Abstract
Background
Genetic studies on chronic inflammatory diseases have resulted in an emphasis on the epithelial interface with the environment and the genes that influence this interaction. This study examines the expression of key epithelial genes implicated in the pathogenesis of other inflammatory disorders for their role in chronic rhinosinusitis (CRS).
Methods
Epithelial cells were collected from the inferior turbinate, middle turbinate, and/or uncinate from 62 subjects undergoing sinonasal surgery. Patient groups included 21 CRS patients with nasal polyposis, 23 CRS patients without nasal polyposis, and 18 controls. Samples were analyzed for S100A7, S100A8, S100A9, SLC9A3R1, G-protein–coupled receptor for asthma, and serine protease inhibitor kazal type 5 (SPINK5) by quantitative real-time polymerase chain reaction. Immunohistochemistry (IHC) was performed to analyze expression of SPINK5 lympho epithelial kazal-type inhibitor (LEKTI) in sinonasal samples.
Results
Expression of S100A7 and S100A8 was significantly decreased in CRS with and without nasal polyps when compared with controls. S100A9 expression was significantly decreased in CRS without nasal polyps, and SPINK5 expression was significantly decreased in CRS with nasal polyps. SPINK5 (LEKTI) protein was detected in sinonasal tissue and was significantly decreased in polyp samples using IHC.
Conclusion
This study shows marked reductions in the level of expression of several genes involved in epithelial barrier maintenance and repair in the inflammatory state of CRS.
Keywords: Chronic rhinosinusitis, epithelial genes, inflammation, LEKTI, nasal polyps, S100A7, S100A8, S100A9, SPINK5
Chronic rhinosinusitis (CRS) is a common clinical syndrome characterized by persistent, symptomatic mucosal inflammation.1 Despite extensive investigation and debate, the etiology of CRS remains elusive. Although complex host genetic factors are widely believed to influence CRS pathogenesis, recent studies have focused on environmental factors such as fungal or bacterial colonization, biofilms, superantigens, osteitis, and allergen exposure. No one causative factor appears to account for all of the manifestations of CRS of which there are two basic phenotypic entities: CRS with nasal polyposis (CRSwNP) and CRS without nasal polyposis (CRSsNP).2 Although clinical symptoms overlap, CRSwNP is associated more strongly with nasal congestion and olfactory loss, and CRSsNP is characterized typically by rhinorrhea and facial pressure/pain. In addition, the histological and molecular profile of CRSwNP is characterized typically by greater tissue eosinophilia and Th2 cytokine skewing and CRSsNP shows a lesser degree of eosinophilia and Th1 predominance.2,3 There has been much debate as to whether these represent a broad spectrum of sinus disease or if these are actually two separate disease entities, each with distinct etiologies.2
Genetic studies of other chronic inflammatory diseases such as atopic dermatitis (AD), psoriasis, asthma, and chronic obstructive pulmonary disease have resulted in a new emphasis on the epithelial interface with the environment and the genes that influence this interaction.4 The epithelium is crucial to both innate and acquired immunity in many varying roles. At the simplest level, the epithelium functions as a physical barrier to foreign pathogens through tight junctions. At a higher level, the epithelium functions in danger recognition through pattern recognition receptors as well as immune defense through the release of cytokines and chemokines, which further regulate innate and adaptive immune responses.5,6
Because the epithelium is now recognized as an important mediator of immune defense, defects in a broad set of epithelial-related genes, theoretically, could render individuals susceptible to a dysfunctional immune response to environmental agents. In patients with psoriasis, an overexpression of members of the S100 protein family of epidermal calcium binding proteins has been shown.7–9 The S100 family regulates the immune system and an overexpression of S100A7, S100A8, and S100A9 may contribute to the proinflammatory state of psoriatic individuals.7 A genetic susceptibility to psoriasis also has been reported in association with SLC9A3R1, another epidermal protein with a role in the regulation of T-cell function.10 Studies on asthma and AD have found additional epithelial genes of interest. G-protein–coupled receptor for asthma (GPRA) susceptibility, present in bronchial epithelium has been identified as an asthma susceptibility gene.11–13 Polymorphisms of serine protease inhibitor kazal type 5 (SPINK5), an epithelial serine protease inhibitor, have been associated with AD.14–17 SPINK5 is believed to have an important role in the balance of inflammatory effects of mast cells on the epithelial barrier. Specifically, allergens often possess serine protease activity, and antiproteases such as SPINK5 may influence susceptibility for atopy.15 The discovery of alterations in the expression of these epithelial genes in asthma and cutaneous disease has led to an increased understanding of the driving forces of chronic inflammation.18 With regard to CRS, data for epithelial dysfunction is scant; however, this disorder frequently is a comorbid condition with asthma and manifests a similar inflammatory response, suggesting potential genetic and pathogenetic overlap. The objective of this study was to determine if the level of expression for genes associated with mechanical barrier and the innate immune response of sinonasal epithelial cells is altered in CRS (both with and without polyps) when compared with a control population.
