and comprehensive phosphopeptide enrichment methodologies are necessary.
Ongoing developments in MS technology will only increase the already exceptionally large sets of phosphoproteomic data. With future instrumentation developments will come increased dynamic range and sensitivity, allowing for identification of even lower-abundance phosphorylation events or the ability to maintain the current level of detection with reduced amounts of sample. One of these areas that stands to impact the field is the MS analysis of phosphopeptides in the opposite polarity – that is, conventional LC-MS/MS-based peptide analysis is conducted using acidic mobile phases and positive electrospray source polarity. Under these conditions, phosphopeptides are subject to significant ion suppression due to their increased acidity, relative to their unmodified peptides (119). Negative-mode ESI holds potential for phosphoproteomics as the acidic nature of phosphopeptides make them more amenable to negative ionization. The main reason this method has not become widespread in application is that collisional activation of peptide anions, during tandem MS, does not produce random backbone fragmentation as in the positive mode so that sequencing is not possible (120–121). Activation of anions with electrons, however, could offer the means to resolve this problem. Electron detachment dissociation (EDD) was the first method for peptide anions and more recently negative ETD (NETD) has been described (122). Preliminary work with NETD for phosphopeptide anions generated predominantly a•-and x-type fragment ions which makes sequence determination more straightforward (123). The latest research with NETD indicates that large-scale phosphopeptide sequencing may be possible with the method and open the door to countless previously invisible phosphopeptides.
Despite these current limitations, application of the technology we already have can yield the identification and quantitation of 15,000 to 20,000 unique phosphorylation sites in roughly two weeks time. Translation of these datasets into biological insight and knowledge is a current obstacle. In our view, the development of informatics tools to sift and winnow this vast sea of data is now the single most important issue facing the field. How do we build biological pathways, connect the information to transcriptomics and proteomics knowledge, or determine which phosphorylation sites are worthy of targeted mutation and molecular biology? These key questions are not simple issues; however, they are critical to continued pursuit of the role of phosphorylation in cellular biology.
Table 1.
Software for phosphorylation localization, data sharing, motif analysis, and site/kinase prediction.
Acknowledgments
We gratefully acknowledge Q. Xia, M. Lee, G. McAlister, J. Brumbaugh, D. Phanstiel, A. Ledvina, J. Russell, A. Peterson, and G. Kreitinger for helpful discussions on the topics reviewed here. We thank the National Science Foundation (0701846), the National Institutes of Health (R01GM080148 and P01GM081629), Thermo Scientific, the American Society of Mass Spectrometry, the Beckman Foundation, and Eli Lilly and Company for financial support of ongoing research projects in the Coon laboratory. Finally, we thank Ryan Lynch and A.J. Bureta for figure illustrations.
ABBREVIATIONS
- GeLC-MS/MS
gel electrophoresis liquid chromatography-tandem mass spectrometry
- IMAC
immobilized metal affinity, chromatography
- PMAC
phosphate metal affinity chromatography
- MOAC
metal oxide affinity chromatography
- SCX
strong cation exchange
- BEMA
(β-elimination/Michael addition)
- MudPIT
multidimensional protein identification technology
- MALDI
matrix-assisted laser desorption ionization
- ECD
electron capture dissociation
- ETD
electron transfer dissociation
- CAD
collision activated dissociation
- iTRAQ
isobaric tags for relative and absolute quantification
- KAYAK
kinase activity assay for kinome profiling
- AQUA
absolute quantification of proteins
- SRM
selected reaction monitoring
- PHOSIDA
phosphorylation site database
- SILAC
stable isotope labeling with amino acids in cell culture
- EDD
electron detachment dissociation
- NETD
negative electron capture dissociation
