Abstract
Most of the bone, cartilage and connective tissue of the lower jaw is derived from cranial neural crest cells (NCCs) arising from the posterior midbrain and hindbrain. Multiple factors direct the patterning of these NCCs, including endothelin-1-mediated endothelin A receptor (Edn1/Ednra) signaling. Loss of Ednra signaling results in multiple defects in lower jaw and neck structures, including homeotic transformation of lower jaw structures into upper jaw-like structures. However, since the Ednra gene is expressed by both migrating and post-migrating NCCs, the actual function of Ednra in cranial NCC development is not clear. Ednra signaling could be required for normal migration or guidance of NCCs to the pharyngeal arches or in subsequent events in post-migratory NCCs, including proliferation and survival. To address this question, we performed a fate analysis of cranial NCCs in Ednra−/− embryos using the R26R;Wnt1-Cre reporter system, in which Cre expression within NCCs results in permanent β-galactosidase activity in NCCs and their derivatives. We find that loss of Ednra does not detectably alter either migration of most cranial NCCs into the mandibular first arch and second arch or their subsequent proliferation. However, mesenchymal cell apoptosis is increased two fold in both E9.5 and E10.5 Ednra−/− embryos, with apoptotic cells being present in and just proximal to the pharyngeal arches. Based on these studies, Ednra signaling appears to be required by most cranial NCCs after they reach the pharyngeal arches. However, a subset of NCCs appear to require Ednra signaling earlier, with loss of Ednra signaling likely leading to premature cessation of migration into or within the arches and subsequent cell death.
Keywords: neural crest, facial morphogenesis, loxP/Cre, fate mapping
Introduction
Most of the bone and cartilage in the craniofacial region arises from transient structures called pharyngeal arches (Fukiishi and Morriss-Kay, 1992, Bronner-Fraser, 1995), which are populated by cranial neural crest cells (NCCs). These NCCs migrate from the neuroectoderm of the posterior midbrain and hindbrain to the arches while undergoing an epithelial to mesenchymal transition (Le Douarin, 1982, Tosney, 1982). This migration occurs in segmental streams that correlate with the axial level of their origin in the midbrain or hindbrain (Trainor, 2005). Once in the arches, NCCs proliferate and differentiate into bone, cartilage and connective tissue of the face and neck. The events regulating NCC development are quite dynamic and perhaps some of the most complex during embryonic development. It is therefore not surprising that craniofacial birth defect syndromes resulting from disruption of cranial NCC migration or differentiation are some of the most common birth defects observed in humans (Wilson et al., 1993, Wilkie and Morriss-Kay, 2001).
While carrying some positional information from their midbrain/hindbrain origins (Noden, 1988), NCCs within pharyngeal arches appear to receive much of their patterning information from molecules secreted by cells in the surrounding environment (Trainor and Krumlauf, 2000, Schilling et al., 2001, Couly et al., 2002, David et al., 2002, Creuzet et al., 2005). One of these factors is endothelin-1 (Edn1), which is expressed on the distocaudal pharyngeal arch ectoderm, the core paraxial mesoderm and the pharyngeal pouch endoderm of the mandibular portion of the first arch as well as in arches 2–6 (Maemura et al., 1996, Clouthier et al., 1998,). Edn1 binds to the endothelin-A receptor (Ednra) found on cranial NCCs and induces downstream signaling events crucial for facial morphogenesis. Targeted inactivation of the genes encoding Edn1, Ednra or endothelin converting enzyme-1 (Ece1) in the mouse results in severe malformation of cranial NCC derivatives in the resulting mice, leading to death shortly after birth (Kurihara et al., 1994, Clouthier et al., 1998, Yanagisawa et al., 1998a). These defects include a homeotic transformation of mandibular arch-derived structures into more proximal-like structures, including the maxilla and palatine bones (Ozeki et al., 2004, Ruest et al., 2004). These changes are accompanied by a loss of normal mandibular arch gene expression and an upregulation in expression of genes normally observed in the proximal mandibular arch and/or maxillary prominences. Thus, in the absence of Ednra signaling, at least some of the NCCs normally destined to form the lower jaw are reprogrammed to form more proximal derivatives.
Similar findings have been observed in zebrafish carrying a strong loss-of-function mutation in the edn1 gene (Miller et al., 2000). These defects could be recapitulated in wild type zebrafish embryos by morpholinos against edn1, with a high concentration of morpholino resulting in defects in dorsal (proximal in the mouse) and ventral (distal in the mouse) arch structures, whereas lower morpholino concentrations primarily only affected ventral arch derivatives (Miller and Kimmel, 2001, Kimmel et al., 2003). These findings suggest that Edn1 may act in a gradient fashion within the arch, with increasing levels in the arch either directing crest cells into the arches or subsequently directing their patterning and differentiation in a concentration-dependent manner. Chimera analysis of Ednra function in mice appears to support this hypothesis. In Ednra mutant chimeras (referred to as Ednra−/− <−> +/+ embryos), Ednra−/− NCCs were excluded from the distal pharyngeal arches at both E9.5 and E10.5, instead accumulating in the proximal arches (Clouthier et al., 2003). The extent of exclusion directly correlated with the percent of wild type cells present. This segregation could result from a migratory defect in Ednra−/− NCCs, including a disruption in the ability of NCCs to move through the arch environment and/or slowed migration. Further, using a short-term in vitro assay system, Fukuhara et al. (2004) have shown that Ednra signaling is essential between E8.5–E9.0. This embryonic stage correlates with late migration of NCCs to the pharyngeal arches and early post-migratory patterning/proliferation.
While the studies listed above are suggestive of a late migratory function for Ednra, previous studies of Ednra−/− embryos have not found defects in the expression of two molecular markers associated with migratory NCCs (Clouthier et al., 2000). Further, injection of human EDN1 into suc/et-1 mutant zebrafish after most NCCs had reached the pharyngeal arches resulted in a rescue of ventral cartilage development (Miller et al., 2000), suggesting that Ednra signaling is not required for NCC migration. These findings also illustrate that function of endothelin signaling in cranial NCC development is conserved in higher vertebrates. To directly assess the movement and fate of NCCs in the absence of Ednra signaling, we have used a two component genetic system to follow the fate of NCCs in Ednra−/− embryos as well as examine early proliferation and apoptosis in these cells. We find that NCC migration to the pharyngeal arches appears normal in Ednra−/− embryos, as does cellular proliferation of most ectomesenchymal cells in the mandibular pharyngeal arch. However, apoptosis in the mandibular arch mesenchyme and overlying ectoderm is locally increased at both E9.5 and E10.5. These findings indicate that while Ednra signaling is not required for overt migration of NCCs to the pharyngeal arches, it may be required for migration of subsets of NCCs, explaining in part some of the craniofacial defects observed in Edn1−/− and Ednra−/− embryos.
Results
Early NCC migration is not altered in Ednra−/− embryos
To analyze the function of Ednra-dependent signaling during development, we first compared the fate of NCCs between wild type and Ednra−/− embryos. To accomplish this, we crossed the Ednra mutant allele into the R26R;Wnt1-Cre background. The R26R strain contains a loxP-flanked lacZ cassette targeted to the ROSA26 locus (Soriano, 1999). In the absence of Cre recombinase, the lacZ gene is inactive. However, crossing with a Cre transgenic mouse strain results in Cre expression in specific cells and hence permanent activation of lacZ expression in those cells and their progeny, which can then be indelibly marked by β-galactosidase staining. In R26R;Wnt1-Cre strain, lacZ expression is observed along the neural tube and in most NCCs emigrating from the neural tube (Chai et al., 2000, Jiang et al., 2000).