MATERIALS AND METHODS
Real-Time Polymerase Chain Reaction (PCR)
Sixty-two patients undergoing nasal surgery at a single institution participated in the study. Twenty-one control, 23 CRSsNP, and 18 CRSwNP gave written informed consent. Subjects classified as CRSsNP met the definition of CRS as defined by the American Academy of Otolaryngology–Head and Neck Surgery Chronic Rhinosinusitis Task Force.19 The CRSwNP group met the aforementioned definition with the additional finding of bilateral polyps present on CT scan and/or endoscopic examination. The control group consisted of patients undergoing nasal surgery (e.g., septoplasty, rhinoplasty, or nasal fracture repair) in which CRS and allergic rhinitis were excluded based on clinical and radiographic analysis. Patients with Samter’s triad (asthma, nasal polyps, and aspirin sensitivity) were not specifically excluded but none were treated during the time frame of this study. Patients with an established immunodeficiency, a diagnosis of classic allergic fungal sinusitis, or cystic fibrosis were, however, excluded from the study.
Under endoscopic guidance, epithelial cells were collected from the inferior turbinate and uncinate process by scraping with a Rhinoprobe (Arlington Scientific, Arlington, TX). The cells were immediately placed in a stabilization reagent (RNAlater; QIAGEN, Valencia, CA).
Total RNA was isolated from the nasal scrapings using the RNeasy Mini kit (QIAGEN, CA). RNA (0.5 mg) was reverse transcribed using a p(dT)15 primer (Roche, Indianapolis, IN) and Moloney murine leukemia virus reverse transcriptase (Applied Biosystems, Foster City, CA) using conditions provided by the manufacturer. One microliter of each cDNA sample, corresponding to 25 ng of total RNA, was used for the real-time PCR. Only isolated mRNA with a concentration of >20.0 μg/mL was used for further analysis. Primers and probes were specifically designed using software (Primer Express; Applied Biosystems) or were predesigned as provided by the manufacturer. The target genes primers and probes are listed in Table 1.
Table 1.
Primers and probes used for Taqman real-time PCR
| Gene | Forward Primers | Reverse Primers | Probe |
|---|---|---|---|
| S100A7 | AACACTCAAGCTGAGAGGTCCAT | TCAGCAGGCTTGGCTTGTC | ACACCAGACGTGATGAC |
| S100A8 | TGGAGAAAGCCTTGAACTCTATCA | GCATGGAAATTCCCCTTTATCA | CGTCTACCACAAGTACT |
| S100A9 | AGAAGGAGAATAAGAATGAAAAGGTCATA | CGCCATCAGCATGATGAACT | AGCAGCTGAGCTTC |
| SLC9A3R1 | CGTGGTGTCCGCCATCA | GTGCTCCTGAGATGGGATCAC | AGTTCTTCAAGAAATGCA |
| GPRA SPINK5 HPRT | Taqman assays on Demand Kit by Applied Biosystems |
All sequences are shown in the 5′ to 3′ orientation.