- HILIC
hydrophilic interaction chromatography
- ICAT
Isotope-coded affinity tags
- ESI
electrospray ionization
- FDR
false discovery rate
- PSMs
peptide–spectrum matches
- ppm
part-per-million
REFERNCES
- 1.Manning G, Plowman GD, Hunter T, Sudarsanam S. Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci. 2002;27:514–520. doi: 10.1016/s0968-0004(02)02179-5. [DOI] [PubMed] [Google Scholar]
- 2.Moorhead GB, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 3.Cohen P. The regulation of protein function by multisite phosphorylation--a 25 year update. Trends Biochem Sci. 2000;25:596–601. doi: 10.1016/s0968-0004(00)01712-6. [DOI] [PubMed] [Google Scholar]
- 4.Sefton BM, Shenolikar S. Overview of protein phosphorylation. Curr Protoc Mol Biol. 2001;Chapter 18(Unit 18):11. doi: 10.1002/0471142727.mb1801s33. [DOI] [PubMed] [Google Scholar]
- 5.Tarrant MK, Cole PA. The chemical biology of protein phosphorylation. Annu Rev Biochem. 2009;78:797–825. doi: 10.1146/annurev.biochem.78.070907.103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagahara H, Latek RR, Ezhevsky SA, Dowdy SF. 2-D phosphopeptide mapping. Methods Mol Biol. 1999;112:271–279. doi: 10.1385/1-59259-584-7:271. [DOI] [PubMed] [Google Scholar]
- 7.Hoffert JD, Knepper MA. Taking aim at shotgun phosphoproteomics. Anal Biochem. 2008;375:1–10. doi: 10.1016/j.ab.2007.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piggee C. Phosphoproteomics: miles to go before it’s routine. Anal Chem. 2009;81:2418–2420. doi: 10.1021/ac802740t. [DOI] [PubMed] [Google Scholar]
- 9.Thingholm TE, Jensen ON, Larsen MR. Analytical strategies for phosphoproteomics. Proteomics. 2009;9:1451–1468. doi: 10.1002/pmic.200800454. [DOI] [PubMed] [Google Scholar]
- 10.Villen J, Beausoleil SA, Gerber SA, Gygi SP. Large-scale phosphorylation analysis of mouse liver. P Natl Acad Sci U S A. 2007;104:1488–1493. doi: 10.1073/pnas.0609836104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia Q, Cheng D, Duong DM, Gearing M, Lah JJ, Levey AI, Peng J. Phosphoproteomic analysis of human brain by calcium phosphate precipitation and mass spectrometry. Journal of Proteome Research. 2008;7:2845–2851. doi: 10.1021/pr8000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grimsrud PA, den Os D, Wenger CD, Swaney DL, Schwartz D, Sussman MR, Ané JM, Coon JJ. Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiology. 2010;152:19–28. doi: 10.1104/pp.109.149625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Swaney DL, Wenger CD, Thomson JA, Coon JJ. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2009;106:995–1000. doi: 10.1073/pnas.0811964106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brill LM, Xiong W, Lee KB, Ficarro SB, Crain A, Xu Y, Terskikh A, Snyder EY, Ding S. Phosphoproteomic analysis of human embryonic stem cells. Cell Stem Cell. 2009;5:204–213. doi: 10.1016/j.stem.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Hoof D, Munoz J, Braam SR, Pinkse MW, Linding R, Heck AJ, Mummery CL, Krijgsveld J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009;5:214–226. doi: 10.1016/j.stem.2009.05.021. [DOI] [PubMed] [Google Scholar]
- 16.Ficarro SB, Zhang Y, Lu Y, Moghimi AR, Askenazi M, Hyatt E, Smith ED, Boyer L, Schlaeger TM, Luckey CJ, Marto JA. Improved electrospray ionization efficiency compensates for diminished chromatographic resolution and enables proteomics analysis of tyrosine signaling in embryonic stem cells. Anal Chem. 2009;81:3440–3447. doi: 10.1021/ac802720e. [DOI] [PubMed] [Google Scholar]
- 17.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MA, Jorgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T. Systematic discovery of in vivo phosphorylation networks. Cell. 2007;129:1415–1426. doi: 10.1016/j.cell.2007.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dengjel J, Akimov V, Olsen JV, Bunkenborg J, Mann M, Blagoev B, Andersen JS. Quantitative proteomic assessment of very early cellular signaling events. Nature Biotechnology. 2007;25:566–568. doi: 10.1038/nbt1301. [DOI] [PubMed] [Google Scholar]