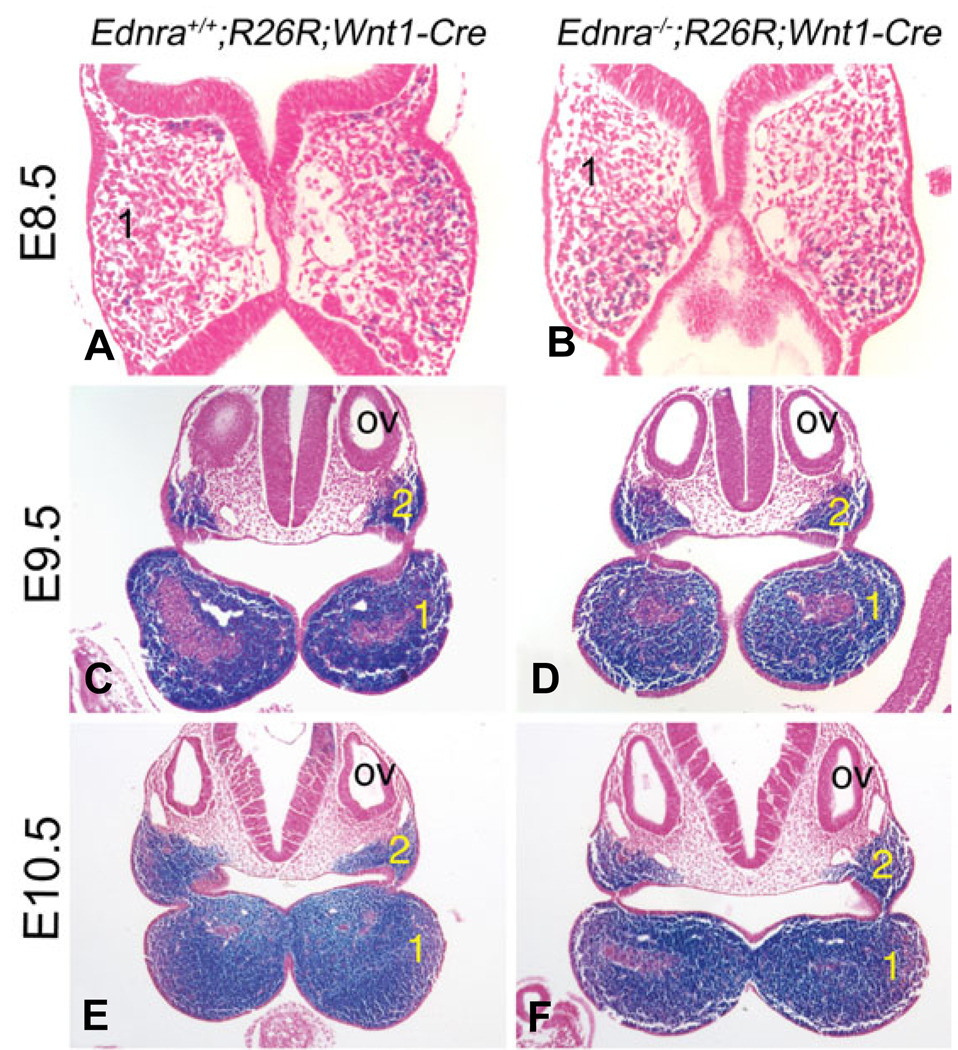
In E8.5 (9–11 somites) Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos, β-gal labeled cells (blue) were observed in the head mesenchyme and in two streams extending ventrally away from the hindbrain towards the pharyngeal arches (NCCs) (Fig. 1A, B). The rostral stream extended into the first pharyngeal arch, which was composed of labeled and unlabeled cells (Fig. 2A, B). In E9.5 (20–25 somites) Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos, labeled cells were observed along the neural tube, head mesenchyme, frontonasal prominence, pharyngeal arches and in cells entering the conotruncus of the heart (Fig. 1C, D). While the shape of the mandibular portion of arch one was altered, the overall staining pattern was similar, with labeled cells observed in the mesenchyme of arches one and two surrounding the unstained core paraxial mesoderm (Fig. 2C, D). In contrast, migration of labeled cells into the heart outflow tract appeared delayed in Ednra−/− embryos (arrows in Fig. 1C, D). At E10.5 (30–35 somites), β-gal labeled cells in Ednra+/+;R26R;Wnt1-Cre and Ednra−/− ;R26R;Wnt1-Cre embryos were observed in a similar distribution to that observed at E9.5, with no notable differences observed in the contribution or distribution of labeled cells in the lateral (Fig. 1E, F) or distal (Fig. 1E’, F’) aspects of the pharyngeal arches. Also as observed at E9.5, labeled cells composed most the mesenchyme in arches one and two excluding the core paraxial mesoderm (Fig. 2C, D), with no differences observed between Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos. In contrast, labeled cells in the outflow tract of the heart in Ednra+/+;R26R;Wnt1-Cre were located more distally than those in Ednra−/−;R26R;Wnt1-Cre embryos (arrows in Fig. 2E, F). While it is possible that sub-populations of NCCs had variations in migration undetectable in our assay, our results indicate overall neural crest cell migration to the pharyngeal arch is not altered by the absence of Ednra signaling.
Fig. 1. Localization of migratory and post-migratory NCCs in Ednra−/−;R26R;Wnt1-Cre embryos.
Lateral (A–F) and ventral (E’,F’) views of Ednra+/+;R26R;Wnt1-Cre (A,C,E,E’) and Ednra−/−;R26R;Wnt1-Cre (B,D,F,F’) embryos. (A,B) At E8.5, β-gal-stained cells (blue) are observed in both Ednra+/+;R26R;Wnt1-Cre (A) and Ednra−/−;R26R;Wnt1-Cre (B) embryos extending from the neural tube toward pharyngeal arch 1 (1) and two (2), with no observed differences present between the two embryos. (C, D) At E9.5, stained cells are observed in Ednra+/+;R26R;Wnt1-Cre (C) and Ednra−/−;R26R;Wnt1-Cre (D) embryos in the frontonasal prominence, maxillary and mandibular portions of the first arch and second, third (3), fourth (4) and sixth (6) arches. No differences are observed in these regions, though stained cells in the outflow tract of wild type embryos appears to extend more distally than those in the outflow of Ednra−/−;R26R;Wnt1-Cre embryos (arrow). (E,F) The staining pattern in E10.5 embryos is similar to that observed at E9.5, with stained cells present throughout the pharyngeal arches (see also ventral views, E’ and F’). However, an apparent delay in NCC migration through the outflow tract in Ednra−/−;R26R;Wnt1-Cre embryos is still apparent (compare arrows in E and F). h, heart.
Fig. 2. Histological analysis of NCCs in the pharyngeal arches of Ednra−/−;R26R;Wnt1-Cre embryos.
Ednra+/+;R26R;Wnt1-Cre (A,C,E) and Ednra−/−;R26R;Wnt1-Cre (B,D,F) embryos were stained in whole mount for β-gal activity and then embedded on a transverse plane in paraffin and sectioned. Sections are counterstained with nuclear fast red. (A,B) In E8.5 embryos, labeled cells are observed in the mesenchyme of the first pharyngeal arch (1) in both Ednra+/+;R26R;Wnt1-Cre (A) and Ednra−/−;R26R;Wnt1-Cre (B) embryos. (C,D) At E9.5, most of the mesenchyme in the mandibular portion of arch one (1) and in arch two (2) in both Ednra+/+;R26R;Wnt1-Cre (C) and Ednra−/−;R26R;Wnt1-Cre (D) embryos is labeled. The unlabeled cells in the center of the arch are part of the core paraxial mesoderm. (E,F) As observed at E9.5, the arch mesenchyme of E10.5 Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos is almost completely composed of labeled cells, excluding the core paraxial mesoderm. ov, otic vesicle.