Real-time PCR was performed in an ABI PRISM 7500 Sequence Detection System thermal cycler (PE Applied Biosystems, Foster City, CA) to quantify mRNA for S100A7, S100A8, S100A9, solute carrier family 9, member 3 regulator 1 (SLC9A3R1), GPRA susceptibility, SPINK5, and hypoxanthine phosphoribosyl transferase (HPRT) by Taqman real-time PCR. Real-time PCR was performed in a total volume of 20 μL with the following components: 1× TaqMan PCR buffer; 5.5 mM of MgCl2, 0.25 mM of dATP, dCTP, and dGTP; 0.5 mM of dUTP; 0.25 U of AmpErase UNG; 0.75 U of Ampli Taq Gold; 0.4 mM of each primer; and 0.2 mM of the Taqman probe. Cycle parameters were 50°C for 2 minutes to activate UNG and 95°C for 10 minutes to activate Taq, followed by 40 cycles of 95°C for 15 seconds and 60°C for 1 minute.
Immunohistochemistry (IHC)
Analysis of SPINK5 protein (also known as lympho epithelial kazal-type inhibitor [LEKTI]) expression was performed using the LEKTI antibody (Invitrogen, South San Francisco, CA), clone 1C11G6, a mouse monoclonal tissue culture supernatant at an initial concentration of 0.1 mg/mL. Data from the manufacturer indicate that specificity for human LEKTI protein has been confirmed by Western blot. Formalin-fixed, 3-μm sections of paraffin-embedded tissue were incubated with LEKTI antibody at a dilution of 1:500 for 1 hour. Specimens included polyp tissue from CRSwNP patients (n = 9) as well as uncinate tissue from control (n = 5), CRSwNP (n = 13), and CRSsNP (n = 9) patients. For a negative control, a matched mouse IgG isotype control antibody, clone 11711 (R & D Systems, Minneapolis, MN), was used at the same dilution as the LEKTI antibody. After rinsing, biotinylated horse anti-mouse secondary antibody (Vector Laboratories, Burlingame, CA) was used for 1 hour at a dilution of 1:500. After a second rinsing, sections were incubated with avidin–peroxidase complex (Vectastain Elite ABC Kit; Vector Laboratories) for 1 hour. After a third rinsing, sections were incubated with diaminobenzidine chromogen (Invitrogen) for 6–10 minutes. Counterstaining with Gill modified hematoxylin (EMD, San Diego, CA) was performed for 10–15 seconds. Sections were then dehydrated, cleared, and mounted.
Semiquantitative analysis was performed using a Zeiss Axiostar Plus light microscope. Slides were blinded, rated on a scale of zero to two in three different locations of tissue structure (epithelium, glands, and subepithelial stroma). A rating of zero was no staining, one was mild to moderate staining, and two was intense staining. Photographs are taken at 400× objective magnification using MicroFire AR digital microscope camera (Optronics, Goleta, CA) and Olympus IX71 Inverted Microscope.
Statistical Analysis
Relative gene expression was calculated by the comparative cycle threshold (Ct) method in which PCR results were normalized to HPRT, a housekeeping gene used for standardization. The fold change of genetic expression was expressed as 2ΔCt (ΔCt = the difference in threshold cycles for the target gene and HPRT). Student’s t-test was applied to determine differences between each CRS group and the control population at the mRNA level. To determine differences in protein IHC staining, the Kruskal-Wallis ANOVA on ranks with Dunnett’s post hoc testing was used. Statistical analysis was performed using SPSS 10.0 for Windows and JMP 5.1 software. A value of p < 0.05 was regarded as statistically significant.
RESULTS
We analyzed mRNA samples from sinonasal epithelial cells in 21 control subjects, 18 subjects with CRSwNP and 23 subjects with CRSsNP. The quantitative gene expression of the target genes listed in Table 1 was determined by real-time PCR and is displayed in Fig. 1. All target genes were detectable in the sinonasal samples. Relatively high levels of expression of mRNA for HPRT, S100A8, S100A9, SLC9A3R1, and SPINK5 were detected whereas lower levels of S100A7 and GPRA were found.