- 19.Kruger M, Kratchmarova I, Blagoev B, Tseng YH, Kahn CR, Mann M. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. P Natl Acad Sci U S A. 2008;105:2451–2456. doi: 10.1073/pnas.0711713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan C, Olsen JV, Daub H, Mann M. Global effects of kinase inhibitors on signaling networks revealed by quantitative phosphoproteomics. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900285-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 2008;31:438–448. doi: 10.1016/j.molcel.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 22.Mertins P, Eberl HC, Renkawitz J, Olsen JV, Tremblay ML, Mann M, Ullrich A, Daub H. Investigation of protein-tyrosine phosphatase 1B function by quantitative proteomics. Molecular & Cellular Proteomics. 2008;7:1763–1777. doi: 10.1074/mcp.M800196-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hilger M, Bonaldi T, Gnad F, Mann M. Systems-wide analysis of a phosphatase knock down by quantitative proteomics and phosphoproteomics. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M800559-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Peck SC. Analysis of protein phosphorylation: methods and strategies for studying kinases and substrates. Plant J. 2006;45:512–522. doi: 10.1111/j.1365-313X.2005.02613.x. [DOI] [PubMed] [Google Scholar]
- 25.Blethrow JD, Glavy JS, Morgan DO, Shokat KM. Covalent capture of kinase-specific phosphopeptides reveals Cdk1-cyclin B substrates. P Natl Acad Sci U S A. 2008;105:1442–1447. doi: 10.1073/pnas.0708966105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt LJ, Tuch BB, Villen J, Johnson AD, Gygi SP, Morgan DO. Global analysis of Cdk1 substrate phosphorylation sites provides insights into evolution. Science. 2009;325:1682–1686. doi: 10.1126/science.1172867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yu YH, Anjum R, Kubota K, Rush J, Villen J, Gygi SP. A site-specific, multiplexed kinase activity assay using stable-isotope dilution and high-resolution mass spectrometry. P Natl Acad Sci U S A. 2009;106:11606–11611. doi: 10.1073/pnas.0905165106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubota K, Anjum R, Yu Y, Kunz RC, Andersen JN, Kraus M, Keilhack H, Nagashima K, Krauss S, Paweletz C, Hendrickson RC, Feldman AS, Wu CL, Rush J, Villen J, Gygi SP. Sensitive multiplexed analysis of kinase activities and activity-based kinase identification. Nature Biotechnology. 2009;27:933–940. doi: 10.1038/nbt.1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bodenmiller B, Mueller LN, Mueller M, Domon B, Aebersold R. Reproducible isolation of distinct, overlapping segments of the phosphoproteome. Nat Methods. 2007;4:231–237. doi: 10.1038/nmeth1005. [DOI] [PubMed] [Google Scholar]
- 30.Heibeck TH, Ding SJ, Opresko LK, Zhao R, Schepmoes AA, Yang F, Tolmachev AV, Monroe ME, Camp DG, Smith RD, Wiley HS, Qian WJ. An Extensive Survey of Tyrosine Phosphorylation Revealing New Sites in Human Mammary Epithelial Cells. Journal of Proteome Research. 2009;8:3852–3861. doi: 10.1021/pr900044c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Peirce MJ, Begum S, Saklatvala J, Cope AP, Wait R. Two-stage affinity purification for inducibly phosphorylated membrane proteins. Proteomics. 2005;5:2417–2421. doi: 10.1002/pmic.200401176. [DOI] [PubMed] [Google Scholar]
- 32.Andersson L, Porath J. Isolation of Phosphoproteins by Immobilized Metal (Fe-3+) Affinity-Chromatography. Anal Biochem. 1986;154:250–254. doi: 10.1016/0003-2697(86)90523-3. [DOI] [PubMed] [Google Scholar]
- 33.Ficarro SB, McCleland ML, Stukenberg PT, Burke DJ, Ross MM, Shabanowitz J, Hunt DF, White FM. Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat Biotechnol. 2002;20:301–305. doi: 10.1038/nbt0302-301. [DOI] [PubMed] [Google Scholar]
- 34.Ndassa YM, Orsi C, Marto JA, Chen S, Ross MM. Improved immobilized metal affinity chromatography for large-scale phosphoproteomics applications. Journal of Proteome Research. 2006;5:2789–2799. doi: 10.1021/pr0602803. [DOI] [PubMed] [Google Scholar]
- 35.Lee J, Xu YD, Chen Y, Sprung R, Kim SC, Xie SH, Zhao YM. Mitochondrial phosphoproteome revealed by an improved IMAC method and MS/MS/MS. Molecular & Cellular Proteomics. 2007;6:669–676. doi: 10.1074/mcp.M600218-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Posewitz MC, Tempst P. Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal Chem. 1999;71:2883–2892. doi: 10.1021/ac981409y. [DOI] [PubMed] [Google Scholar]