Contribution of NCC-derived cells to head and neck structures in wild type and Ednra−/− embryos at E13.5 and E18.5
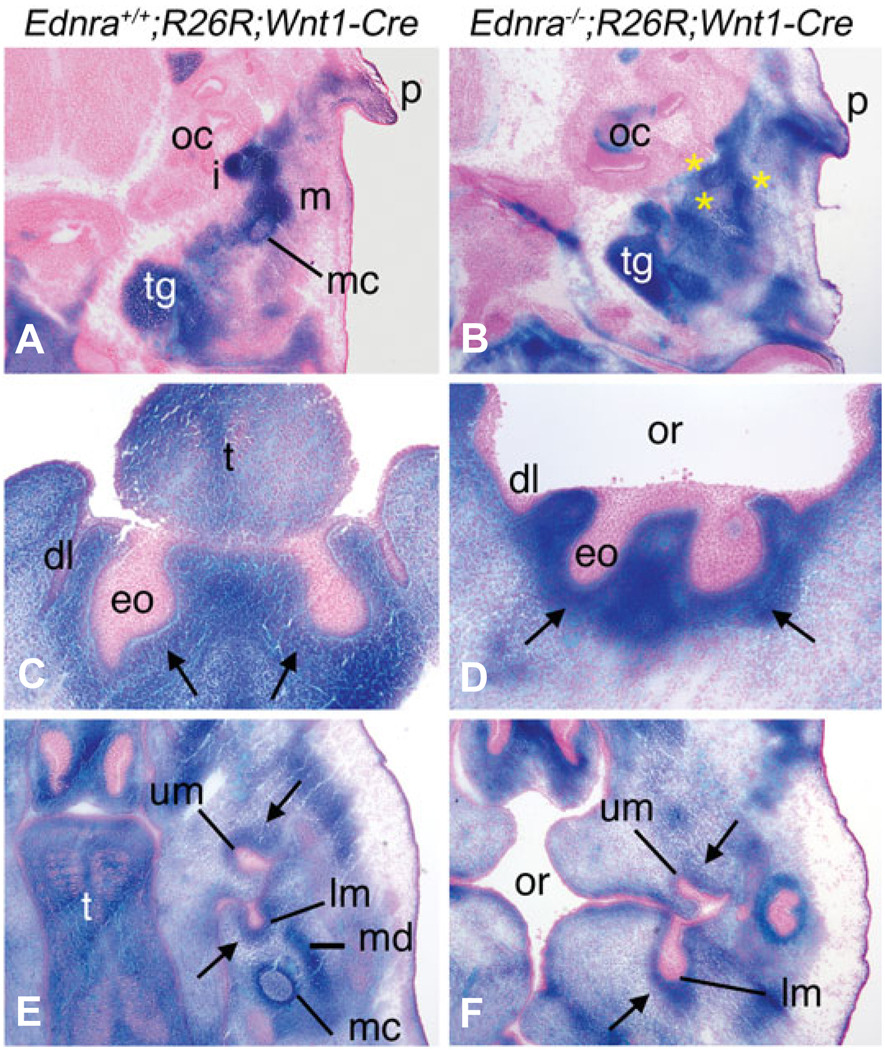
To determine whether loss of Ednra signaling affected the fate of NCCs, we examined the contribution of NCCs to developing structure of the face and neck. In E13.5 Ednra+/+;R26R;Wnt1-Cre embryos, staining was observed in the forming malleus and incus of the middle ear, Meckel’s cartilage, trigeminal ganglion and the ear pinna (Fig. 3A). In E13.5 Ednra−/−;R26R;Wnt1-Cre embryos, the middle ear region was composed of undefined cartilage condensations, though these cartilage structures were composed of labeled cells, as was the ear pinna and trigeminal ganglion (Fig. 3B).
Fig. 3. Contribution of Ednra−/− NCCs to craniofacial structures in E13.5 Ednra−/−;R26R;Wnt1-Cre embryos.
Transverse sections through the head of Ednra+/+;R26R;Wnt1-Cre (A,C,E) and Ednra−/−;R26R;Wnt1-Cre embryos (B,D,F) stained for β-gal activity. Sections were counterstained with nuclear fast red. (A,B) In sections through the lower jaw/middle ear region of Ednra+/+;R26R;Wnt1-Cre embryos, the malleus (m), incus (i) and Meckel’s cartilage (mc) contain labeled cells. Labeled cells are also observed extending toward the ear pinna (p) and in the trigeminal ganglion (tg) (A). In Ednra−/−;R26R;Wnt1-Cre embryos, labeled cells are only observed in unrecognizable cartilaginous condensations (yellow asterisks)(B). Labeled cells are also observed extending toward the pinna and in the trigeminal ganglion. (C,D) In sections through the lower incisor region, labeled cells are observed in the condensing dental mesenchyme (arrows) surrounding the enamel organ (eo) of the lower incisors and in the surrounding mesenchyme in both Ednra+/+;R26R;Wnt1-Cre (C) and Ednra−/−;R26R;Wnt1-Cre (D) embryos. (E,F) In sections through the molar region, labeled cells in Ednra+/+;R26R;Wnt1-Cre embryos compose the condensing dental mesenchyme of the developing upper and lower molars (um and lm, respectively), Meckel’s cartilage (mc) and the condensing mandible (md) (E). In Ednra−/−;R26R;Wnt1-Cre embryos, labeled cells contribute to the dental papilla of the upper and lower molars (arrow) and overall connective tissue of the head, though a definable Meckel’s cartilage and mandible are not present (F). dl, dental lamina; eo, enamel organ; oc, otic capsule; or, oral cavity; t, tongue.
We also examined the contribution of NCC-derived cells to tooth morphogenesis in transverse sections of E13.5 embryos. In the incisor region, labeled cells composed a majority of the condensing dental mesenchyme surrounding the enamels organs in both Ednra+/+;R26R;Wnt1-Cre (arrows in Fig. 3C) and Ednra−/−;R26R;Wnt1-Cre embryos (Fig. 3D), an area populated by NCCs. In the molar region, most of the NCC-derived dental mesenchyme (arrows) surrounding the enamel organs was populated with stained cells in both Ednra+/+;R26R;Wnt1-Cre (Fig.3E) and Ednra−/−;R26R;Wnt1-Cre (Fig.3F) embryos, with no detectable differences observed between the upper and lower molars.
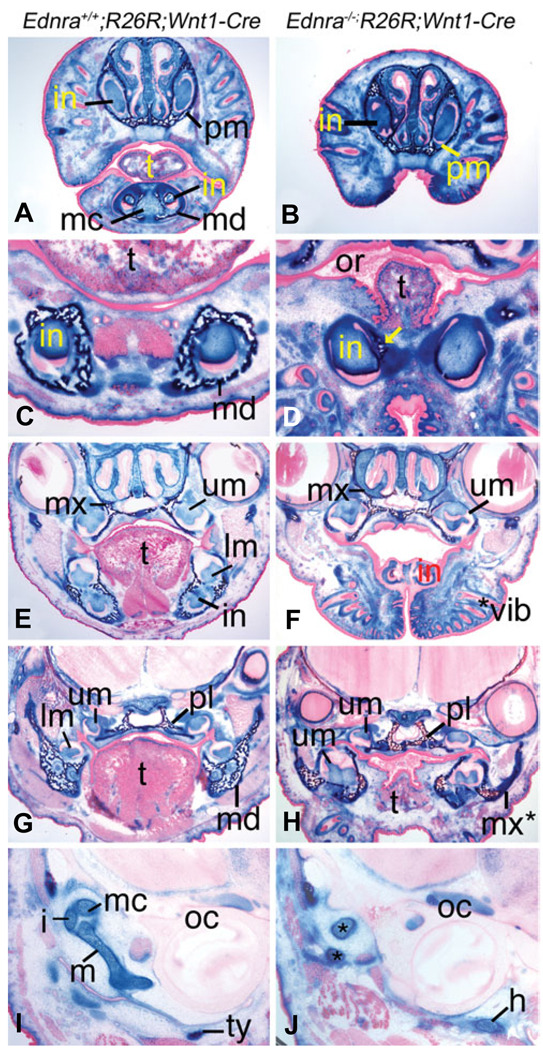
We next examined the contribution of NCC-derived cells to jaw and throat structures of E18.5 embryos in frontal sections stained for β-gal activity. In distal sections, structures in the lower jaw of Ednra+/+;R26R;Wnt1-Cre embryos contained labeled cells, including Meckel’s cartilage, dental pulp of the lower incisors, mandibular bone and connective tissue (Fig. 4A). Due to lower jaw hypoplasia observed in Ednra−/− embryos, these structures were not observed in Ednra−/−;R26R;Wnt1-Cre embryos (Fig. 4B). In more proximal sections, labeled cells composed the mandibular bone, dental pulp of the lower incisors and the surrounding connective tissue of Ednra+/+;R26R;Wnt1-Cre (Fig. 4C) and Ednra−/−;R26R;Wnt1-Cre (Fig. 4D) embryos, though little mandibular bone was present in Ednra−/−;R26R;Wnt1-Cre embryos. More proximally, labeled cells in Ednra+/+;R26R;Wnt1-Cre embryos were observed in the dental pulp of the upper and lower molars and lower incisors, mandible and maxilla bones and surrounding mesenchyme (Fig. 4E). In Ednra−/−;R26R;Wnt1-Cre embryos, labeled cells were apparent in the dental pulp of the upper molars, maxilla and surrounding mesenchyme (Fig. 4F). Bone and lower molars were not observed at this level of section in the lower jaw. Continuing proximally, labeled cells in Ednra+/+;R26R;Wnt1-Creembryos composed the dental pulp of the lower molars, proximal mandible, palatine bones and surrounding mesenchyme (Fig. 4G). In Ednra−/−;R26R;Wnt1-Cre embryos, labeled cells also contributed to the bone in the lower jaw, palatine bones, dental pulp of the upper and lower molars and surrounding mesenchyme. As has been described before (Ozeki et al., 2004, Ruest et al., 2004), the bone in the lower jaw more closely resembled a maxilla than a wild type mandible (compare maxilla in Fig. 4F with the pseudo-maxilla in Fig. 4H). In the middle ear region, labeled cells in Ednra+/+;R26R;Wnt1-Cre embryos were observed in the NCC derived malleus, incus and tympanic ring (Fig. 4I), where as these structures were not apparent in Ednra−/−;R26R;Wnt1-Cre embryos (Fig. 4J). However, labeled cells were present in the aberrant cartilaginous bodies found in the middle ear of mutant embryos, suggesting that as with other head structures, cranial NCC cells were present.