Figure 1.
Average Ct of mRNA of the target genes is shown. The Ct is the cycle at which a noticeable increase in fluorescence above the baseline signal is detected. A lower Ct represents a higher level of mRNA present in the sample. A higher bar, therefore, correlates to a higher level of genetic expression.
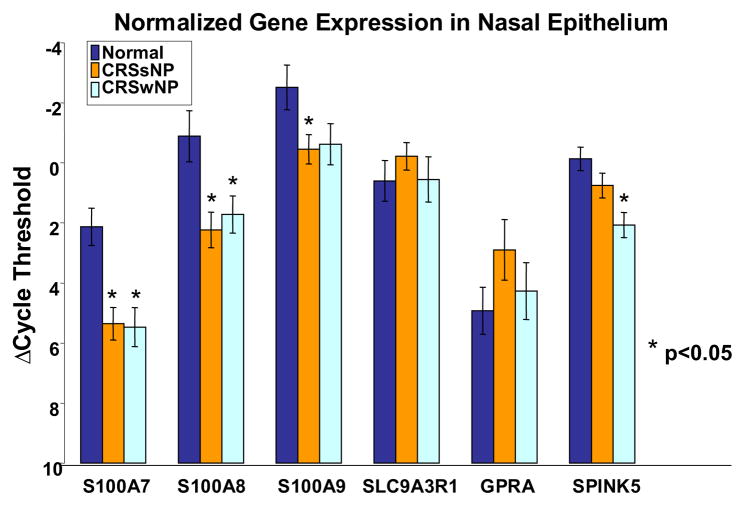
The data were then normalized to the housekeeping gene HPRT as displayed in Fig. 2. Significantly decreased levels of expression in CRS were detected in the S100 family of proteins and SPINK5. S100A7 mRNA expression was significantly decreased in CRSsNP (p < 0.0002) and CRSwNP (p < 0.0004) compared with controls. S100A8 expression also was significantly decreased in CRSsNP (p < 0.004) and CRSwNP (p < 0.016). S100A9 expression was significantly decreased in CRSsNP (p < 0.02), and SPINK5 expression was decreased in CRSsNP and significantly decreased in CRSwNP (p < 0.0003) compared with controls. SLC9A3R1 and GPRA expression did not significantly differ in CRSsNP or CRSwNP compared with controls.
Figure 2.
Real-time PCR levels of expression of target genes in sinonasal tissue normalized to the housekeeping gene HPRT. The significant values are starred and error bars represent SEM.
To further elucidate the significance of the loss of expression of these epithelial genes in CRS, the significant change in genetic expression was calculated as a fold change. S100A7 had the greatest fold change in CRSsNP (9.3-fold) and CRSwNP (10.1-fold). S100A8 showed intermediate fold change differences of 8.6 in CRSsNP and 6.0 in CRSwNP. SPINK5 showed a 4.6-fold down-regulation in nasal polyp subjects and S100A9 showed a 4.2-fold down-regulation in CRSsNP.
A commercial antibody is available to detect LEKTI, the SPINK5 protein. Because there are no reports of expression of this protein in the airways, we performed IHC analysis of surgical samples from normal and CRS patients to determine whether LEKTI expression could be detected. Consistent with the real-time–PCR mRNA data on epithelial cell scrapings, LEKTI protein is present in nasal respiratory epithelium. As seen in Fig. 3, we detected LEKTI staining in both glandular and mucosal epithelial cells in tissue samples. Cellular staining also was observed in the nonglandular subepithelial stroma and the three sites were graded for intensity per methods. As shown in Table 2, a semiquantitative analysis showed significantly more intense LEKTI staining in the epithelium of control tissue when compared with both the polyp and the uncinate tissue from CRSwNP subjects as well as CRSsNP. Composite LEKTI score also was significantly greater in control specimens.