- 37.Wu S, Yang F, Zhao R, Tolic N, Robinson EW, Camp DG, 2nd, Smith RD, Pasa-Tolic L. Integrated workflow for characterizing intact phosphoproteins from complex mixtures. Anal Chem. 2009;81:4210–4219. doi: 10.1021/ac802487q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugiyama N, Masuda T, Shinoda K, Nakamura A, Tomita M, Ishihama Y. Phosphopeptide enrichment by aliphatic hydroxy acid-modified metal oxide chromatography for nano-LC-MS/MS in proteomics applications. Mol Cell Proteomics. 2007;6:1103–1109. doi: 10.1074/mcp.T600060-MCP200. [DOI] [PubMed] [Google Scholar]
- 39.Jensen SS, Larsen MR. Evaluation of the impact of some experimental procedures on different phosphopeptide enrichment techniques. Rapid Commun Mass Sp. 2007;21:3635–3645. doi: 10.1002/rcm.3254. [DOI] [PubMed] [Google Scholar]
- 40.Wang Z, Dong G, Singh S, Steen H, Li J. A simple and effective method for detecting phosphopeptides for phosphoproteomic analysis. J Proteomics. 2009;72:831–835. doi: 10.1016/j.jprot.2009.03.006. [DOI] [PubMed] [Google Scholar]
- 41.Zhou H, Tian R, Ye M, Xu S, Feng S, Pan C, Jiang X, Li X, Zou H. Highly specific enrichment of phosphopeptides by zirconium dioxide nanoparticles for phosphoproteome analysis. Electrophoresis. 2007;28:2201–2215. doi: 10.1002/elps.200600718. [DOI] [PubMed] [Google Scholar]
- 42.Sturm M, Leitner A, Smatt JH, Linden M, Lindner W. Tin dioxide microspheres as a promising material for phosphopeptide enrichment prior to liquid chromatography-(tandem) mass spectrometry analysis. Adv Funct Mater. 2008;18:2381–2389. [Google Scholar]
- 43.Wolters DA, Washburn MP, Yates JR., 3rd An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 44.Zhai B, Villen J, Beausoleil SA, Mintseris J, Gygi SP. Phosphoproteome analysis of drosophila metanogaster embryos. Journal of Proteome Research. 2008;7:1675–1682. doi: 10.1021/pr700696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gauci S, Helbig AO, Slijper M, Krijgsveld J, Heck AJ, Mohammed S. Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal Chem. 2009;81:4493–4501. doi: 10.1021/ac9004309. [DOI] [PubMed] [Google Scholar]
- 46.McNulty DE, Annan RS. Hydrophilic interaction chromatography for fractionation and enrichment of the phosphoproteome. Methods Mol Biol. 2009;527:93–105. x. doi: 10.1007/978-1-60327-834-8_8. [DOI] [PubMed] [Google Scholar]
- 47.Wang JL, Zhang YJ, Jiang H, Cai Y, Qian XH. Phosphopeptide detection using automated online IMAC-capillary LC-ESI-MS/MS. Proteomics. 2006;6:404–411. doi: 10.1002/pmic.200500223. [DOI] [PubMed] [Google Scholar]
- 48.Pinkse MWH, Mohammed S, Gouw LW, van Breukelen B, Vos HR, Heck AJR. Highly robust, automated, and sensitive on line TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. Journal of Proteome Research. 2008;7:687–697. doi: 10.1021/pr700605z. [DOI] [PubMed] [Google Scholar]
- 49.Cantin GT, Shock TR, Park SK, Madhani HD, Yates JR. Optimizing TiO2-based phosphopeptide enrichment for automated multidimensional liquid chromatography coupled to tandem mass spectrometry. Analytical Chemistry. 2007;79:4666–4673. doi: 10.1021/ac0618730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lim KB, Kassel DB. Phosphopeptides enrichment using on-line two-dimensional strong cation exchange followed by reversed-phase liquid chromatography/mass spectrometry. Anal Biochem. 2006;354:213–219. doi: 10.1016/j.ab.2006.04.027. [DOI] [PubMed] [Google Scholar]
- 51.McLachlin DT, Chait BT. Improved beta-elimination-based affinity purification strategy for enrichment of phosphopeptides. Anal Chem. 2003;75:6826–6836. doi: 10.1021/ac034989u. [DOI] [PubMed] [Google Scholar]
- 52.Poot AJ, Ruijter E, Nuijens T, Dirksen EH, Heck AJ, Slijper M, Rijkers DT, Liskamp RM. Selective enrichment of Ser-/Thr-phosphorylated peptides in the presence of Ser-/Thr-glycosylated peptides. Proteomics. 2006;6:6394–6399. doi: 10.1002/pmic.200600373. [DOI] [PubMed] [Google Scholar]
- 53.Arrigoni G, Resjo S, Levander F, Nilsson R, Degerman E, Quadroni M, Pinna LA, James P. Chemical derivatization of phosphoserine and phosphothreonine containing peptides to increase sensitivity for MALDI-based analysis and for selectivity of MS/MS analysis. Proteomics. 2006;6:757–766. doi: 10.1002/pmic.200500073. [DOI] [PubMed] [Google Scholar]