Fig. 4. Fate of cranial NCCs in Ednra−/−;R26R;Wnt1-Cre embryos.
Frontal sections through the head of E18.5 Ednra+/+;R26R;Wnt1-Cre (A,C,E,G) and Ednra−/−;R26R;Wnt1-Cre (B,D,F,H) embryos. (A,B) Labeled cells are observed in the head mesenchyme, premaxilla (pm), mandible (md), Meckel’s cartilage (mc) and dental papilla of the upper and lower incisors (in) of Ednra+/+;R26R;Wnt1-Cre embryos (A). A similar pattern is observed in Ednra−/−;R26R;Wnt1-Cre embryos, though the recessed lower jaw is not observed (B). (C,D) In sections through the lower incisors, labeled cells are observed in the dental papilla and mandible of Ednra+/+;R26R;Wnt1-Cre embryos (C). In Ednra−/− ;R26R;Wnt1-Cre embryos (D), labeled cells are present in the dental papilla and small amount of mandible that is present (arrow). (E,F) Labeled cells in Ednra+/+;R26R;Wnt1-Cre embryos are observed in the maxilla (mx), dental papilla of upper (um) and lower (lm) molars and lower incisor (in) and surrounding mesenchyme (E). A similar pattern is observed in Ednra−/−;R26R;Wnt1-Cre embryos (F), though the lower molars are proximal to this plane of section due to the mandibular hypoplasia. (G,H) In sections proximal to those in (E,F), labeled cells in wild type embryos are observed in the mandible and palatine bones (pl), dental papilla of the upper and lower molars and surrounding mesenchyme (G). In Ednra−/−;R26R;Wnt1-Cre embryos, a similar contribution of labeled cells is observed, including the dysmorphic bone in the lower jaw that has previously been shown to have undergone homeosis to a maxilla-like structure (pseudo-maxilla, mx*). (I,J) In Ednra+/+;R26R;Wnt1-Cre embryos, the malleus (m), incus (i), Meckel’s cartilage (mc) and tympanic ring (ty) are all populated by labeled cells (I). In Ednra−/−;R26R;Wnt1-Cre embryos, aberrant cartilage (*) containing labeled cells have replaced the malleus and incus (J). Labeled cells are also present in the greater horns (h) of the hyoid, which are aberrantly present near the otic capsule (oc). or, oral cavity; t tongue; *vib, ectopic vibrissae.
Early apoptosis in pharyngeal arch mesenchyme of Ednra−/− embryos
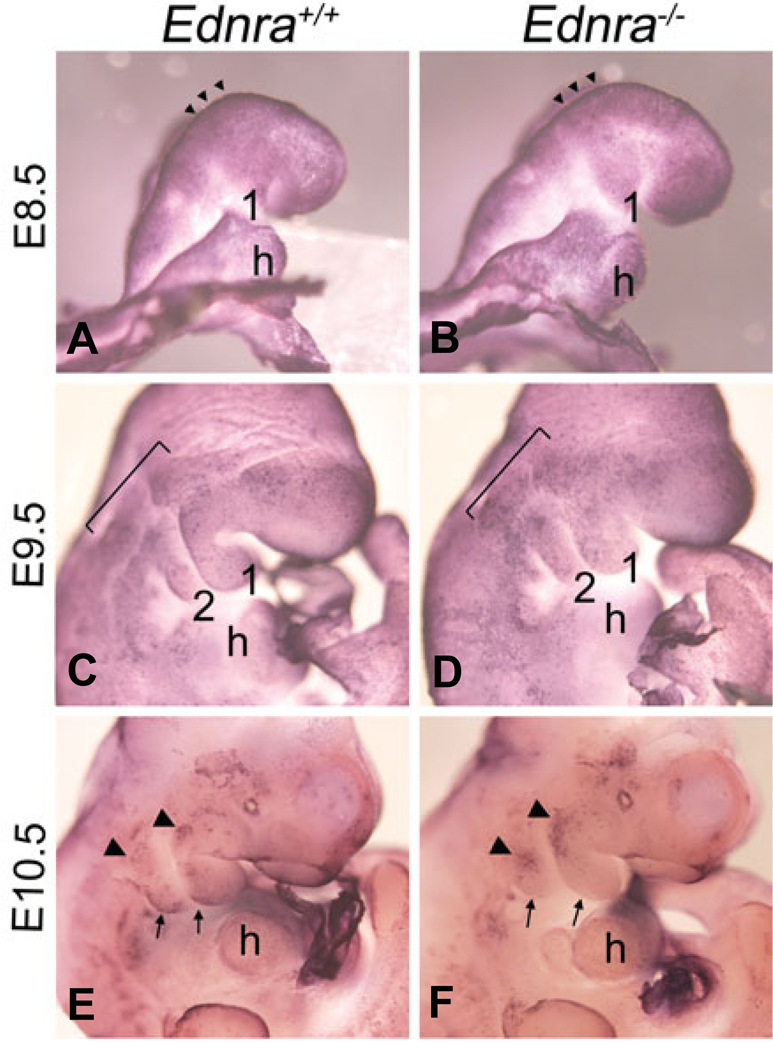
While distal mandibular bone is reduced in E18.5 Ednra−/− and Edn1−/− embryos, our current data suggests that overall migration of NCCs to the pharyngeal arches is normal. One explanation for this is that Ednra signaling is crucial for either proliferation or survival of a subpopulation of migratory or post-migratory NCCs. We have previously shown that there is decreased proliferation and increased apoptosis in the first pharyngeal arch of E11.5 Ednra−/− embryos (Clouthier et al., 2000). However, these changes could be secondary to earlier defects in NCC development. To look at more primary changes, we first examined the incidence of cell death in E8.5–E10.5 embryos in whole mount using the TdTmediated dUTP nick end labeling (TUNEL) technique. At E8.5, normal apoptosis was observed along the neural folds, though very little apoptosis was observed between the midbrain/hindbrain and pharyngeal arches in either wild type (Fig. 5A) or Ednra−/− (Fig. 5B) embryos. In E9.5 wild type embryos, TUNEL-positive cells were observed in otic placode and in cells proximal to the pharyngeal arches (bar). Scattered labeled cells were also present in the mandibular first arch and arches 2–4 (Fig. 5C). In E9.5 Ednra−/− embryos, apoptosis proximal to and within the arches appeared to be increased (Fig. 5D). In E10.5 wild type embryos, apoptosis in wild type embryos was observed in the frontonasal prominence, optic vesicle, temporal region of the head and the proximal (arrowheads) and caudal (arrows) aspects of the first pharyngeal arch (Fig. 5E). In Ednra−/− embryos, the incidence of apoptosis was similar in the frontonasal prominence, optic vesicle and head (Fig. 5F). However, increased apoptosis was observed in the proximal region of the first and second pharyngeal arches (arrowheads in Fig. 5F) while apoptosis in the caudal region of the mandibular arch was not observed (arrows in Fig. 5F).
Fig. 5. Whole mount apoptosis analysis in Ednra−/− embryos.
Lateral and ventral views of wild type (A,C,E) and Ednra−/− (B,D,F) embryos at E8.5 (A,B), E9.5 (C,D) and E10.5 (E,F) following whole mount TUNEL analysis, with NBT/BCIP as the substrate, which produces a blue deposit. (A,B) At E8.5, apoptosis is confined to the neural fold in the midbrain and hindbrain regions (arrowheads) of both wild type (A) and Ednra−/− (B) embryos. (C,D) In E9.5 wild type embryos, scattered apoptosis is observed proximal to and within pharyngeal arches one and two (C). In E9.5 Ednra−/− embryos, apoptosis is observed in a similar pattern to that of wild type embryos, though the qualitative level of apoptosis appears higher, especially in the arches (D). (E,F) In the pharyngeal arch region of E10.5 wild type embryos, apoptosis is observed in cells at the proximal aspect of arches one and two (arrowheads) and in cells along the caudal aspects of both arches (arrows) (E). In E10.5 Ednra−/− embryos, apoptosis is more prominent in the proximal aspects of the mandibular and second arches (arrowheads) but is not apparent in the caudal aspects of arches one and two (arrows) (F). h, heart.