Figure 3.
Immunohistochemical staining for LEKTI in epithelial cells from the uncinate process and polyp tissue at 400× magnification. (A) Control IgG of uncinate from CRSwNP subject does not stain for LEKTI. (B) LEKTI staining of uncinate from a control subject shows intense staining in the epithelial and glandular tissue, whereas light to moderate staining of LEKTI is seen in uncinate samples from (C) CRSsNP and (D) CRSwNP. (E) Minimal LEKTI staining in epithelial and glandular cells is seen in polyp tissue. Glandular staining reveals similar trends. (F) Glandular staining in the uncinate of a control subject with intense staining for LEKTI. Less staining is again seen in the uncinate from (G) CRSsNP and (H) CRSwNP.
Table 2.
Mean semiquantitative scores of LEKTI staining in epithelium, glands, and subepithelial stroma
| CRSwNP (n = 13) | CRSsNP (n = 9) | Polyp (n = 13 | Control (n = 5) | Kruskal-Wallis ANOVA p Value | |
|---|---|---|---|---|---|
| Epithelium | 0.88 | 0.67 | 0.36 | 1.63 | 0.0015 |
| (scale = 0–2) | p < 0.05 | p < 0.012 | p < 0.0002 | ||
| Glands | 0.58 | 0.25 | 0.20 | 1.05 | 0.028 |
| (scale = 0–2) | p < 0.498 | p < 0.099 | p < 0.01 | ||
| Subepithelial stroma | 0.19 | 0.06 | 0.00 | 0.80 | 0.020 |
| (scale = 0–2) | p < 0.028 | p < 0.011 | p < 0.002 | ||
| Total | 1.65 | 0.98 | 0.56 | 3.48 | 0.0004 |
| (scale = 0–6) | p < 0.237 | p < 0.028 | p < 0.0002 |
The difference between the groups is displayed in the Kruskal-Wallis ANOVA on ranks. The p values in italics reflect post hoc comparison with control samples, with statistically significant values in bold. LEKTI staining was significantly decreased in epithelium and subepithelial stroma in CRSwNP, CRSsNP, and polyp tissue compared with control samples. Among the groups, polyp tissue showed the least staining in all areas.
DISCUSSION
In this study, members of the S100 family of proteins and the serine protease inhibitor SPINK5 were found at decreased levels of expression in CRS with and without nasal polyps when compared with controls. These are key epithelial genes involved in barrier function and raise a potentially novel model for the pathogenesis of CRS in which alterations in epithelial barrier function may play a role in the chronic inflammatory state. The S100 family of proteins is part of the epidermal differentiation complex on chromosome 1q21, which is a collection of epithelial genes important in both antimicrobial and immunoregulatory functions.20 These actions have been studied extensively in the skin where S100A7, S100A8, and S100A9 are believed to work as calcium sensor proteins; expression influences the delicate balance between keratinocyte proliferation and differentiation, a balance that is frequently altered in wound healing.7 In psoriasis, an overexpression of these genes is believed to contribute to the hyper-proliferation and abnormal differentiation of keratinocytes seen in the disease. In contrast, we demonstrate a down-regulation of mRNA for S100A7, S100A8, and S100A9 in CRS. Diminished S100 protein expression could theoretically result in a decreased proliferative or aberrant regenerative capacity of the respiratory epithelium, particularly in response to injury. In CRS, the presence of a relatively immature or poorly reparative epithelium would hypothetically decrease the mechanical barrier effect and accentuate both sensitization and the inflammatory response to a variety of exogenous agents. In addition, S100A7 has been shown to possess intrinsic antimicrobial activity against Escherichia coli, thought to be important in the local innate defense of healthy skin.21 Analogous decreases in expression in the respiratory epithelium could play a role in the suggested infectious component of CRS inflammation. In addition, diminished innate defense could result in a more robust acquired immune response characteristic of CRS histopathology.