- 54.Tao WA, Wollscheid B, O’Brien R, Eng JK, Li XJ, Bodenmiller B, Watts JD, Hood L, Aebersold R. Quantitative phosphoproteome analysis using a dendrimer conjugation chemistry and tandem mass spectrometry. Nature Methods. 2005;2:591–598. doi: 10.1038/nmeth776. [DOI] [PubMed] [Google Scholar]
- 55.Napper S, Kindrachuk J, Olson DJH, Ambrose SJ, Dereniwsky C, Ross ARS. Selective extraction and characterization of a histidine-phosphorylated peptide using immobilized copper(II) ion affinity chromatography and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Analytical Chemistry. 2003;75:1741–1747. doi: 10.1021/ac026340f. [DOI] [PubMed] [Google Scholar]
- 56.Besant PG, Attwood PV. Detection and analysis of protein histidine phosphorylation. Mol Cell Biochem. 2009;329:93–106. doi: 10.1007/s11010-009-0117-2. [DOI] [PubMed] [Google Scholar]
- 57.Sleno L, Volmer DA. Ion activation methods for tandem mass spectrometry. J Mass Spectrom. 2004;39:1091–1112. doi: 10.1002/jms.703. [DOI] [PubMed] [Google Scholar]
- 58.Olsen JV, Macek B, Lange O, Makarov A, Horning S, Mann M. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 59.Zhang Y, Ficarro SB, Li S, Marto JA. Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J Am Soc Mass Spectrom. 2009;20:1425–1434. doi: 10.1016/j.jasms.2009.03.019. [DOI] [PubMed] [Google Scholar]
- 60.Palumbo AM, Reid GE. Evaluation of Gas-Phase Rearrangement and Competing Fragmentation Reactions on Protein Phosphorylation Site Assignment Using Collision Induced Dissociation-MS/MS and MS(3) Anal Chem. 2008 doi: 10.1021/ac801768s. [DOI] [PubMed] [Google Scholar]
- 61.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. Journal of the American Chemical Society. 1998;120:3265–3266. [Google Scholar]
- 62.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci U S A. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Coon JJ, Syka JEP, Schwartz JC, Shabanowitz J, Hunt DF. Anion dependence in the partitioning between proton and electron transfer in ion/ion reactions. Int J Mass Spectrom. 2004;236:33–42. [Google Scholar]
- 64.Coon JJ. Collisions or electrons? Protein sequence analysis in the 21st century. Anal Chem. 2009;81:3208–3215. doi: 10.1021/ac802330b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Swaney DL, McAlister GC, Coon JJ. Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat Methods. 2008;5:959–964. doi: 10.1038/nmeth.1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 2007;104:2199–2204. doi: 10.1073/pnas.0611217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sweet SMM, Bailey CM, Cunningham DL, Heath JK, Cooper HJ. Large Scale Localization of Protein Phosphorylation by Use of Electron Capture Dissociation Mass Spectrometry. Molecular & Cellular Proteomics. 2009;8:904–912. doi: 10.1074/mcp.M800451-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Boersema PJ, Mohammed S, Heck AJ. Phosphopeptide fragmentation and analysis by mass spectrometry. J Mass Spectrom. 2009;44:861–878. doi: 10.1002/jms.1599. [DOI] [PubMed] [Google Scholar]
- 69.Hoffert JD, Pisitkun T, Wang GH, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: Regulation of aquaporin-2 phosphorylation at two sites. P Natl Acad Sci U S A. 2006;103:7159–7164. doi: 10.1073/pnas.0600895103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang F, Jaitly N, Jayachandran H, Luo QZ, Monroe ME, Du XX, Gritsenko MA, Zhang R, Anderson DJ, Purvine SO, Adkins JN, Moore RJ, Mottaz HM, Ding SJ, Lipton MS, Camp DG, Udseth HR, Smith RD, Rossie S. Applying a targeted label-free approach using LC-MS AMT tags to evaluate changes in protein phosphorylation following phosphatase inhibition. Journal of Proteome Research. 2007;6:4489–4497. doi: 10.1021/pr070068e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nita-Lazar A, Saito-Benz H, White FM. Quantitative phosphoproteomics by mass spectrometry: past, present, and future. Proteomics. 2008;8:4433–4443. doi: 10.1002/pmic.200800231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schreiber TB, Mausbacher N, Breitkopf SB, Grundner-Culemann K, Daub H. Quantitative phosphoproteomics--an emerging key technology in signal-transduction research. Proteomics. 2008;8:4416–4432. doi: 10.1002/pmic.200800132. [DOI] [PubMed] [Google Scholar]