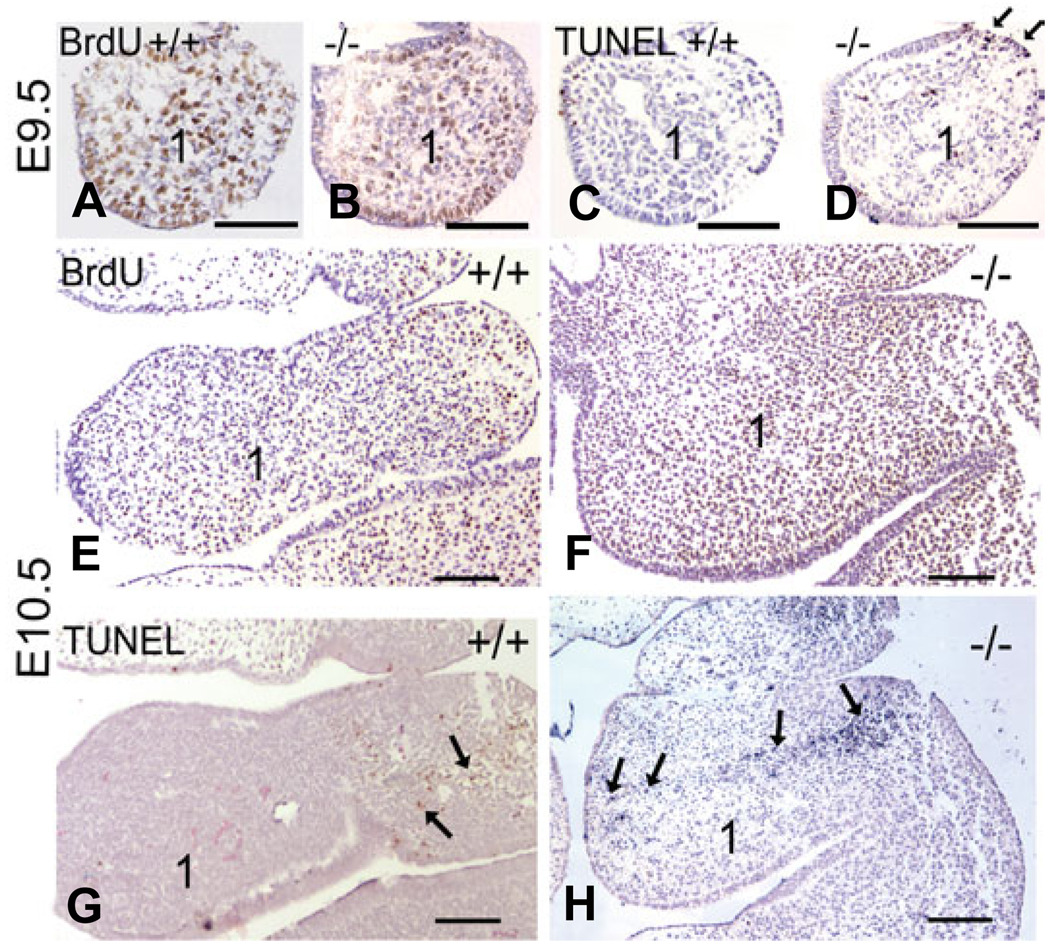
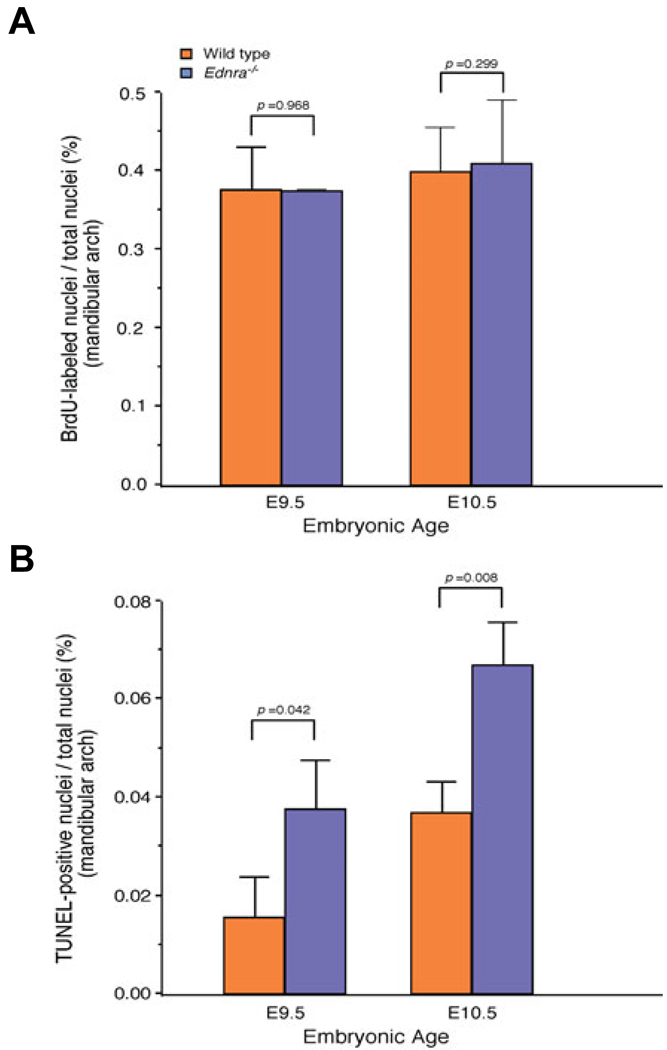
To examine these changes on a cellular level, we performed TUNEL analysis on sections through the mandibular arch of wild type and Ednra−/− embryos and compared these findings with changes in cellular proliferation determined by bromodeoxy uridine (BrdU) incorporation analysis on consecutive slides. For each analysis, three sections through the mandibular arch of three different embryos were used (see Materials and Methods for a detailed description). At E9.5, overall mesenchymal cell proliferation in the mandibular arch was similar in both wild type and Ednra−/− embryos (Fig. 6A, B and Fig. 7A). In contrast, TUNEL analysis of consecutive sections to those used for BrdU analysis showed that apoptosis was increased two fold in the first arch mesenchyme of Ednra−/− embryos (Fig. 6D and Fig 7B) compared to that found in wild type embryos (Fig. 6C and Fig. 7B). There was also a consistent increase in cell death in a localized area of the proximal arch ectoderm (arrows in Fig. 6D), though overall ectodermal apoptosis was not significantly different when compared to the ectoderm of wild type embryos (data not shown). At E10.5, proliferation in the mandibular arch was similar between wild type and Ednra−/− embryos (Fig. 6E, F and Fig. 7A). In contrast, there was a two-fold increase in cell death within the mandibular arch mesenchyme of Ednra−/− embryos (Fig. 6H and Fig. 7B). Most of the cell death was observed in the proximal arch, with TUNEL-positive cells extending into the distal arch (Fig. 6H). This pattern was very similar to that observed in the whole mount TUNEL assays.
Fig. 6. Comparison of proliferation and cell death in Ednra−/− embryos.
Transverse sections through the mandibular arch one and arch two of wild type (A,C,E,G) and Ednra−/− (B,D,F,H) following BrdU (A,B,E,G) or TUNEL (C,D,F,H) analysis. (A,B) BrdU labeling in the mandibular arch (1) appears similar between wild type (A) and Ednra−/− (B) embryos. (C,D) In consecutive sections to those in (A,B), few TUNEL-positive cells are observed in the mesenchyme of E9.5 wild type embryos (C), though numerous labeled nuclei (red) are observed in the mesenchyme of Ednra−/− embryos (D). There is also a local increase in TUNEL-positive cells in the ectoderm of the proximal arch (arrows in D). (E,F) At E10.5, BrdU staining appears similar between wild type (E) and Ednra−/− (F) embryos in the first arch. (G,H) In consecutive sections to those in (E,F), TUNEL-positive cells (red) in wild type embryos are confined to the proximal aspects of arches one and two (arrows in G). In Ednra−/− embryos, TUNEL-positive cells (blue) extend from this proximal region into the distal arch as denoted by arrows (H; the difference in color between labeled cells in G and H is due to different chromagens. This does not affect the outcome or sensitivity of the experiments). The magnification bar in each panel represents 100 µm.
Fig. 7. Quantification of proliferation and cell death in Ednra−/− embryos.
Incidence of proliferating (A) and apoptotic (B) mesenchymal cells in the mandibular arch of wild type (orange) and Ednra−/− (blue) embryos at both E9.5 and E10.5. Values represent the number of labeled nuclei as a percentage of total nuclei. p values comparing both genotypes are listed above each embryonic age.