Exposure to proteases derived from allergens, bacteria, and fungi occurs with regularity. SPINK5 is a known protease inhibitor with a role on skin barrier function but has an unknown role in the airways. Diminished expression of anti-proteases such as SPINK5 at this site may render the sinus epithelium more susceptible to fungal or bacterial proteases resulting in a generalized increase in antigen exposure as thought to occur in AD.22 Defects in protease inhibition also have been suspected to facilitate bacterial colonization. Mutations of SPINK5 have been identified in Netherton’s syndrome, a disease characterized by ichthyosiform erythroderma (a hair-shaft defect) and a susceptibility to develop AD and Th2 responses. It is well established that the skin of >90% of AD patients is colonized with Staphylococcus aureus,23 an organism frequently cultured from nasal airways of patients with CRSwNP.24,25 Additionally, SPINK5 has been proposed to be an inhibitory regulator of desquamation.26 Decreased levels of SPINK5, therefore, may lead to increased cell turnover and generalized epithelial barrier dysfunction, which may influence susceptibility to colonization. It is possible that the diminished SPINK5 expression that we observe in CRSwNP may contribute to an excessive TH2 response to colonized staphylococci and/or fungi. In the airways, polymorphisms in SPINK5 have been associated with asthma, but this is the first report to our knowledge that indicates expression of SPINK5 (LEKTI) protein in airway epithelial cells and reduced expression of SPINK5 mRNA in CRSwNP.14,15
The disease entities in this study, CRS with and without nasal polyps, were both characterized by diminished expression of genes involved in epithelial barrier function when compared with control tissue. Prior studies of sinonasal mucosa have shown expression of host defense molecules, such as toll-like receptors, in the sinonasal epithelium27,28 but their contributions to epithelial damage in tissue from patients with CRS is unknown and likely to be indirect. Similar to findings in the bronchial epithelium in asthma, the epithelium in nasal polyps has shown varying degrees of desquamation, unrelated to the presence of asthma.29 In addition, a loss of desmosomal length, important for adhesion and barrier function of epithelium, has been shown in nasal polyps.30 The significant reduction in expression of epithelial factors known to be involved in mechanical and innate immune barrier functions, S100A7 and SPINK5, raises potential mechanisms for the epithelial damage previously reported in nasal polyposis. As established by IHC, the protein encoded by SPINK5 expression is present in the upper airways and is reduced in CRSwNP, in particular within nasal polyps themselves. With respect to absolute protein quantity in tissue, graded immunohistochemical data must always be interpreted with caution; however, these results indicate that mRNA quantified by PCR is translated into protein. Taken together, these data suggest that the pathophysiology of polyposis may be associated with diminished tissue protease activity, specifically decreased LEKTI. Additional studies will be required to determine whether the reduced expression of mRNA for S100A7 is reflected by reduced expression of the respective protein.
Although data regarding specific genetic variations is scant, CRS is believed to result from complex host–environment interactions involving multiple genetic loci. The current results suggest significant genetic overlap between CRS with and without nasal polyps. Furthermore, the current gene expression data suggest that, broadly speaking, CRS is one of several disorders including AD, psoriasis, inflammatory bowel disease, asthma, and chronic obstructive pulmonary disease, wherein the interface between self and the environment is the site of disease pathogenesis mediated by a dysfunctional host response. Variations in the mechanical and innate immune barrier of the host have been proposed to play a major role in disease expression in the lung, gut, and skin. The current report suggests the hypothesis that a defective barrier also may be associated with CRS. Viewed in this light, the various etiologic factors thus far implicated in CRS (e.g., bacteria, fungi, biofilms, allergens, etc.) then would be viewed as disease modifiers rather than discrete etiologic agents. Future studies focused on genes that maintain the mechanical and innate immune barrier of the nasal epithelium may be of significant value in the elucidation of CRS etiology and pathogenesis.