- 73.Zhu H, Pan S, Gu S, Bradbury EM, Chen X. Amino acid residue specific stable isotope labeling for quantitative proteomics. Rapid Commun Mass Spectrom. 2002;16:2115–2123. doi: 10.1002/rcm.831. [DOI] [PubMed] [Google Scholar]
- 74.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 75.Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics. 2005;4:310–327. doi: 10.1074/mcp.M400219-MCP200. [DOI] [PubMed] [Google Scholar]
- 76.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 77.Oda Y, Huang K, Cross FR, Cowburn D, Chait BT. Accurate quantitation of protein expression and site-specific phosphorylation. P Natl Acad Sci U S A. 1999;96:6591–6596. doi: 10.1073/pnas.96.12.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pasa-Tolic L, Jensen PK, Anderson GA, Lipton MS, Peden KK, Martinovic S, Tolic N, Bruce JE, Smith RD. High throughput proteome-wide precision measurements of protein expression using mass spectrometry. Journal of the American Chemical Society. 1999;121:7949–7950. [Google Scholar]
- 79.Cantin GT, Venable JD, Cociorva D, Yates JR., 3rd Quantitative phosphoproteomic analysis of the tumor necrosis factor pathway. J Proteome Res. 2006;5:127–134. doi: 10.1021/pr050270m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lecchi S, Nelson CJ, Allen KE, Swaney DL, Thompson KL, Coon JJ, Sussman MR, Slayman CW. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J Biol Chem. 2007;282:35471–35481. doi: 10.1074/jbc.M706094200. [DOI] [PubMed] [Google Scholar]
- 81.McClatchy DB, Dong MQ, Wu CC, Venable JD, Yates JR. N-15 metabolic labeling of mammalian tissue with slow protein turnover. J Proteome Res. 2007;6:2005–2010. doi: 10.1021/pr060599n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu CC, MacCoss MJ, Howell KE, Matthews DE, Yates JR. Metabolic labeling of mammalian organisms with stable isotopes for quantitative proteomic analysis. Anal Chem. 2004;76:4951–4959. doi: 10.1021/ac049208j. [DOI] [PubMed] [Google Scholar]
- 83.Kruger M, Moser M, Ussar S, Thievessen I, Luber CA, Forner F, Schmidt S, Zanivan S, Fassler R, Mann M. SILAC mouse for quantitative proteomics uncovers kindlin-3 as an essential factor for red blood cell function. Cell. 2008;134:353–364. doi: 10.1016/j.cell.2008.05.033. [DOI] [PubMed] [Google Scholar]
- 84.Lee HJ, Na K, Kwon MS, Kim H, Kim KS, Paik YK. Quantitative analysis of phosphopeptides in search of the disease biomarker from the hepatocellular carcinoma specimen. Proteomics. 2009;9:3395–3408. doi: 10.1002/pmic.200800943. [DOI] [PubMed] [Google Scholar]
- 85.Sachon E, Mohammed S, Bache N, Jensen ON. Phosphopeptide quantitation using amine-reactive isobaric tagging reagents and tandem mass spectrometry: application to proteins isolated by gel electrophoresis. Rapid Commun Mass Sp. 2006;20:1127–1134. doi: 10.1002/rcm.2427. [DOI] [PubMed] [Google Scholar]
- 86.Phanstiel D, Zhang Y, Marto JA, Coon JJ. Peptide and protein quantification using iTRAQ with electron transfer dissociation. J Am Soc Mass Spectrom. 2008;19:1255–1262. doi: 10.1016/j.jasms.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang F, Wu S, Stenoien DL, Zhao R, Monroe ME, Gritsenko MA, Purvine SO, Polpitiya AD, Tolic N, Zhang Q, Norbeck AD, Orton DJ, Moore RJ, Tang K, Anderson GA, Pasa-Tolic L, Camp DG, 2nd, Smith RD. Combined pulsed-Q dissociation and electron transfer dissociation for identification and quantification of iTRAQ-labeled phosphopeptides. Anal Chem. 2009;81:4137–4143. doi: 10.1021/ac802605m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Thompson A, Schafer J, Kuhn K, Kienle S, Schwarz J, Schmidt G, Neumann T, Johnstone R, Mohammed AK, Hamon C. Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 89.Gygi SP, Rist B, Gerber SA, Turecek F, Gelb MH, Aebersold R. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nature Biotechnology. 1999;17:994–999. doi: 10.1038/13690. [DOI] [PubMed] [Google Scholar]