Discussion
We have shown in this study that cranial NCC migration appears normal in Ednra−/− embryos, even though this knockout mouse shows aberrant craniofacial bone and cartilage development. However, while overall proliferation in the NCC-derived ectomesenchyme of the mandibular arch was not altered, apoptosis was increased two fold. Together, these findings explain in part the structural defects that exist in Ednra−/− and Edn1−/− embryos.
Absence of Ednra signaling does not disrupt migration of NCCs to the pharyngeal arches
Precise cranial NCC movement from the hindbrain to the pharyngeal arches is an important developmental event, with defects in emigration, migration or survival leading to craniofacial dysmorphologies and possible death (Depew and Simpson, 2006, Walker and Trainor, 2006). However, our analysis of NCC development and distribution in Ednra−/− embryos suggests that most NCCs emigrate properly from the neural tube and reach the pharyngeal arches in the absence of Ednra signaling, arguing against gross defects in NCC migration as a cause of missing or malformed structures in Ednra−/− mice. Further, these observations are consistent with previous findings showing normal expression of migratory NCC markers in Ednra−/− mouse embryos (Clouthier et al., 2000) and in suc/et1 mutant zebrafish embryos (Miller et al., 2000).
One caveat of this interpretation is that we cannot exclude mistargeting of subsets of NCCs into or within the pharyngeal arches, as NCCs from all midbrain/hindbrain regions are marked by strong β-gal staining in R26R;Wnt1-Cre embryos (Chai et al., 2000). This makes it virtually impossible to determine whether there is mis-targeting of cells within the pharyngeal arches of Ednra−/−;R26R;Wnt1-Cre embryos. However, skeletal elements derived from NCCs in the mandibular arch have a very specific contribution of NCCs, with most distal jaw structures (such as the mandible and Meckel’s cartilage) being composed of posterior midbrain NCCs while more proximal structures (such as the malleus) are a mixture of midbrain and hindbrain NCCs (Couly et al., 1996, Kontges and Lumsden, 1996, Couly et al., 1998). Since Ednra−/− embryos lack the distal portion of the mandible, it is plausible that loss of Ednra signaling could disrupt targeted cell movement of posterior NCCs into the distal arch. We have previously shown using chimera analysis that Ednra−/− NCCs appear to migrate more poorly into the distal arches than wild type NCCs in Ednra−/− <−> +/+ chimeras (Clouthier et al., 2003). This could reflect slower migration compared to wild type NCCs or a loss of path finding ability in Ednra−/− NCCs when mixed with wild type NCCs. In the future, expression analysis of guidance and migratory markers within the arches of Ednra−/− embryos may shed additional light on the role of Ednra signaling in intra-arch movement of cranial NCCs.
Ednra signaling has also been associated with inhibition of cell migration via c-Jun N-terminal kinase (JNK) in human epithelial-like 293 cells (Yamauchi et al., 2002), which appears to be mediated by Nck1 (Miyamoto et al., 2004). While it is possible that Ednra signaling directs NCC targeting within the arches by acting as a stop signal to subsets of migrating NCCs, Ednra expression occurs in migrating NCCs and in post-migratory NCCs within the pharyngeal arches (Clouthier et al., 1998, Yanagisawa et al., 1998a). This would seem to argue against Ednra playing an inhibitory role in NCC movement. Further, the poor migration of NCCs in Ednra−/− <−> +/+ chimeras (Clouthier et al., 2003) suggests that Ednra signaling does not result in cessation of NCC migration.
Increased ectomesenchymal cell apoptosis in Ednra−/− embryos
We have previously shown that loss of Ednra signaling causes both a decrease in proliferation and an increase in apoptosis of pharyngeal arch ectomesenchymal cells at E11.5 (Clouthier et al., 2000). While these changes could partially explain why duplicated structures in Ednra−/− embryos appear smaller than their maxillary counterparts, it is not clear whether this late apoptosis represented the only apoptosis, whether earlier changes in cell survival were present or whether this apoptosis was restricted to the mesenchyme. Here we have shown that while mesenchymal proliferation is unchanged in E9.5 and E10.5 Ednra−/− embryos, increased mesenchymal apoptosis is evident in E9.5 Ednra−/− embryos. Increased apoptosis in E10.5 embryos is also observed in the mandibular arch, extending proximally to distally into the arches. Regulation of NCC patterning and development relies in part from signals originating from the overlying ectoderm (Cobourne and Sharpe, 2003, Clouthier and Schilling, 2004), with removal of the ectoderm in cultured mandibular arches disrupting gene expression required for normal arch development (for review, see (Cobourne and Sharpe, 2003)). Implanting beads soaked with Fgf8 (Tucker et al., 1999), Bmp4 (Tucker et al., 1998), or Edn1 (Tucker et al., 1999), three of the primary ectodermal signals regulating arch development, into these explants can partially restore normal gene expression. A role for Fgf8 and Bmp4 in cell survival has also been demonstrated using tissue-specific inactivation of Fgf8 (Trummp et al., 1999, Macatee et al., 2003) and Bmp4 (Liu et al., 2005) in the mandibular arch ectoderm. In these experiments, wide spread mesenchymal apoptosis is observed, which results in failure of lower jaw formation. As both Bmp4 and Fgf signaling is abrogated in Ednra−/− and Edn1−/− embryos (Ozeki et al., 2004, Ruest et al., 2004), it is tempting to speculate that loss of Ednra signaling in the ectomesenchyme results in loss of patterning cues required by the ectoderm. This would lead to loss of normal ectodermal gene expression, followed by loss of ectomesenchymal gene expression and subsequent ectomesenchymal cell death. Such a circular pathway is likely quite complicated, since the level of cell death is much lower and more focused in Ednra−/− embryos. However, these findings emphasize the significance of Ednra-mediated epithelial-mesenchymal signaling cascades during facial development.
Increased apoptosis is also observed in the mesenchyme just proximal to the arch in E10.5 Ednra−/− embryos. This area, extending distally into the arch, could reflect a subpopulation of NCCs that did not properly reach their intended destinations within the pharyngeal arches. As described above, Ednra−/− cells appear to accumulate in the proximal aspects of mandibular arch one in Ednra−/− <−> +/+ chimeric embryos, with the remainder of the arch composed of wild type cells (Clouthier et al., 2003). While our current results indicate that a similar event may happen in Ednra−/− embryos, the identity of this population is not known. Since Ednra−/− and Edn1−/− embryos lack most distal bone in the lower jaw (Kurihara et al., 1994, Clouthier et al., 1998, Ozeki et al., 2004, Ruest et al., 2004), perhaps this group of apoptotic cells reflects NCCs destined for the distal arch. Following the fate of single cells or groups of cells from the midbrain and hindbrain (Trainor et al., 2003, Kulesa et al., 2004) may help identify the source of these cells and further our understanding of the roles of Ednra signaling in NCC development.
Materials and Methods
Generation of Wnt1-Cre/R26R/Ednra−/− mouse embryos
Generation and genotyping of Ednra−/− (Clouthier et al., 1998), R26R (Soriano, 1999) and Wnt1-Cre (Danielian et al., 1998) mouse strains have been previously published. Genotyping was performed by PCR using genomic DNA isolated from a small tail biopsy or embryonic yolk sac.
β-galactosidase staining
To examine β-gal staining in whole embryos, E8.5 to E10.5 Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos were collected and fixed for 1 hour in 4% paraformaldehyde. Embryo staining and photography was performed as previously described (Ruest et al., 2003). Embryos were analyzed and photographed on an Olympus SZX12 stereomicroscope fitted with a DP11 digital camera. To observe β-gal staining at the cellular level, stained embryos were dehydrated through graded ethanols and cedarwood oil (Polysciences, Inc.) and then embedded in paraffin and sectioned at 10 microns on a Leica RM2135 microtome. Sections were collected on Superfrost Plus microscope slides (Fisher Scientific), rehydrated, counterstained with nuclear fast red, dehydrated and coverslipped with DPX mounting media (BDH). Analysis of stained sections was performed using a Nikon E600 microscope with an attached SPOT RT digital camera.
To analyze β-gal staining in embryo sections, E13.5 and E18.5 Ednra+/+;R26R;Wnt1-Cre and Ednra−/−;R26R;Wnt1-Cre embryos were collected, snap-frozen in OCT freezing media in a dry ice / ethanol bath, sectioned and stained. Sections were counterstained with nuclear fast red and coverslipped in DPX mounting media.