References
- 1.Benninger MS, Ferguson BJ, Hadley JA, et al. Adult chronic rhinosinusitis: Definitions, diagnosis, epidemiology, and pathophysiology. Otolaryngol Head Neck Surg. 2003;129(suppl 3):S1–S32. doi: 10.1016/s0194-5998(03)01397-4. [DOI] [PubMed] [Google Scholar]
- 2.Meltzer EO, Hamilos DL, Hadley JA, et al. Rhinosinusitis: Establishing definitions for clinical research and patient care. Otolaryngol Head Neck Surg. 2004;131(Suppl 6):S1–S62. doi: 10.1016/j.otohns.2004.09.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Zele T, Claeys S, Gevaert P, et al. Differentiation of chronic sinus diseases by measurement of inflammatory mediators. Allergy. 2006;61:1280–1289. doi: 10.1111/j.1398-9995.2006.01225.x. [DOI] [PubMed] [Google Scholar]
- 4.Cookson W. The immunogenetics of asthma and eczema: A new focus on the epithelium. Nat Rev Immunol. 2004;4:978–988. doi: 10.1038/nri1500. [DOI] [PubMed] [Google Scholar]
- 5.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1:135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 6.Yang D, Biragyn A, Kwak LW, et al. Mammalian defensins in immunity: More than just microbicidal. Trends Immunol. 2002;23:291–296. doi: 10.1016/s1471-4906(02)02246-9. [DOI] [PubMed] [Google Scholar]
- 7.Broome AM, Ryan D, Eckert RL. S100 protein subcellular localization during epidermal differentiation and psoriasis. J Histochem Cytochem. 2003;51:675–685. doi: 10.1177/002215540305100513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semprini S, Capon F, Tacconelli A, et al. Evidence for differential S100 gene over-expression in psoriatic patients from genetically heterogeneous pedigrees. Hum Genet. 2002;111:310–313. doi: 10.1007/s00439-002-0812-5. [DOI] [PubMed] [Google Scholar]
- 9.Madsen P, Rasmussen HH, Leffers H, et al. Molecular cloning, occurrence, and expression of a novel partially secreted protein “psoriasin” that is highly up-regulated in psoriatic skin. J Investig Dermatol. 1991;97:701–712. doi: 10.1111/1523-1747.ep12484041. [DOI] [PubMed] [Google Scholar]
- 10.Helms C, Cao L, Krueger JG, et al. A putative RUNX1 binding site variant between SLC9A3R1 and NAT9 is associated with susceptibility to psoriasis. NatGenet. 2003;35:349–356. doi: 10.1038/ng1268. [DOI] [PubMed] [Google Scholar]
- 11.Melen E, Bruce S, Doekes G, et al. Haplotypes of G protein-coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med. 2005;171:1089–1095. doi: 10.1164/rccm.200410-1317OC. [DOI] [PubMed] [Google Scholar]
- 12.Kormann MS, Carr D, Klopp N, et al. G-Protein-coupled receptor polymorphisms are associated with asthma in a large German population. Am J Respir Crit Care Med. 2005;171:1358–1362. doi: 10.1164/rccm.200410-1312OC. [DOI] [PubMed] [Google Scholar]
- 13.Laitinen T, Polvi A, Rydman P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–304. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 14.Walley AJ, Chavanas S, Moffatt MF, et al. Gene polymorphism in Netherton and common atopic disease. Nat Genet. 2001;29:175–178. doi: 10.1038/ng728. [DOI] [PubMed] [Google Scholar]
- 15.Kabesch M, Carr D, Weiland SK, et al. Association between polymorphisms in serine protease inhibitor, kazal type 5 and asthma phenotypes in a large German population sample. Clin Exp Allergy. 2004;34:340–345. doi: 10.1111/j.1365-2222.2004.01860.x. [DOI] [PubMed] [Google Scholar]