- 90.Hill JJ, Callaghan DA, Ding W, Kelly JF, Chakravarthy BR. Identification of okadaic acid-induced phosphorylation events by a mass spectrometry approach. Biochem Biophys Res Commun. 2006;342:791–799. doi: 10.1016/j.bbrc.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 91.Platt MD, Salicioni AM, Hunt DF, Visconti PE. Use of Differential Isotopic Labeling and Mass Spectrometry To Analyze Capacitation-Associated Changes in the Phosphorylation Status of Mouse Sperm Proteins. Journal of Proteome Research. 2009;8:1431–1440. doi: 10.1021/pr800796j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Huang SY, Tsai ML, Wu CJ, Hsu JL, Ho SH, Chen SH. Quantitation of protein phosphorylation in pregnant rat uteri using stable isotope dimethyl labeling coupled with IMAC. Proteomics. 2006;6:1722–1734. doi: 10.1002/pmic.200500507. [DOI] [PubMed] [Google Scholar]
- 93.Ding SJ, Wang Y, Jacobs JM, Qian WJ, Yang F, Tolmachev AV, Du X, Wang W, Moore RJ, Monroe ME, Purvine SO, Waters K, Heibeck TH, Adkins JN, Camp DG, 2nd, Klemke RL, Smith RD. Quantitative phosphoproteome analysis of lysophosphatidic acid induced chemotaxis applying dual-step (18)O labeling coupled with immobilized metal-ion affinity chromatography. J Proteome Res. 2008;7:4215–4224. doi: 10.1021/pr7007785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Shi Y, Yao X. Oxygen isotopic substitution of peptidyl phosphates for modification-specific mass spectrometry. Anal Chem. 2007;79:8454–8462. doi: 10.1021/ac7017052. [DOI] [PubMed] [Google Scholar]
- 95.Barr JR, Maggio VL, Patterson DG, Jr, Cooper GR, Henderson LO, Turner WE, Smith SJ, Hannon WH, Needham LL, Sampson EJ. Isotope dilution--mass spectrometric quantification of specific proteins: model application with apolipoprotein A-I. Clin Chem. 1996;42:1676–1682. [PubMed] [Google Scholar]
- 96.Gerber SA, Kettenbach AN, Rush J, Gygi SP. The absolute quantification strategy: application to phosphorylation profiling of human separase serine 1126. Methods Mol Biol. 2007;359:71–86. doi: 10.1007/978-1-59745-255-7_5. [DOI] [PubMed] [Google Scholar]
- 97.Traweger A, Wiggin G, Taylor L, Tate SA, Metalnikov P, Pawson T. Protein phosphatase 1 regulates the phosphorylation state of the polarity scaffold Par-3. P Natl Acad Sci U S A. 2008;105:10402–10407. doi: 10.1073/pnas.0804102105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 99.Bakalarski CE, Haas W, Dephoure NE, Gygi SP. The effects of mass accuracy, data acquisition speed, and search algorithm choice on peptide identification rates in phosphoproteomics. Anal Bioanal Chem. 2007;389:1409–1419. doi: 10.1007/s00216-007-1563-x. [DOI] [PubMed] [Google Scholar]
- 100.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 101.Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. Journal of Proteome Research. 2008;7:3054–3059. doi: 10.1021/pr800169k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bailey CM, Sweet SM, Cunningham DL, Zeller M, Heath JK, Cooper HJ. SLoMo: Automated Site Localization of Modifications from ETD/ECD Mass Spectra. Journal of Proteome Research. 2009;8:1965–1971. doi: 10.1021/pr800917p. [DOI] [PubMed] [Google Scholar]
- 103.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho. ELM: a database of phosphorylation sites--update 2008. Nucleic Acids Res. 2008;36:D240–244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bodenmiller B, Campbell D, Gerrits B, Lam H, Jovanovic M, Picotti P, Schlapbach R, Aebersold R. PhosphoPep-a database of protein phosphorylation sites in model organisms. Nature Biotechnology. 2008;26:1339–1340. doi: 10.1038/nbt1208-1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Gnad F, Ren SB, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8 doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 107.Obenauer JC, Cantley LC, Yaffe MB. Scansite 2.0: proteome-wide prediction of cell signaling interactions using short sequence motifs. Nucleic Acids Research. 2003;31:3635–3641. doi: 10.1093/nar/gkg584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics. 2004;4:1633–1649. doi: 10.1002/pmic.200300771. [DOI] [PubMed] [Google Scholar]
- 109.Kim JH, Lee J, Oh B, Kimm K, Koh IS. Prediction of phosphorylation sites using SVMs. Bioinformatics. 2004;20:3179–3184. doi: 10.1093/bioinformatics/bth382. [DOI] [PubMed] [Google Scholar]