Analysis of cellular proliferation and apoptosis
The detection of the proliferating cells was performed through bromodeoxyuridine (BrdU) incorporation analysis, which detects cells in S-phase, as previously described (Clouthier et al., 2000). Briefly, pregnant Ednra+/− mice at E9.5 and E10.5 were injected i.p. with 200 mg/kg body weight of BrdU one hour before embryo collection. Three embryos of each genotype were fixed in 4% paraformaldehyde on ice for 1 hour, rinsed in PBS, dehydrated and then embedded in paraffin and sectioned at 7 microns along a transverse plane. Three sections from each of the three embryos of each genotype were chosen for analysis, with sections separated by 50–90 microns. BrdU incorporation was detected using the BrdU Immunohistochemistry System (Oncogene Research Products). Both labeled nuclei and total nuclei in each section were counted, with counting aided by magnifying images in Photoshop. The overall level of proliferation is presented as the number of total labeled nuclei as a percentage of total nuclei. Statistical analysis was performed using Excel. This included a test for statistical significance between wild type and Ednra−/− sections, which was performed using a two-tailed t-test.
For sectional analysis of apoptosis at E9.5 and E10.5, consecutive sections to those used for BrdU analysis were selected. Detection of apoptotic cells was performed using DeadEnd Labeling Kit (Promega), a kit that uses TdT-mediated dUTP nick end labeling (TUNEL). For most sections, diamiobenzadiene was used as the substrate, which produces a brown/red color. Detection of apoptotic cells in sections from E10.5 Ednra−/− embryos was performed using the substrates nitroblue tetrasolium salt (NBT) and 5-bromo-4-chloro-3-indolyl phosphate (BCIP), which produces a blue color. The results of the experiments were not affected by the choice of the substrate. To quantify the incidence of apoptosis, labeled and total nuclei were counted, with the incidence of apoptosis expressed as the number of apoptotic nuclei as a percentage of total nuclei. Statistical analysis and significance were calculated as described above.
For whole-mount TUNEL analysis of both wild type and Ednra−/− embryos, E8.5, E9.5 and E10.5 embryos were collected and fixed in 4% paraformaldehyde for 16 hours at 4°C, rinsed, washed 3 times with PBS containing 0.1% Tween-20 (PBT) and transferred to 100% methanol. Embryos were bleached with 5% hydrogen peroxide in methanol for 5 hours at room temperature. After rinsing and rehydrating, embryos were treated with 5 µg/ml proteinase K (Roche) for 1 minute (E8.5), 2 minutes (E9.5) and 3.5 minutes (E10.5) and then re-fixed with 0.2% glutaraldehyde/4% paraformaldehyde in PBT at room temperature for 20 minutes. After being treated for 20 minutes 0.1% sodium borohydride (Sigma) in PBT, embryos were rinsed in PBT and then immersed in 1x TdT buffer (30 mM Tris (pH 7.5), 140 mM sodium cacodylate (pH 7.2), 1 mM CoCl2) for 10 minutes. Embryos were then placed in TdT reaction mix (1x TdT buffer, 20 µM digoxigenin-dUTP, 20 µM dTTP, 0.3U TdT (Promega)) and incubated at 37°C for 2 hours with occasional mixing. After washing with TBST (TBS plus 0.1% Tween-20), embryos were heated at 70°C for 20 minutes and then blocked with 0.5% blocking reagent (Roche) in TBST for 1 hour at room temperature. Embryos were incubated with an anti-DIG antibody linked to alkaline phosphates (Roche; pre-absorbed with inactivated mouse embryo powder in blocking reagent) at 1:15,000 (E8.5 and E9.5), or 1:5000 (E10.5) overnight at 4°C. After washing embryos 5 times for 1 hour each in TBST containing 2 mM levamisole and rinsing with NTMT (100 mM NaCl, 100 mM Tris-HCl (pH 0.5), 50 mM MgCl2, 0.1% Tween-20) twice for 20 minutes each, color development was performed using NBT and BCIP in NTMT in the dark until the desired staining was obtained.
Acknowledgements
The authors would like to thank Tinisha Taylor, Akiko Abe and Kelly Lee for technical assistance and Kristin Artinger for suggestions, discussions and critical review of the manuscript. This work was supported in part by grants from the Kentucky Excellence in Education Research Trust Fund, the American Heart Association Ohio Valley Affiliate (0160184B and 0355379B) and the NIH (DE 014181) to D.E.C. D.E.C. is a recipient of a Career Development Award from the NIH/NIDCR (DE 014765). L.B.R. is a recipient of a Research Fellowship from the Heart and Stroke Foundation of Canada.
Abbreviations used in this paper
- Ece
endothelin converting enzyme
- Edn
endothelin-1
- Ednra
endothelin-A receptor
- NCC
neural crest cell
References
- Bronner-Fraser M. Origins and developmental potential of the neural crest. Exp. Cell Res. 1995;218:405–417. doi: 10.1006/excr.1995.1173. [DOI] [PubMed] [Google Scholar]
- Chai Y, Jiang X, Ito Y, Bringas P, Han J, Rositch DH, Soriano P, Mcmahon AP, Sucov HM. Fate of the mammalian cranial neural crest during tooth and mandibular morphogenesis. Development. 2000;127:1671–1679. doi: 10.1242/dev.127.8.1671. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Hosoda K, Richardson JA, Williams SC, Yanagisawa H, Kuwaki T, Kumada M, Hammer RE, Yanagisawa M. Cranial and cardiac neural crest defects in endothelin-A receptor-deficient mice. Development. 1998;125:813–824. doi: 10.1242/dev.125.5.813. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Schilling TF. Understanding endothelin-1 function during craniofacial development in the mouse and zebrafish. Birth Defects Res. (Part C) 2004;72:190–199. doi: 10.1002/bdrc.20007. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Hammer RE, Richardson JA, Yanagisawa H. Cell-autonomous and non-autonomous actions of endothelin-A receptor signaling in craniofacial and cardiovascular development. Dev. Biol. 2003;261:506–519. doi: 10.1016/s0012-1606(03)00128-3. [DOI] [PubMed] [Google Scholar]
- Clouthier DE, Williams SC, Yanagisawa H, Wieduwilt M, Richardson JA, Yanagisawa M. Signaling pathways crucial for craniofacial development revealed by endothelin-A receptor-deficient mice. Dev. Biol. 2000;217:10–24. doi: 10.1006/dbio.1999.9527. [DOI] [PubMed] [Google Scholar]
- Cobourne MT, Sharpe PT. Tooth and jaw: Molecular mechanisms of patterning in the first branchial arch. Arch. Oral Biol. 2003;48:1–14. doi: 10.1016/s0003-9969(02)00208-x. [DOI] [PubMed] [Google Scholar]
- Couly GF, Creazzo TL, Bennaceur S, Vincent C, Le Douarin NM. Interactions between Hox-negative cephalic neural crest cells and the foregut endoderm in patterning the facial skeleton in the vertebrate head. Development. 2002;129:1061–1073. doi: 10.1242/dev.129.4.1061. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Le Douarin NM. The regeneration of the cephalic neural crest, a problem revisited: The regenerating cells originate from the contralateral or from the anterior and posterior neural fold. Development. 1996;122:3393–3407. doi: 10.1242/dev.122.11.3393. [DOI] [PubMed] [Google Scholar]
- Couly GF, Grapin-Botton A, Coltey P, Ruhin B, Le Douarin NM. Determination of the identity of the derivatives of the cephalic neural crest: Incompatibility between Hox gene expression and lower jaw development. Development. 1998;125:3445–3459. doi: 10.1242/dev.125.17.3445. [DOI] [PubMed] [Google Scholar]
- Creuzet S, Couly G, Le Douarin NM. Patterning the neural crest derivatives during development of the vertebrate head: Insights from avian studies. J. Anat. 2005;207:447–459. doi: 10.1111/j.1469-7580.2005.00485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, Muccino D, Rowitch DH, Michael SK, Mcmahon AP. Modification of gene activity in mouse embryos in utero by a tamoxifen-inducible form of Cre recombinase. Curr. Biol. 1998;8:1323–1326. doi: 10.1016/s0960-9822(07)00562-3. [DOI] [PubMed] [Google Scholar]