- 16.Kusunoki T, Okafuji I, Yoshioka T, et al. SPINK5 polymorphism is associated with disease severity and food allergy in children with AD. J Allergy Clin Immunol. 2005;115:636–638. doi: 10.1016/j.jaci.2004.12.1114. [DOI] [PubMed] [Google Scholar]
- 17.Kato A, Fukai K, Oiso N, et al. Association of SPINK5 gene polymorphisms with AD in the Japanese population. Br J Dermatol. 2003;148:665–669. doi: 10.1046/j.1365-2133.2003.05243.x. [DOI] [PubMed] [Google Scholar]
- 18.Hudson TJ. Skin barrier function and allergic risk. Nat Genet. 2006;38:399–400. doi: 10.1038/ng0406-399. [DOI] [PubMed] [Google Scholar]
- 19.Lanza DC, Kennedy DW. Adult rhinosinusitis defined. Otolaryngol Head Neck Surg. 1997;117:S1–S7. doi: 10.1016/S0194-59989770001-9. [DOI] [PubMed] [Google Scholar]
- 20.Marenholz I, Heizmann CW, Fritz G. S100 proteins in mouse and man: From evolution to function and pathology (including an update of the nomenclature) Biochem Bio-phys Res Commun. 2004;322:1111–1122. doi: 10.1016/j.bbrc.2004.07.096. [DOI] [PubMed] [Google Scholar]
- 21.Glaser R, Harder J, Lange H, et al. Antimicrobial psoriasin (S100A7) protects human skin from E. coli infection. Nat Immunol. 2005;6:57–64. doi: 10.1038/ni1142. [DOI] [PubMed] [Google Scholar]
- 22.Moffatt MF. SPINK5: A gene for AD and asthma. Clin Exp Allergy. 2004;34:325–327. doi: 10.1111/j.1365-2222.2004.01915.x. [DOI] [PubMed] [Google Scholar]
- 23.Breuer K, Kapp A, Werfel T. Bacterial infections and AD. Allergy. 2001;56:1034–1041. doi: 10.1034/j.1398-9995.2001.00146.x. [DOI] [PubMed] [Google Scholar]
- 24.Busaba NY, Siegel NS, Salman SD. Microbiology of chronic ethmoid sinusitis: Is this a bacterial disease? Am J Otolaryngol. 2004;25:379–384. doi: 10.1016/j.amjoto.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 25.Van Zele T, Gevaert P, Watelet JP, et al. Staphylococcus aureus colonization and IgE antibody formation to enterotoxins is increased in nasal polyposis. J Allergy Clin Immunol. 2004;114:981–983. doi: 10.1016/j.jaci.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 26.Komatsu N, Takata M, Otsuki N, et al. Elevated stratum corneum hydrolytic activity in Netherton syndrome suggests an inhibitory regulation of desquamation by SPINK5-derived peptides. J Investig Dermatol. 2002;118:436–443. doi: 10.1046/j.0022-202x.2001.01663.x. [DOI] [PubMed] [Google Scholar]
- 27.Lane AP, Truong-Tran QA, Myers A, et al. Serum amyloid A, properdin, complement 3, and toll-like receptors are expressed locally in human sinonasal tissue. Am J Rhinol. 2006;20:117–123. [PMC free article] [PubMed] [Google Scholar]
- 28.Dong Z, Yang Z, Wang C. Expression of TLR2 and TLR4 messenger RNA in the epithelial cells of the nasal airway. Am J Rhinol. 2005;19:236–239. [PubMed] [Google Scholar]
- 29.Wladislavosky-Waserman P, Kern EB, Holley KE, et al. Epithelial damage in nasal polyps. Clin Allergy. 1984;14:241–247. doi: 10.1111/j.1365-2222.1984.tb02203.x. [DOI] [PubMed] [Google Scholar]
- 30.Shahana S, Jaunmuktane Z, Asplund M, Roomans G. Ultrastructural investigation of epithelial damage in asthmatic and non-asthmatic polyps. Respir Med. 2006;100:2018–2028. doi: 10.1016/j.rmed.2006.02.012. [DOI] [PubMed] [Google Scholar]