- 110.Xue Y, Zhou F, Zhu M, Ahmed K, Chen G, Yao X. GPS: a comprehensive www server for phosphorylation sites prediction. Nucleic Acids Res. 2005;33:W184–187. doi: 10.1093/nar/gki393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wong YH, Lee TY, Liang HK, Huang CM, Wang TY, Yang YH, Chu CH, Huang HD, Ko MT, Hwang JK. KinasePhos 2.0: a web server for identifying protein kinase-specific phosphorylation sites based on sequences and coupling patterns. Nucleic Acids Res. 2007;35:W588–594. doi: 10.1093/nar/gkm322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Xue Y, Li A, Wang L, Feng H, Yao X. PPSP: prediction of PK-specific phosphorylation site with Bayesian decision theory. BMC Bioinformatics. 2006;7:163. doi: 10.1186/1471-2105-7-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nature Biotechnology. 2007;25:285–286. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 114.Medzihradszky KF, Darula Z, Perlson E, Fainzilber M, Chalkley RJ, Ball H, Greenbaum D, Bogyo M, Tyson DR, Bradshaw RA, Burlingame AL. O-sulfonation of serine and threonine: mass spectrometric detection and characterization of a new posttranslational modification in diverse proteins throughout the eukaryotes. Mol Cell Proteomics. 2004;3:429–440. doi: 10.1074/mcp.M300140-MCP200. [DOI] [PubMed] [Google Scholar]
- 115.Bossio RE, Marshall AG. Baseline resolution of isobaric phosphorylated and sulfated peptides and nucleotides by electrospray ionization FTICR ms: another step toward mass spectrometry-based proteomics. Anal Chem. 2002;74:1674–1679. doi: 10.1021/ac0108461. [DOI] [PubMed] [Google Scholar]
- 116.Medzihradszky KF, Guan S, Maltby DA, Burlingame AL. Sulfopeptide fragmentation in electron-capture and electron-transfer dissociation. J Am Soc Mass Spectrom. 2007;18:1617–1624. doi: 10.1016/j.jasms.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 117.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 118.Boersema PJ, Foong LY, Ding VM, Lemeer S, van Breukelen B, Philp R, Boekhorst J, Snel B, den Hertog J, Choo AB, Heck AJ. In depth qualitative and quantitative profiling of tyrosine phosphorylation using a combination of phosphopeptide immuno-affinity purification and stable isotope dimethyl labeling. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900291-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Gropengiesser J, Varadarajan BT, Stephanowitz H, Krause E. The relative influence of phosphorylation and methylation on responsiveness of peptides to MALDI and ESI mass spectrometry. J Mass Spectrom. 2009;44:821–831. doi: 10.1002/jms.1581. [DOI] [PubMed] [Google Scholar]
- 120.Witze ES, Old WM, Resing KA, Ahn NG. Mapping protein post-translational modifications with mass spectrometry. Nat Methods. 2007;4:798–806. doi: 10.1038/nmeth1100. [DOI] [PubMed] [Google Scholar]
- 121.Old WM, Shabb JB, Houel S, Wang H, Couts KL, Yen CY, Litman ES, Croy CH, Meyer-Arendt K, Miranda JG, Brown RA, Witze ES, Schweppe RE, Resing KA, Ahn NG. Functional proteomics identifies targets of phosphorylation by B-Raf signaling in melanoma. Mol Cell. 2009;34:115–131. doi: 10.1016/j.molcel.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Budnik BA, Haselmann KF, Zubarev RA. Electron detachment dissociation of peptide di-anions: an electron-hole recombination phenomenon. Chem Phys Lett. 2001;342:299–302. [Google Scholar]
- 123.Coon JJ, Shabanowitz J, Hunt DF, Syka JE. Electron transfer dissociation of peptide anions. J Am Soc Mass Spectrom. 2005;16:880–882. doi: 10.1016/j.jasms.2005.01.015. [DOI] [PubMed] [Google Scholar]
- 124.Alpert AJ, Andrews PC. Cation-Exchange Chromatography of Peptides on Poly(2-Sulfoethyl Aspartamide)-Silica. J Chromatogr. 1988;443:85–96. doi: 10.1016/s0021-9673(00)94785-x. [DOI] [PubMed] [Google Scholar]
- 125.Larsen MR, Thingholm TE, Jensen ON, Roepstorff P, Jorgensen TJD. Highly selective enrichment of phosphorylated peptides from peptide mixtures using titanium dioxide microcolumns. Molecular & Cellular Proteomics. 2005;4:873–886. doi: 10.1074/mcp.T500007-MCP200. [DOI] [PubMed] [Google Scholar]
- 126.Ross PL, Huang YN, Marchese JN, Williamson B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, Purkayastha S, Juhasz P, Martin S, Bartlet-Jones M, He F, Jacobson A, Pappin DJ. Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol Cell Proteomics. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]