- David NB, Saint-Etienne L, Tsang M, Schilling TF, Rosa FM. Requirement for endoderm and FGF3 in ventral head skeleton formation. Development. 2002;129:4457–4468. doi: 10.1242/dev.129.19.4457. [DOI] [PubMed] [Google Scholar]
- Depew MJ, Simpson CA. Dev. Dyn; 21st century neonatology and the comparative development of the vertebrate skull; 2006. pp. 1256–1291. [DOI] [PubMed] [Google Scholar]
- Fukiishi Y, Morriss-Kay GM. Migration of cranial neural crest cells to the pharyngeal arches and heart in rat embryos. Cell Tissue Res. 1992;268:1–8. doi: 10.1007/BF00338048. [DOI] [PubMed] [Google Scholar]
- Fukuhara S, Kurihara Y, Arima Y, Yamada N, Kurihara H. Temporal requirement of signaling cascade involving endothelin-1/endothelin receptor type A in branchial arch development. Mech. Dev. 2004;121:1223–1233. doi: 10.1016/j.mod.2004.05.014. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, Mcmahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ullmann B, Walker M, Miller CT, Crump JG. Endothelin 1-mediated regulation of pharyngeal bone development in zebrafish. Development. 2003;130:1339–1351. doi: 10.1242/dev.00338. [DOI] [PubMed] [Google Scholar]
- Kontges G, Lumsden A. Rhombencephalic neural crest segmentation is preserved throughout craniofacial ontogeny. Development. 1996;122:3229–3242. doi: 10.1242/dev.122.10.3229. [DOI] [PubMed] [Google Scholar]
- Kulesa P, Ellies DL, Trainor PA. Comparative analysis of neural crest cell death, migration and function during vertebrate embryogenesis. Dev. Dyn. 2004;229:14–29. doi: 10.1002/dvdy.10485. [DOI] [PubMed] [Google Scholar]
- Kurihara Y, Kurihara H, Suzuki H, Kodama T, Maemura K, Nagai R, Oda H, Kuwaki T, Cao W-H, Kamada N, et al. Elevated blood pressure and craniofacial abnormalities in mice deficient in endothelin-1. Nature. 1994;368:703–710. doi: 10.1038/368703a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin NM. The neural crest. Cambridge: Cambridge Univ. Press; 1982. [Google Scholar]
- Liu W, Selever J, Murali D, Sun X, Brugger SM, Ma L, Schwartz RJ, Maxson R, Furuta Y, Martin JF. Threshold-specific requirements for Bmp4 in mandibular development. Dev. Biol. 2005;283:282–293. doi: 10.1016/j.ydbio.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Macatee TL, Hammond BP, Arenkiel BR, Francis L, Frank DU, Moon AM. Ablation of specific expression domains reveals discrete functions of ectoderm- and endoderm-derived FGF8 during cardiovascular and pharyngeal development. Development. 2003;130:6361–6374. doi: 10.1242/dev.00850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maemura K, Kurihara H, Kurihara Y, Oda H, Ishikawa T, Copeland NG, Gilbert DJ, Jenkins NA, Yazaki Y. Sequence analysis, chromosomal location and developmental expression of the mouse preproendothelin-1 gene. Genomics. 1996;31:177–184. doi: 10.1006/geno.1996.0029. [DOI] [PubMed] [Google Scholar]
- Miller CT, Kimmel CB. Morpholino phenocopies of endothelin 1 (sucker) and other anterior arch class mutations. Genesis. 2001;30:186–187. doi: 10.1002/gene.1061. [DOI] [PubMed] [Google Scholar]
- Miller CT, Schilling TF, Lee K-H, Parker J, Kimmel CB. Sucker encodes a zebrafish Endothelin-1 required for ventral pharyngeal arch development. Development. 2000;127:3815–3838. doi: 10.1242/dev.127.17.3815. [DOI] [PubMed] [Google Scholar]
- Miyamoto Y, Yamauchi J, Mizuno N, Itoh H. The adaptor protein Nck1 mediates endothelin A receptor-regulated cell migration through the Cdc42-dependent c-Jun N-terminal kinase pathway. J. Biol. Chem. 2004;279:34336–34342. doi: 10.1074/jbc.M402767200. [DOI] [PubMed] [Google Scholar]
- Noden DM. Interactions and fates of avian craniofacial mesenchyme. Development. 1988;103:121–140. doi: 10.1242/dev.103.Supplement.121. [DOI] [PubMed] [Google Scholar]
- Ozeki H, Kurihara Y, Tonami K, Watatani K, Kurihara H. Endothelin-1 regulates the dorsoventral branchial arch patterning in mice. Mech. Dev. 2004;121:387–395. doi: 10.1016/j.mod.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Ruest L-B, Dager M, Yanagisawa H, Charité J, Hammer RE, Olson EN, Yanagisawa M, Clouthier DE. dHAND-Cre transgenic mice reveal specific potential functions of dHAND during craniofacial development. Dev. Biol. 2003;257:263–277. doi: 10.1016/s0012-1606(03)00068-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruest L-B, Xiang X, Chew-Lim K, Levi G, Clouthier DE. Endothelin-A receptor-dependent and independent signaling pathways in establishing mandibular identity. Development. 2004;131:4413–4423. doi: 10.1242/dev.01291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling TF, Prince V, Ingham PW. Plasticity in zebrafish hox expression in the hindbrain and cranial neural crest. Dev. Biol. 2001;231:201–216. doi: 10.1006/dbio.2000.9997. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacz expression with the ROSA26 cre reporter strain. Nat. Gen. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Tosney KW. The segregation and early migration of cranial neural crest cells in the avian. Dev. Biol. 1982;89:13–24. doi: 10.1016/0012-1606(82)90289-5. [DOI] [PubMed] [Google Scholar]
- Trainor P, Krumlauf R. Plasticity in mouse neural crest cells reveals a new patterning role for cranial mesoderm. Nat. Cell Biol. 2000;2:96–102. doi: 10.1038/35000051. [DOI] [PubMed] [Google Scholar]
- Trainor PA. Specification of neural crest cell formation and migration in mouse embryos. Semin. Cell Dev. Biol. 2005;16:683–693. doi: 10.1016/j.semcdb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Trainor PA, Melton KR, Manzanares M. Origins and plasticity of neural crest cells and their roles in jaw and craniofacial evolution. Int. J. Dev. Biol. 2003;47:541–553. [PubMed] [Google Scholar]
- Trummp A, Depew MJ, Rubenstein JLR, Bishop JM, Martin GR. Cre-mediated gene inactivation demonstrates that FGF8 is required for cell survival and patterning of the first branchial arch. Genes Dev. 1999;13:3136–3148. doi: 10.1101/gad.13.23.3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS, Al Khamis A, Sharpe PT. Interactions between Bmp-4 and Msx1 act to restrict gene expression to odontogenic mesenchyme. Dev. Dyn. 1998;212:533–539. doi: 10.1002/(SICI)1097-0177(199808)212:4<533::AID-AJA6>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Tucker SA, Yamada G, Grigoriou M, Pachnis V, Sharpe PT. Fgf-8 determines rostral-caudal polarity in the first branchial arch. Development. 1999;126:51–61. doi: 10.1242/dev.126.1.51. [DOI] [PubMed] [Google Scholar]
- Walker M, Trainor P. Craniofacial malformations: Intrinsic vs extrinsic neural crest cell defects in Treacher Collins and 22q11 deletion syndromes. Clin. Genet. 2006;69:471–479. doi: 10.1111/j.0009-9163.2006.00615.x. [DOI] [PubMed] [Google Scholar]
- Wilkie AO, Morriss-Kay GM. Genetics of craniofacial development and malformation. Nat. Rev. Gen. 2001;2:458–468. doi: 10.1038/35076601. [DOI] [PubMed] [Google Scholar]
- Wilson DI, Burn J, Scambler P, Goodship J. DiGeorge syndrome: Part of CATCH 22. J. Med. Genet. 1993;30:852–856. doi: 10.1136/jmg.30.10.852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Miyamoto Y, Kokubu H, Nishii H, Okamoto M, Sugawara Y, Hirasawa A, Tsujimoto G, Itoh H. Endothelin suppresses cell migration via the Jnk signaling pathway in a manner dependent upon Src kinase, Rac1 and Cdc42. FEBS Let. 2002;527:284–288. doi: 10.1016/s0014-5793(02)03231-3. [DOI] [PubMed] [Google Scholar]
- Yanagisawa H, Yanagisawa M, Kapur RP, Richardson JA, Williams SC, Clouthier DE, De Wit D, Emoto N, Hammer RE. Dual genetic pathways of endothelin-mediated intercellular signaling revealed by targeted disruption of endothelin converting enzyme-1 gene. Development. 1998a;125 doi: 10.1242/dev.125.5.825. [DOI] [PubMed] [Google Scholar]